Abstract
Traditional Chinese Medicines (TCMs) have been historically used to treat bacterial infections. However, the molecules responsible for these anti-infective properties and their potential mechanisms of action have remained elusive. Using a high-throughput assay for type III protein secretion in Salmonella enterica serovar Typhimurium, we discovered that several TCMs can attenuate this key virulence pathway without affecting bacterial growth. Amongst the active TCMs, we discovered that baicalein, a specific flavonoid from Scutellaria baicalensis, targets S. Typhimurium pathogenicity island-1 (SPI-1) type III secretion system (T3SS) effectors and translocases to inhibit bacterial invasion of epithelial cells. Structurally related flavonoids present in other TCMs, such as quercetin also inactivated the SPI-1 T3SS and attenuated S. Typhimurium invasion. Our results demonstrate that specific plant metabolites from TCMs can directly interfere with key bacterial virulence pathways and reveals a previously unappreciated mechanism of action for anti-infective medicinal plants.
INTRODUCTION
Medicinal plants have provided fertile ground for modern drug discovery1. Nonetheless, these plant-derived extracts remain a poorly characterized reservoir of anti-infective agents2,3. For example, a variety of TCMs have been reported to have protective effects against bacterial and viral infections, but the specific molecules responsible for these antimicrobial activities and their mechanism(s) of action are largely unknown4. The molecules and mechanisms of TCMs used to treat infectious gastroenteritis have been particularly elusive, since plant extracts and metabolites attenuate infection but do not exhibit significant bactericidal activity5. These observations suggest that specific compounds from TCMs may target virulence factors rather than the growth of enteric pathogens.
To explore the anti-infective activity of TCMs on enteric pathogens, we focused on T3SSs as these protein translocation needle-like complexes are key virulence mechanisms for Gram-negative bacterial pathogens responsible for gastroenteritis. T3SSs are multiprotein machines evolved to inject bacterial effector proteins into host cells to modulate key pathways for infection. Salmonella enterica serovar Typhimurium (S. Typhimurium) encodes two of these protein secretion systems within its pathogenicity islands to mediate bacterial entry into non-phagocytic cells as well as intracellular survival and replication6. Combined, these T3SSs deliver up to 50 bacterial effector proteins that can target a diverse array of cellular activities such as cytoskeleton dynamics, membrane trafficking and transcription6. Given the essential and conserved roles of T3SSs amongst many enteric bacterial pathogens7 and the uncharacterized anti-infective activity of medicinal plants4, we screened a collection of TCMs and discovered that specific flavonoids from TCMs, such as baicalein and quercetin, can covalently label protein substrates of the SPI-1 T3SS and inhibit S. Typhimurium invasion of host cells. Our results suggest that specific flavonoids from TCMs inhibit bacterial virulence rather than targeting growth and reveals a key mechanism by which select TCMs can attenuate bacterial pathogenesis.
RESULTS
High-throughput screen of TCMs reveals specific flavonoid inhibitors of S. Typhimurium type III protein secretion
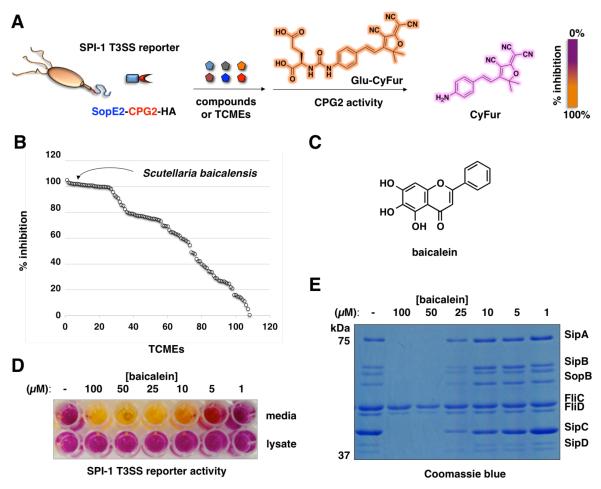
To evaluate the activity of TCMs on type III protein secretion in S. Typhimurium, we screened 120 TCMs with previously suggested anti-infective activity4 using a high-throughput fluorescence previously developed by our laboratory (Figure 1A)8. The type III protein secretion assay employs carboxypeptidase G2 (CPG2) as an enzyme reporter that it is fused to the C-terminus of a known S. Typhimurium T3SS protein effector (SopE2). SopE2-CPG2-HA is secreted and specifically reports on type III protein secretion through cleavage of fluorogenic substrates (Glu-CyFur), which provides a sensitive method for monitoring the inhibitory activity of small molecules targeted at T3SSs (Figure 1A)8. Our screen of anti-infective TCMs at 5 mg/mL revealed 30 medicinal plant extracts with significant S. Typhimurium pathogenicity island-1 (SPI-1) T3SS inhibitory activity (Figure 1B and Table S1). None of these TCMs had significant inhibitory activity on S. Typhimurium growth in culture medium (Table S1). Of the most active TCMs (Figure S1A), we focused on Scutellaria baicalensis (Huángqín) extracts (Figure 1B) as this medicinal plant is a key component of “Scutellaria and Coptis Decoction”, an herbal remedy that has been historically used to treat infectious gastroenteritis4,9. In accordance to the reported isolation of flavonoids from the medicinal plant Scutellaria baicalensis10, our analysis of the major metabolites from Scutellaria baicalensis and other active TCMs demonstrated that only specific flavonoids could inhibit the SPI-1 T3SS reporter levels in the S. Typhimurium culture supernatant (Figure S1B). Notably, baicalein (Figure 1C) from Scutellaria baicalensis reduced the levels of the SPI-1 T3SS reporter (Figure 1D) as well as endogenously expressed SPI-1 T3SS substrates (SipA, SipB, SopB, SipC and SipD) (Figure 1E and Table S2) in S. Typhimurium culture supernatant in dose-dependent manner, without affecting the bacterial growth (Figure S1C) or the recombinant CPG2 reporter activity (Figure S1D). Although baicalein-treated S. Typhimurium showed reduced levels of secreted SPI-1 T3SS reporter, the expression level of the reporter was unaffected (Figure 1C). In contrast, chrysin and baicalin, a deoxygenated and a glycosylated derivative, found in Scutellaria baicalensis extracts were both inactive (Figure S1A). These results demonstrated that a specific flavonoid from the TCM Scutellaria baicalensis, baicalein, does not interfere with bacterial growth but can target secreted SPI-1 T3SS substrates.
Figure 1.
