Abstract
Introduction
High-throughput loss-of-function genetic screening tools in yeast or other model systems except in mammalian cells have been implemented to study human susceptibility to chemical toxicity. Here, we employed a newly developed human haploid cell (KBM7)-based mutagenic screening model (KBM7-mu cells) and examined its applicability in identifying genes whose absence allows cells to survive and proliferate in the presence of chemicals.
Methods
KBM7-mucells were exposed to 200 µM Chlorpyrifos (CPF), a widely used organophosphate pesticide, a dose causing approximately 50% death of cells after 48 h of treatment. After a 2–3 week period of continuous CPF exposure, survived single cell colonies were recovered and used for further analysis. DNA isolated from these cells was amplified using Splinkerette PCR with specific designed primers, and sequenced to determine the genomic locations with virus insertion and identify genes affected by the insertion. Quantitative realtime reverse transcription PCR (qRT-PCR) was used to confirm the knockdown of transcription of identified target genes.
Results
We identified total 9 human genes in which the cells carrying these genes conferred the resistance to CPF, including AGPAT6, AIG1, ATP8B2, BIK, DCAF12, FNBP4, LAT2, MZF1-AS1 and PPTC7. MZF1-AS1 is an antisense RNA and not included in the further analysis. qRT-PCR results showed that the expression of 6 genes was either significantly reduced or completely lost. There were no changes in the expression of DCAF12 and AGPAT6 genes between the KBM7-mu and the control KBM7 cells.
Discussion
The KBM7-mu genetic screening system can be modified and applied to identify novel susceptibility genes in response to environmental toxicants, which could provide valuable insights into potential mechanisms of toxicity.
Keywords: Chlorpyrifos, KBM7-mutated haploid cells, Loss-of-function genetic screening, Susceptibility to environmental chemical exposure
1. Introduction
Loss-of-function genetic screening models have become popular tools for phenotype selection, identification of genetic susceptibility genes and understanding molecular mechanism of toxic chemical exposures. Mammalian cells have a diploid genome, which has restricted the use of loss-of-function genetic screening models in these higher organisms (Kotecki, Reddy, & Cochran, 1999). Recently, the human haploid genetic screening system (KBM7-mu) was developed from a derivative of a chronicmyeloid leukemia (CML) cell line (KBM7) of hematopoietic origin by a group at MIT (Carette et al., 2009). This system was developed utilizing gene-trap retroviruses that contain a strong adenoviral splice acceptor site and a marker (green fluorescent protein; GFP) in reverse orientation of the retroviral backbone. The integrated virus DNA sequences disrupted the expression of the localized gene, and also provided a convenient molecular tag to identify the location of insertion in the genome. The KBM7-mu cell model shares the same ideology and approach as yeast or other model systems-based loss-of-function genetic screening tools. However, given its human cell origin, the KBM7-mu loss-of-function model could potentially be a more powerful tool and used in the identification of null susceptibility genes following exposure to various environmental toxicants.
Organophosphate pesticides (OPs) are one of the main classes of insecticides in use both in farmlands and households. Acute exposure to all OPs causes a similar neurobehavioral syndrome through a common mechanism of cholinesterase inhibition (Rusyniak & Nanagas, 2004). Chronic exposure to OPs has also associated with other neurobehavioral deficits, however, the mechanism is apparently not related with cholinesterase inhibition (Kamel & Hoppin, 2004; Kamel et al., 2005; Moser et al., 2005; Ray & Richards, 2001). The modes of action behind these neurotoxic symptoms as well as other adverse outcomes caused by chronic OP exposure are elusive. Additionally, humans vary in their responses to chronic OP exposure, and paraoxonase 1 (PON1) gene is the only known genetic factor that has been shown to be associated with differential susceptibility to OP induced neurotoxicity (Albers, Garabrant, Berent, & Richardson, 2010; Holland et al., 2006; Searles Nielsen et al., 2005). Chlorpyrifos (CPF) is one of the most used OPs worldwide (USEPA, 2002). Studies have shown that chronic CPF exposure could affect the neurodevelopment, particularly for children (USEPA, 2000). The available data indicated that this effect is likely unrelated to cholinesterase inhibition, (Potera, 2012; Song et al., 1997).
In the present study, we aim to examine the applicability of this KBM7-mu screening model to identify human susceptible genes to toxicant exposure, establish and optimize the experimental protocol. Furtherer more, we are interested in identifying human susceptibility genes to CPF and exploring potential mechanisms of CPF-induced toxicities. Thus, here we use KBM7-Mu cells and perform screens for genes conferring resistance to CPF.