Baicalein from Scutellaria baicalensis (Huángqín) extract inhibits SPI-1 T3SS. (A) Scheme for SopE2-CPG2-HA:Glu-CyFur reporter system for monitoring type III protein secretion. SopE2-CPG2-HA (SPI-1 T3SS) reporter encodes an enzyme activity of carboxypeptidase G2 (CPG2) that when fused to the C-terminus of a known S. Typhimurium T3SS bacterial effector (SopE2) can be secreted and specifically report on type III protein secretion through cleavage of fluorogenic substrates (Glu-CyFur). (B) Screen of TCM activity at 5 mg/mL using SopE2-CPG2-HA:Glu-CyFur reporter system. TCMs were incubated with bacteria for 4 hr and growth media was collected and analyzed for CPG2 activity using Glu-CyFur. CyFur fluorescence was monitored at 610 nm with excitation at 563 nm. (C) Structure of baicalein. (D) Dose-dependent activity of baicalein on SPI-1 T3SS (SopE2-CPG2-HA) reporter in S. Typhimurium growth media and cell lysate. (E) Dose-dependent effect of baicalein on the levels of SPI-1 T3SS secreted proteins (SipA, SipB, SopB, SipC, and SipD) and flagella components levels in S. Typhimurium growth media. The identities of the polypeptides were confirmed by in-gel digestion and LC-MS/MS analysis.
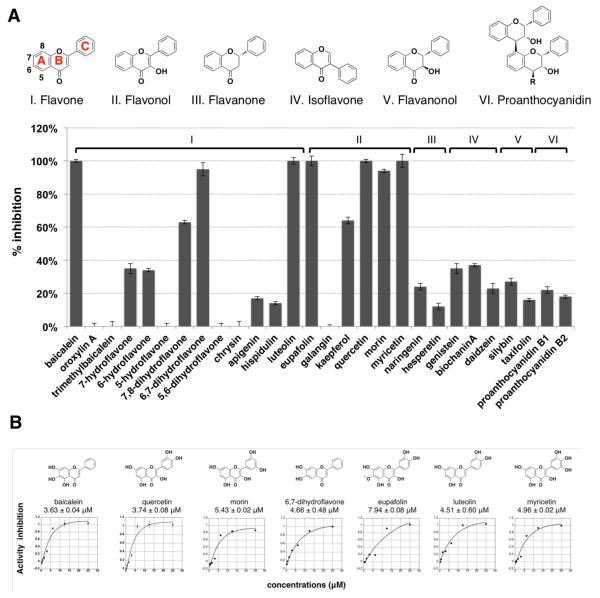
To understand structural features and scope of flavonoid activity on SPI-1 T3SS, we evaluated additional baicalein derivatives and other naturally occurring flavonoids. The analysis of baicalein derivatives showed that the 1,2-catechol moiety and flavone scaffold are important for full activity, since methylated or truncated forms of baicalein were inactive (Figure S2). Our analysis of other naturally occurring flavonoid sub-classes at 25 μM revealed that only flavones and flavonols with 1,2-catechol functionality exhibited inhibitory activity greater than 50 percent (Figure 2A). IC50 measurements of the active flavonoids showed that quercetin had similar activity to that of baicalein (Figure 2B). Interestingly, quercetin is reported in most of the active TCMs we evaluated (Figure S3) and is also a major component of many human dietary supplements. Like baicalein, quercetin reduced the levels of SP1-T3SS reporter and endogenously expressed bacterial effectors in S. Typhimurium media (Figures S4A, B), but had no effect on bacterial growth (Figure S4C). Based on the analysis from SopE2-CPG2-HA fluorescence reporter assay and the secretion of endogenous SPI-1 T3SS substrates, the active flavonoids have comparable or better activity than the previously reported synthetic T3SS inhibitors, INP-000711 and TTS2912 (Figures S4D, E). These results demonstrate that in addition to baicalein, other naturally occurring flavonoids with specific 1,2-catechol motifs, such as quercetin, can also antagonize SPI-1 T3SS substrates.
Figure 2.
Survey of naturally occuring flavonoids against type III protein secretion reporter. (A) Structures of six major classes of flavonoid family: I, flavone (showing the positions of A, B, and C ring), II, flavonol, III, flavanone, IV, isoflavone, V, flavanonol, and VI, proanthocyanidin. Profile of T3SS inhibitory activity of 28 compounds representing 6 different classes of flavonoids at 25 μM using SopE2-CPG2-HA reporter system. Mean ± s.d., n = 3. (B) Chemical structures and IC50 values of most active T3SS inhibitors measured with the SopE2-CPG2-HA reporter fluorescence assay. Mean ± s.d., n = 3.
Flavonoids from medicinal plants inhibit S. Typhimurium invasion of epithelial cells
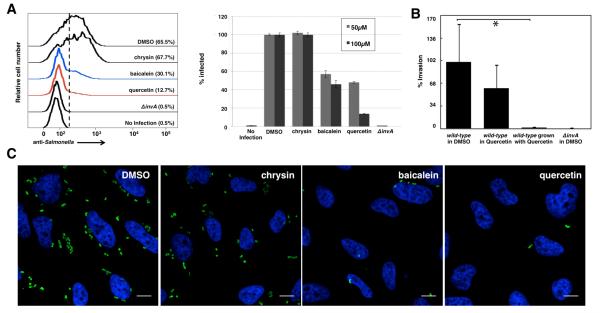
We then tested the ability of the active flavonoids to inhibit S. Typhimurium invasion of cultured epithelial cells, a phenotype strictly dependent on the activity of the SPI-1 T3SS13. Bacteria grown in the presence of baicalein or quercetin showed significant reduction in invasion of cultured HeLa cells (Figure 3A). In contrast, bacteria grown in the presence of chrysin, an inactive flavonoid (Figure S2), were able to enter cultured cells comparable to untreated bacteria (Figure 3A). The inhibitory effect of quercetin at 100 μM was almost complete since the internalization levels of quercetin-treated bacteria phenocopied the levels of a non-invasive S. Typhimurium invA mutant, which lacks an essential component of the SPI-1 T3SS14 (Figures 3B). The inhibition of S. Typhimurium invasion could also be readily observed by immunofluorescence analysis of intracellular bacteria (Figure 3C). Preincubation of the bacteria with active flavonoids was important, as treatment of HeLa cells with quercetin prior to or during bacterial infection alone did not significantly affect S. Typhimurium invasion (Figure 3B). These results demonstrate that active flavonoids, baicalein and quercetin, do not affect bacterial growth and but can inhibit the SPI-1 T3SS-dependent S. Typhimurium invasion of host cells.
Figure 3.
Active flavonoids inhibit SPI-1 T3SS-mediated bacterial invasion of epithelial cells. (A) Left: Flow cytometry analysis of flavonoids at 100 μM on S. Typhimurium invasion of HeLa cells judged by anti-S. Typhimurium antibody staining. MOI = 10, 30 minute infection. Experiment was done in triplicate and similar results were seen in two independent runs. Right: Baicalein and quercetin blocked the invasion of S. Typhimurium into HeLa cells in a dose-dependent manner (darker color: 100 μM, lighter color: 50 μM). Mean ± s.d., n = 3. The values have been normalized to those of DMSO treated, which were considered 100 %. (B) Growth of S. Typhimurium in quercetin abolishes bacterial invasion of cultured epithelial cells. S. Typhimurium was grown in the presence of quercetin (labeled “wt grown in quercetin”) or, alternatively, quercetin was added shortly (30 min) before bacteria were added to cultured epithelial cells (labeled “wt in quercetin). The ability of S. Typhimurium to enter into cultured epithelial cells was evaluated by the gentamicin protection assay13 as described in the Supplemental Methods. Values are the mean ± standard deviation of three independent measurements and represent the % of the inoculum that survive the gentamicin treatment because of their intracellular location. The values have been standardized to those of wild type, which were considered 100 %. The indicated p values were determined by the standard Student t test. * indicates p <0.001 (change panel C for the revised version, which includes *). (C) Immunofluorescence analysis of intracellular S. Typhimurium in HeLa cells with 100 μM flavonoids. Scale bar = 10 μm.
Flavonoids from medicinal plants target S. Typhimurium type III secretion substrates
The observation that inhibition of bacterial invasion by quercetin required preincubation with S. Typhimurium suggested that these flavonoids may block the synthesis or assembly of the SPI-1 T3SS needle complex rather than protein secretion itself. Indeed, before the bacteria encounters mammalian cells, the SPI-1 T3SS is deployed and ready for protein delivery but contact with mammalian cells is required for its full activation15. However, we found that S. Typhimurium grown in the presence of baicalein or quercetin had wild type levels of SPI-1 T3SS effectors (SipB, SptP, SipC, SipA, SipD) needle components (InvG, PrgH, PrgK, PrgI) and fully assembled needle complexes (Figures S5A, B). Since the assembly of the needle complex requires type III protein secretion function, these results suggested that baicalein and quercetin must inhibit S. Typhimurium invasion by a mechanism that did not involve complete block of protein secretion complex. Consistent with this hypothesis, we observed that growth of S. Typhimurium in the presence of quercetin led to reduced levels of some but not all SPI-1 T3SS secreted proteins in culture supernatants (Figure S5C). For example, the levels of secreted SipD were unaffected by addition of quercetin (Figure S5C). Incubation of baicalein or quercetin with S. Typhimurium resulted in the formation of a brown precipitate in the growth media that was not observed with inactive controls (Figure S5D). Western blot analyses of this precipitate material revealed the presence of wild type levels of SPI-1 T3SS secreted proteins (Figure S5C). These results indicated that active flavonoids do not inhibit type III protein secretion directly, but may affect the folding, stability or activity of endogenous SPI-1 T3SS protein substrates as well as effector-enzyme reporters such as SopE2-CPG2.
Although the active flavonoids do not appear to inhibit type III secretion per se, baicalein and quercetin exhibited a strong inhibitory effect on S. Typhimurium invasion (Figure 3), a phenotype strictly dependent on SPI-1 T3SS effector function. This observation combined with the fact that the active flavonoids specifically affected the behavior of type III secreted proteins indicated that these compounds might directly target protein effectors or the protein translocases, which are type III secreted proteins required for the passage of effectors through the eukaryotic cell membrane16. Based on the reactive 1,2-catechol motifs present in the active flavonoids (Figure 2), we hypothesized that these compounds may covalently modify SPI-1 T3SS substrates, which could result in their functional inactivation.
To test this hypothesis, we synthesized alkyne-flavonoid probes for activity-based protein profiling studies using click chemistry17,18 (Figures 4A and S6). As the phenolic hydroxyl groups appeared to be important for baicalein activity (Figure S2), an alkyne tag was installed on the C-ring of baicalein (alk-baicalein) which retained activity compared to inactive alk-Me3-baicalein probe (Figure 4B). These alkyne-flavonoid probes were then incubated with S. Typhimurium under the same conditions of T3SS inhibition and assayed for protein labeling in the cell lysate. Reaction of the S. Typhimurium cell lysates with azide-rhodamine (az-rho) dye using click chemistry showed that alk-baicalein could covalently modify bacterial proteins compared to the inactive probes (Figures 4C and S7A). To determine whether quercetin could also covalently modify S. Typhimurium proteome in a similar manner to baicalein, we performed competition assays with the alk-baicalein probe. Addition of either baicalein or quercetin, but not inactive chrysin, reduced the labeling of alk-baicalein in a dose-dependent manner (Figure S7B).
Figure 4.
Chemical proteomic analysis of alk-baicalein labeled proteins in S. Typhimurium. (A) Alkyne-flavonoid probes for activity-based protein profiling studies. (B) Effect of alkyne-flavonoid probes on the levels of SPI-1 T3SS secreted proteins in S. Typhimurium culture growth media. (C) In-gel fluorescence analysis of alkyne-flavonoid probe labeling of S. Typhimurium proteins following click chemistry reaction with az-rho fluorescence dye.
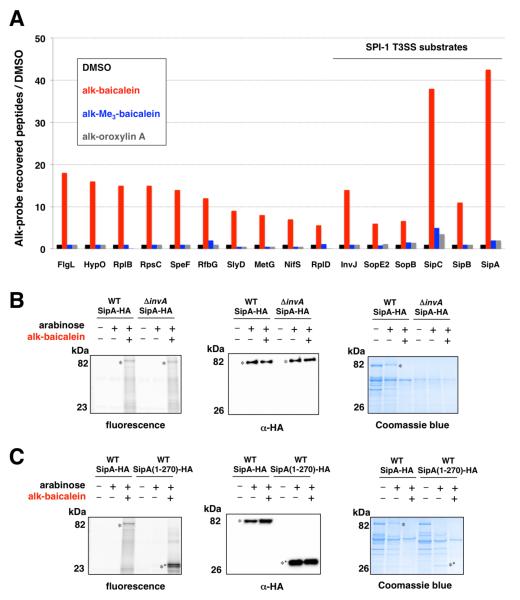
To identify the protein targets of the alk-baicalein probe, we performed chemical proteomics using a cleavable azide-functionalized biotinylated affinity tag followed by gel-based protein identification19. Compared to the control samples, alk-baicalein preferentially labeled 16 S. Typhimurium proteins with more than 5-fold enrichment, including SPI-1 T3SS protein translocases (SipC and SipB) and effectors (SipA, SopB and SopE2) as well as the secreted needle-assembly regulatory protein InvJ (Figure 5A and Table S3). To validate the covalent labeling of these SPI-1 T3SS substrates, we focused on the most prominent alk-baicalein labeled protein SipA, an injected S. Typhimurium protein effector involved actin remodeling and membrane ruffling required for bacterial invasion of epithelial cells20. Indeed, expression and affinity enrichment of C-terminal HA-tagged SipA (SipA-HA) in S. Typhimurium (Table S4) confirmed that alk-baicalein covalently labels this SPI-1 T3SS substrate, as judged by in-gel fluorescence detection (Figure 5B). Alk-baicalein also labeled SipA-HA in SPI-1 T3SS mutant strain (invA) (Figure 5B), demonstrating flavonoid labeling of SPI-1 T3SS protein substrates does not require protein secretion and can occur within S. Typhimurium. One conserved feature of SPI-1 T3SS substrates is a hydrophobic N-terminal domain that interacts with chaperones involved in protein secretion21. We hypothesized that the active flavonoids may bind this conserved hydrophobic motif within SipA and react with nucleophilic amino acids, as previously suggested for α-synuclein22. Analysis of an N-terminal SipA-HA construct (1-270) expressed in S. Typhimurium demonstrated that a truncated mutant contain this conserved hydrophobic motif is sufficient for covalent flavonoid labeling (Figure 5C). Although alk-baicalein also covalently labeled other S. Typhimurium proteins (Figure 5A and Table S3), these proteins are not associated with SPI-1 T3SS-dependent invasion of host cells and are unlikely to be functionally relevant. These results demonstrate that active flavonoids can covalently modify a conserved chaperone-binding motif S. Typhimurium SPI-1 T3SS substrates and may inactivate these secreted effectors and translocases to prevent bacterial invasion.