2. Materials and methods
2.1. Chemicals and reagents
Chlorpyrifos (CPF) was purchased from Chem Service Inc. (West Chester, PA). CPF solutions were freshly prepared in 0.05% DMSO (Sigma-Aldrich, St. Louis, MO). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) was obtained from Sigma-Aldrich (St. Louis, MO). Iscove's Modified Dulbecco's Medium (IMDM) with l-Glutamine and 25 mM HEPES was acquired from Life Tech (Grand Island, NY). Fetal Bovine Serum (FBS) was obtained from Lonza Group Ltd. (Walkersville, MD); and Penicillin-Streptomycin and 1× trypsin-EDTA (0.25%) were from Cellgro Mediatech Inc. (Manassas, VA). Restriction enzymes, Sau3A1 and NheI, were ordered from New England BioLabs Inc. (Ipswich, MA). Pfu Turbo HotStart DNA Polymerase was ordered from Agilent (Santa Clara, CA).
2.2. Cell culture and initial sorting
An insertional mutagenized loss-of-function model was created using a human haploid cell line, KBM7, and named as KBM7-mu (Carette et al., 2009). Dr. Brummelkamp from Massachusetts Institute of Technology generously provided us with this novel model system. Briefly, an insertional mutagenized loss-of-function model was created using a human haploid cell line, KBM7. This model was developed utilizing gene-trap retroviruses that contain a strong adenoviral splice acceptor site and a marker (green fluorescent protein; GFP) in reverse orientation of the retroviral backbone. The integrated virus DNA sequences disrupted the expression of the localized gene, and also provided a convenient molecular tag to identify the location of insertion in the genome. The KBM7-mu cells were maintained in IMDM with l-Glutamine and 25 mM HEPES, containing 10% Fetal Bovine Serum and 1% penicillin/streptomycin, and incubated at 37 °C in a fully humidified atmosphere containing 5% CO2. Cell sorting was conducted using a Becton Dickinson FACSAria cell sorter to obtain only GFP-positive KBM7-mu cells, guaranteeing viral incorporation in the sorted cell population (KBM7-mu).
2.3. MTT assay and CPF treatment of KBM7-mu cells
The MTT assay was performed to assess cell viability after CPF treatment. Cells were cultured in phenol red free IMDM medium in 96well plates in a volume of 100 µl medium/well at a density of 5 × 104 cells/ml. Two days after incubation with CPF (six replicates/CPF concentration and one DMSO control), 10 µl sterile MTT dye was added to each well and plates were incubated at 37 °C for 4 h. 200 µl dimethyl sulfoxide was added and thoroughly mixed for 10 min. Spectrophotometric absorbance at 570 nm was measured in a microplate reader. Based on the cytotoxicity data observed from MTT assays, exposure to 150–250µM CPF caused an approximately 35–70% cell death over a 2 days culture period. Thus, 200 µM CPF was selected as the desired treatment dose, and KBM7-mu cells were exposed to this concentration of CPF over a period of approximately 21 days. Cell culture medium containing 200 µM CPF was refreshed approximately every 3–4 days. At the end of CPF treatment, a single cell was sorted into each well of three 96-well culture plates using a Becton Dickinson FACSAria cell sorter. The cells were again cultured in the IMDM medium containing 200 µM CPF over a period of 14 days. The survived single cell colony wells were gradually transferred to larger wells over another 7 days culture period, followed by 7 days of culture in a flask with continuing CPF exposure.