Figure 5.
Validation of alk-baicalein labeled SPI-T3SS substrate, SipA. (A) Summary of S. Typhimurium proteins that were labeled by alkyne-flavonoid probes. The total number of peptide-sequence matches (PSM) between alk-baicalein and the inactive probes, alk-trimethylbaicalein, alk-oroxylin A were compared to DMSO control. The graph shows the 16 proteins that were labeled by alk-baicalein with more than 5-fold PSM enrichment compared to control samples. (B) Alk-baicalein labeling of SipA-HA in wild-type (wt) and T3SS-deficient (invA) S. Typhimurium. SipA-HA was immunopurified from alk-baicalein treated S. Typhimurium, reacted with az-rho and analyzed by in-gel fluorescence and anti-HA western blot. Coomassie blue staining of bacterial culture supernatants was performed in parallel to assay on protein secretion. (C) SipA-HA and SipA(1-270)-HA was immunopurified from alk-baicalein treated S. Typhimurium and analyzed as described in (B).
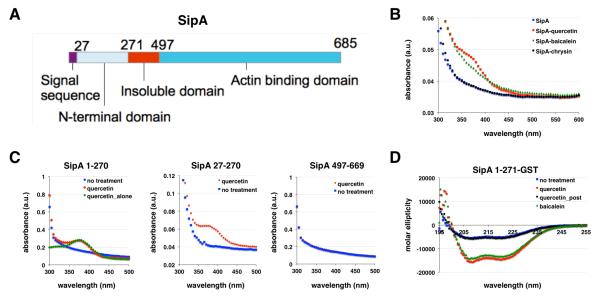
To independently validate the covalent interaction of specific flavonoids with SPI-1 T3SS substrates, we evaluated the binding of these compounds with SipA-glutathione-S-transferase (GST) fusions in bacteria and in vitro using the intrinsic UV-absorption properties of flavonoids at 370 nm (Figure 6). The SipA-GST constructs were also expressed in E. coli to evaluate whether flavonoid covalent modification was specific to incubation with S. Typhimurium (Figure 6A). Indeed, expression of SipA-GST in E. coli incubated with baicalein or quercetin yielded altered UV-absorbance profiles from 350 – 400 nm of purified SipA-GST compared to untreated or inactive flavonoid (chrysin)-treated samples (Figures 6B and S8). We then evaluated different SipA-GST constructs to determine the domain of flavonoid labeling in E. coli, which demonstrated that active flavonoids (quercetin) primarily associates with the N-terminal chaperone binding domain of SipA, independent of signal sequence (residues 1-27) and did not occur within the C-terminal actin binding domain of SipA (Figure 6C), which are consistent with our analysis of alk-baicalein labeling of SipA-HA constructs in S. Typhimurium (Figure 5).
Figure 6.
Binding and circular dichroism analysis (CD) of SipA-flavonoid interactions. (A) SipA domains. (B) Spectra of SipA-GST fusion purified from the E. coli treated with flavonoids. Quercetin and baicalein treated SipA-GST had a shoulder at the 370 nm region, while the inactive flavonoid chrysin and untreated SipA did not show this spectral feature. (C) Spectra of the N- and C-terminal domains of SipA purified with and without quercetin. Quercetin-treated SipA N-terminal construct exhibits a peak at 370 nm, while the signal sequence mutant (27-271) and C-terminal domain-GST fusion did not. (D) CD spectra of SipA untreated and expressed in the presence of flavonoids. CD spectra of SipA-1-271-GST expressed without the presence of flavonoids showed a characteristic α-helical pattern. Treatment with 50 μM quercetin and baicalein induced significant spectra change at 208 nm and 222 nm. However, addition of quercetin to purified SipA-1-271-GST in vitro did not induce changes in CD spectra.
To monitor conformational changes due to flavonoid binding interactions, we performed circular dichroism (CD) experiments on SipA-1-271-GST constructs from E. coli incubated with quercetin or baicalein. In agreement with the published crystal structure of SipA21,23, the CD results showed that SipA-1-271-GST retained an α-helical character with negative bands at 222 nm and 208 nm and a positive band at 193 nm23 (Figure 6D). Treatment with baicalein or quercetin induced significant CD spectra changes at the characteristic α-helical bands, which suggests association with the active flavonoids altered the secondary structure of SipA N-terminal domain (Figure 6D). Incubation of purified SipA N-terminal domain (1-271)-GST fusion with quercetin in vitro did not enhance UV-absorbance at 370 nm or alter the CD spectra (Figure 6D). These results suggest that the active flavonoids baicalein and quercetin bind to the N-terminal domain (1-271) of SipA inside bacteria, but not in vitro and affect the secondary structure of this chaperone-binding domain in SPI-1 T3SS substrates.
DISCUSSION
TCMs have been used to treat infections by enteric bacterial pathogens, but the molecular mechanisms by which these medicinal plant extracts attenuate bacterial pathogenesis have been unclear. By focusing on type III protein secretion responsible for bacterial virulence, we have discovered specific compounds within TCM extracts and their mechanism of action that provides an explanation for some of the previously proposed anti-infective properties of these ancient medicines4,24. While flavonoids exhibit diverse pharmacological activities in mammalian cells25, are potential antiviral agents26 and have minimal activity on bacteria growth as antibiotics27,28, we demonstrated that these plant-derived metabolites can attenuate infection by targeting virulence factors such as protein substrates delivered by T3SSs. Our subsequent chemical proteomic studies demonstrated that the most active flavonoids, contained reactive 1,2 catechol motifs that covalently modify secreted bacterial effectors and translocases required bacterial invasion of host cells. The active flavonoid covalent modification of conserved hydrophobic domains of T3SS substrates and may interfere with the folding, secretion or activity of these key virulence factors. Our observations may explain some of the previously reported anti-infective properties of TCM extracts that produce active flavonoids4,24. As activated phenolic chemotypes present in baicalein and quercetin are also found in previous reported T3SS inhibitors (INP-000711 and TTS2912) and other active plant metabolites from TCMs associated with protein aggregation29, the covalent modification of proteins described here may provide a general mechanism for inactivating proteins. Since many plant bacterial pathogens use type III secretion systems to cause disease and sense flavonoids30, it is possible that some of these compounds may have evolved as a consequence host-pathogen interactions. Moreover, flavonoids have also been reported to target other anti-infective properties such as expression of virulence genes31,32, toxin activity33-35, quorum sensing36-39 and biofilm formation40,41 in bacterial pathogens as well as attenuate destructive host inflammation42-44. Given the broad pharmacological activities of flavonoids, these plant metabolites may affect microbial virulence mechanisms, including type III protein secretion substrates shown here, in addition to host pathways for their anti-infective properties in vivo. With the resurgence in covalent drugs45, the development of more potent and bioavailable flavonoid antagonists of T3SSs should yield new anti-infective agents to selectively combat bacterial pathogens.
EXPERIMENTAL SECTION
High-throughput type III secretion assay for screening of extracts and compounds
S. Typhimurium containing SopE2-CPG2-HA plasmid was grown overnight at 37 °C in LB containing 50 μg/mL ampicillin and was diluted by 1:30. In each well of the sterile Nunc 96 deep-well plates (2 mL), 400 μL of the diluted culture was then grown for 4 hours in the presence of compounds or TCMs at indicated concentrations with DMSO or H2O as controls. All added volumes of inhibitors or DMSO were <1% of the total volume in each experiment. After 4 hours, the OD600 was measured to confirm comparable bacterial growth. 250 μL of each sample was transferred into a 96-well plate and bacterial cells were spun down at 5,250 g for 6 minutes. 20 μL of supernatant from each sample was then transferred into a 384-well costar black-bottom plate. 80 μL of CPG2 buffer (50 mM Tris, 0.1 mM ZnCl2, pH 7.4) containing 0.1% Brij-97 to aid in solubility of CyFur8 and 10 μM Glu-CyFur was added, and fluorescence readings (λex= 563 nm, λem= 610 nm with no cutoff) were started immediately and monitored as kinetic readings for 2 hours. Absorbance and fluorescence data were collected on SpectraMax M2 multi-detection reader (Molecular Devices). The percent inhibition by chemical inhibitors or TCMs was determined by calculating the rates of change in fluorescence of the CPG2-expressing strain in the presence of compound in comparison to the DMSO or H2O control. IC50 values of the inhibitors were then determined using KaleigaGraph version 4.1 and fitting to the exponential decay equation: y = m1 + m2*exp(−m3*x).
Plasmids, bacterial strains, and growth conditions
S. Typhimurium strain expressing SopE2-CPG2-HA fusion protein8 and the S. Typhimurium strains SB90546, SB30046, IR71547 and the T3SS-defective invA mutant14 have been previously described. All LB media used were made from BD Difco™ LB (Luria-Bertani) Broth Miller, which contains 10 g/L NaCl.
Profiling of secreted proteins in bacterial culture supernatants
1:30 dilutions of overnight cultures of S. Typhimurium were grown in LB for 4 h in the presence of compounds at indicated concentrations. Secreted proteins from 1 mL culture were precipitated overnight with a final concentration of 10% TCA at 4 °C. Secreted effectors were pelleted at 14,000 rpm for 30 minutes and washed with 250 μL ice-chilled acetone. This procedure was repeated 2 times and the precipitates were allowed to dry for 15 minutes before the addition of 4% SDS. Secreted proteins were then separated by a 4-20% SDS-PAGE and stained with coomassie blue.
Cell culture, bacterial infection, flow cytometry and immunofluorescence
HeLa cells were cultured in 12-well plates in DMEM supplemented with 10% FBS (Gemini Bio-Products) at 37 °C in a humidified incubator with an atmosphere of 5% CO2. S. Typhimurium strains IR715, SB300, and the invA deletion mutant were grown overnight in LB at 37 °C in a shaker set at 250 rpm. Bacteria were diluted 1/33 in LB containing DMSO or inhibitors, and were grown for an additional 4 hours. Optical density readings were used to determine an MOI of 10 and bacteria was added to cells in a total volume of 500 uL DMEM/10% FBS containing inhibitors or DMSO as a control. Plates containing cells and bacteria were then centrifuged at room temperature for 5 min at 1000 x g. Plates were then transferred back to the 37 °C incubator for 30 min to allow infection to proceed. Cells were then washed three times with room temperature PBS containing 100 ug/mL gentamycin and were then incubated with DMEM/10% FBS containing 100 ug/mL gentamycin and inhibitors or DMSO as a control at 37 °C for an additional 30 min. Cells were then washed an additional three times with room temperature PBS containing 100 ug/mL gentamycin to remove any remaining extracellular bacteria. Cells were trypsinized and then fixed with ice cold 3.7% paraformaldehyde in PBS for 10 minutes followed by permeabilization with ice cold 0.2% saponin in PBS for 10 min. Cells were then blocked with ice cold 2% FBS in PBS for 10 min. All antibody stainings and washes were performed with ice-cold 0.2% saponin in PBS. Cells were stained for bacterial antigens with anti-Salmonella rabbit serum (Biodesign International, 1/250 dilution) for 1 h and washed three times. Goat anti-rabbit secondary antibody conjugated to AlexaFluor 488 (Invitrogen) was used at a 1/1000 dilution for 30 min. Cells were then washed 3 times. Flow cytometry was performed using a Becton Dickinson LSRII machine and FlowJo software was used for analysis. 30,000 cells were analyzed for each sample. Cellular debris was eliminated from analysis of all samples based on forward and side scatter measurements and never constituted more than 10% of collected events. Gates drawn to illustrate infected cells were based on a value close to 0% for non-infected samples. For the CFU-based infection studies, a protocol was used based on a previous report13. For microscopy, HeLa cells were grown on glass coverslips. Infections and staining were performed using the same protocol as described above for flow cytometry. For the imaging the extracellular S. Typhimurium, the scrapped-harvest HeLa cells were not permeablized and there is no gentamycin added. Cells were treated with TOPRO-3 (1/1000 dilution, Invitrogen) as a final step to stain nuclei and were mounted using Prolong Gold Antifade Reagent (Invitrogen).
Analysis of endogenously expressed T3SS needle components and substrates
S. Typhimurium supernatants and cell lysates were prepared as previously described and analyzed by western blot by pooling specific antibodies for T3SS substrates48. Needle complexes were purified from S. Typhimurium strain SB905 as previously described49, and subsequently analyzed by western blot using a previously described antibody specific for the needle complex components PrgH, PrgK, and InvG50. Purified needle complexes were applied to glow discharged carbon coated Cu-grids and stained with 2% phosphotungstic acid, pH 7.0 and imaged by electron microscopy. Images were acquired using a Tecnai Biotwin TEM (FEI Company, Hillsboro) at 80 kV using Morada Soft Imaging system and 6 M pixel CCD camera (Olympus, Munster, Germany).
Coomassie staining of secreted proteins
1:30 dilutions of overnight cultures of S. typhimurium were grown in LB for 4 h in the absence or presence of compounds at indicated concentrations. Secreted proteins from 1 mL culture were precipitated overnight with a final concentration of 10% TCA at 4 °C. Secreted effectors were pelleted at 14,000 rpm for 30 minutes and washed with 250 μL ice-chilled acetone. This procedure was repeated 2 times and the precipitates were allowed to dry for 15 minutes before the addition of Laemmli buffer. Secreted proteins were then separated by 4-20% SDS-PAGE and stained with SimplyBlue™ SafeStain (Invitrogen). Gels were imaged with ChemiDoc™ XRS+ System and Image Lab™ Software (Bio-Rad).
Preparation of bacterial total cell lysates
S. Typhimurium strain 14028 WT and ΔinvA mutant carrying pBAD-SipA-HA or pBAD-SipA(1-270)-HA were cultured in the absence or presence of 0.2% arabinose and labeled with alk-baicalein or DMSO control in 4 mL LB at 37 °C with 220 rpm shaking for 4 hr. S. Typhimurium cells were pelleted at 15000 g for 1 min, and pellets were lysed with 200 μL CelLytic B 2X Cell Lysis Reagent (Sigma) containing 1X EDTA-free protease inhibitor cocktail (Roche), 0.5 mg/ml lysozyme (in ddH2O) (Sigma), and 0.2 μL S16 benzonase (Sigma). After re-suspension, pellets were sonicated for 10 sec for 3 times, then were incubated on ice for 30 min. Cell lysates were centrifuged at 15000 g for 1 min to remove cell debris and supernatants were collected. Protein concentration was estimated by BCA assay with BCA Protein Assay Kit (Thermo).
Chemical proteomic analysis with alkyne-flavonoid probes
S. Typhimurium strain (IR715) were labeled with alk-baicalein, alk-Me3-baicalein, alk-oroxylin A or DMSO control in LB at 37 °C for 4 hr. S. Typhimurium pellets were lysed with 0.1% SDS buffer (50 mM triethanolamine (TEA), 150 mM NaCl, 0.1% SDS, 1X EDTA-free protease inhibitor cocktail (Roche), 5 mM PMSF from a 250 mM stock in EtOH, 0.1 μL S16 benzonase (Sigma)) by sonicating pellet for 10 sec, then incubating on ice for 10 min. After which 5 μL of 10 μg/mL lysozyme (in ddH2O) (Sigma) was added to each sample and incubated on ice for 30 min. SDS buffer (12% w/v in 50 mM TEA, 150 mM NaCl) was then added to bring final SDS concentration to 4%. Cells were sonicated once more for 5 sec. Cell lysates were centrifuged at 1000 g for 5 min to remove cell debris and supernatant was collected. From these alkyne-flavonoid or DMSO-treated samples, 50 μg of total cell lysates was suspended in 44.5 μL of 4% SDS buffer. CuAAC reactants 1 μL of 5 mM az-rho (100 μM final concentration), 1 μL of freshly prepared 50 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (final concentration 1 mM), 2.5 μL 2 mM tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (final concentration 100 μM) and 1 μL 50 mM CuSO4·5H2O (final concentration 1 mM), were then added for a final volume of 50 μL. CuAAC reactions were allowed to proceed for 1 h at room temperature and a chloroform-methanol precipitation of the proteins was performed before SDS-PAGE. In-gel fluorescence scanning was performed using a Typhoon 9400 imager (Amersham Biosciences).
For proteomic experiments, cell lysates (5-10 mg) were brought up to a volume of 8.9 mL with a final SDS concentration of 2%. To this, 200 μL of 5 mM azido-azo-biotin19; 200 μL of 50 mM TCEP; 500 μL of 2 mM TBTA; and 200 μL of 50 mM CuSO4 were added, and samples were incubated for 1.5 h at room temperature. After incubation, proteins were precipitated by chloroform/methanol precipitation as described: 4x MeOH, 1.5X chloroform, and 3X ddH2O were added to reaction, and samples were vortexed vigorously. Samples were centrifuged at 5200 g for 30 min, and the aqueous layer was removed. Samples were washed two times with 4X methanol, by vortexing vigorously, centrifuging at 5200 g for 30 min, then decanting off the supernatant each time. Protein pellets were air-dried, then resuspended in 1 mL 4% SDS buffer and 20 μL 0.5 M EDTA pH 8. Protein concentration was estimated by BCA assay.
Cell lysates (1-5 mg) after CuAAC reaction were added to PBS-washed high-capacity streptavidin agarose beads (Thermo scientific) (beads in a 50% Brij-97 buffer slurry, 100 μL per 5 mg protein). Beads were incubated with cell lysates for 1 h at room temperature with rocking. Beads were then washed 2X with 10 mL of 0.1% SDS in PBS, 3X with 10 mL PBS, 2X with 10 mL 50 mM ABC, then incubated for 30 min with freshly made 8 M urea solution (500 μL 8M urea, 25 μL 200 mM TCEP, 25 μL 400 mM iodoacetamide). Beads were washed 2X with 10 mL 50 mM ABC, then transferred to a dolphin tube with 1 mL ABC. Proteins were eluted off beads in two steps with a total of 400 μL freshly prepared elution buffer (0.5% SDS, 25 mM Na2S2O4 in 50 mM ABC) by rocking for 1 h at room temperature, then for 30 m. Eluant was concentrated using a Microcon YM-10 centrifugal filter (Millipore) as per manufacturers protocol. Microcon filters were rinsed twice with 50 μL 1% SDS/75 mM BME to collect all sample, and samples were dried by vacuum at room temperature. Samples were resuspended in 25 μL LDS buffer (1X LDS (Invitrogen), 5% BME), boiled for 5 min at 95 °C, spun at 1000 g for 1 min, then separated by SDS-PAGE. Gels were stained with Coomassie blue and destained overnight (40% methanol, 10% acetic acid, 50% water). Each gel lane was cut into 8 slices. These slices were washed once in 50 mM ABC, twice in 1:1 50 mM ABC to acetonitrile (ACN), and once in ACN. Slices were dried by vacuum. 2 μg of trypsin in 50 mM ABC was added to each gel slice, and slices were incubated overnight at 37 °C. Proteins released from the gel were collected, and gel slices were washed in 1:1 50 mM ABC:ACN + 0.1% TFA twice, collecting the supernatant each time. The pooled supernatant was dried by vacuum then resuspended in 25 μL 0.1% TFA in water for analysis by LCMS/MS. Proteomics data analysis was performed at Rockefeller University Proteomics Research Center using Mascot through Proteome Discoverer (Thermo Proteome Discoverer version 1.3.0.339.).
Construction of pBAD-SipA-HA and pBAD-SipA(1-270)-HA
SipA-HA were amplified from genome of S. Typhimurium strain 14028 WT with primers ZZ91 and ZZ92. PCR product was digested with EcoRI and XbaI and ligated to pBAD vector to yield pBAD-SipA-HA. pBAD-SipA(1-270)-HA were made through site-directed mutagenesis on pBAD-SipA-HA with primers ZZ115 and ZZ116, and QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) according to manufacturer’s manual.
In-gel fluorescence analysis of SipA-HA and SipA(1-270)-HA
From the alk-baicalein or DMSO-treated total cell lysates prepared as described above, 250 μg of each total cell lysates were incubated with 20 μL PBS-T-washed EZview™ Red Anti-HA Affinity Gel (Sigma) at 4 °C for 1 hour with end-to-end rotation. Samples were washed with 200 μL PBS-T for 3 times, and 36 μL of PBS was added to each sample. 4 μL of click chemistry reagents were added to each sample as a master mix (az-rho: 0.1 mM, 10 mM stock solution in DMSO; tris(2-carboxyethyl)phosphine hydrochloride (TCEP): 1 mM, 50 mM freshly prepared stock solution in ddH2O; tris[(1-benzyl- 1H-1,2,3-triazol-4-yl)methyl]amine (TBTA): (0.1 mM, 2 mM stock in 4:1 t-butanol: DMSO); CuSO4 (1 mM, 50 mM freshly prepared stock in ddH2O). Samples were mixed well and incubated at room temperature for 1 h. After incubation, samples were washed with 200 μL PBS-T for 3 times, and were boiled with Laemmli buffer 95 °C for 5 min before being loaded onto a 4-20% gel (Bio-Rad) for SDS-PAGE. In-gel fluorescence scanning was performed using a Typhoon 9400 imager (Amersham Biosciences. Anti-HA western blotting was performed in parallel to confirm proper expression of HA-tagged proteins, with monoclonal anti-HA-HRP antibody HA-7 (Sigma) at 1:5000 dilution.
Analysis of SipA-GST interaction with flavonoids
The SipA-glutathione S-transferase (GST) fusion constructs21,51 were expressed in E. coli BL21 (DE3) containing and induced at OD 0.8 with 1 mM IPTG for 16 hrs at 20 °C. Flavonoids (100 μM) were added 1 hour before IPTG induction. Cells collected via centrifugation and lysed in Wash Buffer (1XPBS, 1 mM DTT, 1 mM EDTA) with 0.1% Triton-X and a protease inhibitor cocktail (Roche). The clarified cell lysates were placed on pre-equilibrated 1 mL Glutathione columns (Qiagen) and washed with 10 column volumes of Wash Buffer. Protein was eluted with Elution Buffer (50 mM Tris pH 8.0, 0.4 M NaCl, 50 mM reduced glutathione, 0.1% Triton X-100, 1 mM DTT) and 1 mL fractions were collected. All fractions were tested for protein purity via SDS-PAGE gels. Fractions containing SipA were pooled, concentrated, and buffer exchanged into 1XPBS. The SipA-GST constructed purified in this way were SipA 1-270, SipA 27-270, and SipA 497-669. The protein samples were then characterized with UV-visible (Molecular Devices SpectraMax M2) and circular dichroism (Aviv model 202 CD spectrometer) spectroscopy to determine if any structural changes occurred in the flavonoid bound proteins. Experiments were run at 0.25 mg/mL with proteins purified from cultures treated either with quercetin, baicalein, or chrysin. In addition, SipA from untreated cultures was tested with the addition of 25 μM and 50 μM exogenous quercetin. The spectra window was set between 180 and 275 nm. The CD spectra of SipA were shown to be mostly α-helical, which agrees with previous crystallography studies.51
Supplementary Material
ACKNOWLEDGEMENTS
We thank The Rockefeller Proteomics Resource Center for mass spectrometry analysis. We thank the Stebbins laboratory (Rockefeller University) for SipA-GST plasmids. P.D. was graduate student in Weill Cornell-Rockfeller-Sloan Kettering Tri-Institutional Training Program in Chemical Biology. J.R. was supported NIH F32 postdoctoral fellowship AI096890. Y-C.C. is a fellow of the National Foundation for Cancer Research. This work was supported by grants to Y-C.C. (NCI PO1CA154295), J.E.G. (NCCAM R01AT007671 and NIAID AI30492) and H.C.H (NCCAM R01AT007671, Starr Cancer Consortium and Lerner Trust).
Footnotes
Supporting information. Supporting information is available free of charge on ACS Publications website at DOI: TBD. Details on screening and characterization of TCMs and flavonoids as well as synthesis, characterization and proteomic analysis of flavonoid probes are included.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).White NJ. Science. 2008;320:330. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- (2).Corson TW, Crews CM. Cell. 2007;130:769. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lewis K, Ausubel FM. Nat. Biotechnol. 2006;24:1504. doi: 10.1038/nbt1206-1504. [DOI] [PubMed] [Google Scholar]
- (4).Bensky DC, S., Stoger E. Chinese Herbal Medicine: Materia Medica. Third ed Eastland Press; 2004. [Google Scholar]
- (5).Lewis K, Ausubel FM. Nat Biotechnol. 2006;24:1504. doi: 10.1038/nbt1206-1504. [DOI] [PubMed] [Google Scholar]
- (6).Haraga A, Ohlson MB, Miller SI. Nat. Rev. Microbiol. 2008;6:53. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- (7).Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Annu Rev Microbiol. 2014;68:415. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yount JS, Tsou LK, Dossa PD, Kullas AL, van der Velden AW, Hang HC. J. Am. Chem. Soc. 2010;132:8244. doi: 10.1021/ja102257v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen JK, a. C., T. T. Chinese Herbal Formulas and Applications. First ed Art of Medicine Press; 2008. [Google Scholar]
- (10).Li HB, Chen F. J. Chromatogr. A. 2005;1074:107. doi: 10.1016/j.chroma.2005.03.088. [DOI] [PubMed] [Google Scholar]
- (11).Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. Infect. Immun. 2005;73:3104. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, Bronstein PA, Kline T, Miller SI. Cell Host Microbe. 2008;4:325. doi: 10.1016/j.chom.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Galan JE, Curtiss R., 3rd Proc. Natl. Acad. Sci. U. S. A. 1989;86:6383. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Galan JE, Ginocchio C, Costeas P. J. Bacteriol. 1992;174:4338. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zierler MK, Galan JE. Infect. Immun. 1995;63:4024. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Collazo CM, Galan JE. Mol. Microbiol. 1997;24:747. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- (17).Speers AE, Adam GC, Cravatt BF. J. Am. Chem. Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- (18).Speers AE, Cravatt BF. Chem. Biol. 2004;11:535. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- (19).Yang YY, Grammel M, Raghavan AS, Charron G, Hang HC. Chem. Biol. 2010;17:1212. doi: 10.1016/j.chembiol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhou D, Mooseker MS, Galan JE. Science. 1999;283:2092. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- (21).Lilic M, Vujanac M, Stebbins CE. Mol. Cell. 2006;21:653. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- (22).Meng X, Munishkina LA, Fink AL, Uversky VN. Biochemistry. 2009;48:8206. doi: 10.1021/bi900506b. [DOI] [PubMed] [Google Scholar]
- (23).Holzwarth G, Doty P. J. Am. Chem. Soc. 1965;87:218. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- (24).Xie Y, Yang W, Tang F, Chen X, Ren L. Curr Med Chem. 2015;22:132. doi: 10.2174/0929867321666140916113443. [DOI] [PubMed] [Google Scholar]
- (25).Pietta PG. J Nat Prod. 2000;63:1035. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- (26).Sun F, Huang R. Curr Drug Targets. 2014;15:175. doi: 10.2174/13894501113146660203. [DOI] [PubMed] [Google Scholar]
- (27).Cushnie TP, Lamb AJ. Int. J. Antimicrob. Agents. 2011;38:99. doi: 10.1016/j.ijantimicag.2011.02.014. [DOI] [PubMed] [Google Scholar]
- (28).Daglia M. Curr Opin Biotechnol. 2012;23:174. doi: 10.1016/j.copbio.2011.08.007. [DOI] [PubMed] [Google Scholar]
- (29).Duan D, Doak AK, Nedyalkova L, Shoichet BK. ACS Chem. Biol. 2015;10:978. doi: 10.1021/cb5009487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Deakin WJ, Broughton WJ. Nat Rev Microbiol. 2009;7:312. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- (31).Vikram A, Jesudhasan PR, Jayaprakasha GK, Pillai SD, Jayaraman A, Patil BS. Int J Food Microbiol. 2011;145:28. doi: 10.1016/j.ijfoodmicro.2010.11.013. [DOI] [PubMed] [Google Scholar]
- (32).Vargas P, Farias GA, Nogales J, Prada H, Carvajal V, Baron M, Rivilla R, Martin M, Olmedilla A, Gallegos MT. Environmental microbiology reports. 2013;5:841. doi: 10.1111/1758-2229.12086. [DOI] [PubMed] [Google Scholar]
- (33).Lee JH, Park JH, Cho MH, Lee J. Curr Microbiol. 2012;65:726. doi: 10.1007/s00284-012-0229-x. [DOI] [PubMed] [Google Scholar]
- (34).Dong J, Zhang Y, Chen Y, Niu X, Zhang Y, Yang C, Wang Q, Li X, Deng X. Antimicrob Agents Chemother. 2015;59:7054. doi: 10.1128/AAC.01416-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tombola F, Campello S, De Luca L, Ruggiero P, Del Giudice G, Papini E, Zoratti M. FEBS Lett. 2003;543:184. doi: 10.1016/s0014-5793(03)00443-5. [DOI] [PubMed] [Google Scholar]
- (36).Vandeputte OM, Kiendrebeogo M, Rasamiravaka T, Stevigny C, Duez P, Rajaonson S, Diallo B, Mol A, Baucher M, El Jaziri M. Microbiology. 2011;157:2120. doi: 10.1099/mic.0.049338-0. [DOI] [PubMed] [Google Scholar]
- (37).Lee KM, Lim J, Nam S, Yoon MY, Kwon YK, Jung BY, Park Y, Park S, Yoon SS. FEMS Microbiol Lett. 2011;321:67. doi: 10.1111/j.1574-6968.2011.02311.x. [DOI] [PubMed] [Google Scholar]
- (38).Bhargava N, Singh SP, Sharma A, Sharma P, Capalash N. Future Microbiol. 2015;10:1953. doi: 10.2217/fmb.15.107. [DOI] [PubMed] [Google Scholar]
- (39).Gopu V, Meena CK, Shetty PH. PLoS One. 2015;10:e0134684. doi: 10.1371/journal.pone.0134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Rajendran N, Subramaniam S, Christena LR, Muthuraman MS, Subramanian NS, Pemiah B, Sivasubramanian A. Nat Prod Res. 2015:1. doi: 10.1080/14786419.2015.1104673. [DOI] [PubMed] [Google Scholar]
- (41).Ding X, Yin B, Qian L, Zeng Z, Yang Z, Li H, Lu Y, Zhou S. J Med Microbiol. 2011;60:1827. doi: 10.1099/jmm.0.024166-0. [DOI] [PubMed] [Google Scholar]
- (42).Sugiyama T, Kawaguchi K, Dobashi H, Miyake R, Kaneko M, Kumazawa Y. FEMS Immunol Med Microbiol. 2008;53:306. doi: 10.1111/j.1574-695X.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- (43).Li-Weber M. Cancer Treat Rev. 2009;35:57. doi: 10.1016/j.ctrv.2008.09.005. [DOI] [PubMed] [Google Scholar]
- (44).Peluso I, Miglio C, Morabito G, Ioannone F, Serafini M. Critical reviews in food science and nutrition. 2015;55:383. doi: 10.1080/10408398.2012.656770. [DOI] [PubMed] [Google Scholar]
- (45).Singh J, Petter RC, Baillie TA, Whitty A. Nat. Rev. Drug. Discov. 2011;10:307. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- (46).Kaniga K, Bossio JC, Galan JE. Mol Microbiol. 1994;13:555. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- (47).Stojiljkovic I, Baumler AJ, Heffron F. J Bacteriol. 1995;177:1357. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kato J, Lefebre M, Galan JE. J Bacteriol. 2015;197:3007. doi: 10.1128/JB.00434-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Science. 1998;280:602. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- (50).Sukhan A, Kubori T, Wilson J, Galan JE. J Bacteriol. 2001;183:1159. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lilic M, Galkin VE, Orlova A, VanLoock MS, Egelman EH, Stebbins CE. Science. 2003;301:1918. doi: 10.1126/science.1088433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.