2.4. Determine the genomic segment disrupted by virus insertion
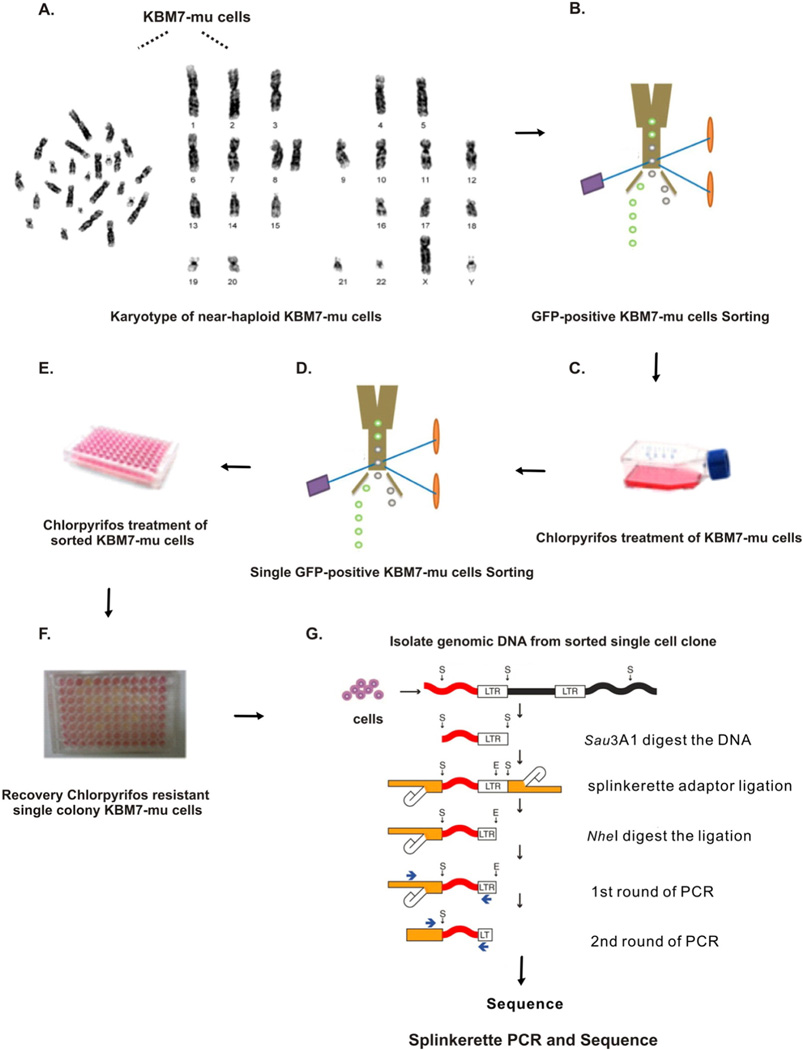
Trapped genes in the CPF-resistant cell colonies were identified by utilizing the Splinkerette PCR (Uren et al., 2009) protocol with modifications to fit for the KBM7 mutational loss-of-function model (Fig. 1). The detailed methods as following:
Fig. 1.
Illustration of the methods used in identifying CPF resistance genes using KBM7 cells mutagenesis model. (A) G-banding analysis to confirm karyotypes of near-haploid KBM7-mu cells; (B) enrich the GFP-positive KBM7-mu cells; (C) CPF treatment: cells receive multiple treatments of 200 µM CPF over a period of approximately 2–3 weeks; (D) single cell sorting and collection after CPF treatment; (E) continuing 200 µM CPF treatment for additional 2–3 weeks; (F) cells show color change (from pink to orange/yellow) indicate cell survival and growth, representing CPF resistance. (G) Splinkerette-PCR method used to isolate retroviral insertion sites for sequencing. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Modified from Uren et al. (2009).
I. DNA isolation and digestion
DNA was collected from the recovered single cell colonies using the FlexiGene® DNA kit (Qiagen, Valencia, CA) and quantified using a NanoDrop. Two microgram DNA for each cell colony was digested with the restriction enzyme Sau3A1, and the reaction was incubated at 37 °C for 16 h, and then stored at −20 °C (Table 1).
Table 1.
Sau3A1 enzyme digestion reaction setup.
| Component | Quantity/tube (µl) |
|---|---|
| CPF-resistant cellular DNA | 2 µg calculations (Fig. 2) |
| Sau3A1 enzyme (20 U/µl) | 2 |
| 10× NEBuffer 1 | 3 |
| 100× BSA (10 mg/ml) | 1 |
| Double-distilled water | Bring final volume to 30 |
| Final volume | 30 |
II. Adaptor ligation
The adaptor mix consisted of a short- and long-strand oligonucleotide (Table 4). The stock concentrations (200 µM) prepared in NEBuffer 2, for both the long- and short-stand adaptors, were used to prepare 50 µM dilutions in double distilled water. In a 1.5 ml tube, the adaptor mix was prepared by adding 50 µl of short-strand adaptor (50 µM) with 50 µl of long-strand adaptor (50 µM), creating a final concentration of 25 µM. The adaptor oligonucleotides and mix were stored frozen at −20 °C. Before each ligation reaction the adaptor mix is thawed on ice, and then 25 µl are transferred into a PCR tube. Using the PCR machine, the mix is denatured by heating to 95 °C for 5 min and then annealed by slowly cooling to room temperature, dropping approximately 1 °C every 15 s. The tube is placed directly on ice upon completion of the cooling cycle. Immediately before setting up the ligation reaction, the Sau3A1 digested DNA was thawed and heated in a block at 65 °C for 20 min, then put on ice. The ligation reaction was carried out in a 1.5 ml tube using T4 DNA ligase (Table 2). The reaction was incubated at 4 °C for 16 h. Immediately following incubation, the T4 DNA ligase was inactivated by heating the reaction tube at 65 °C for 20 min in a heating block. The ligation reaction was then stored at −20 °C.
Table 4.
Oligonucleotides and primers used.
| Sequence (5′–3′) | |
|---|---|
| Adaptor ligation | |
| Short-Strand Adaptor |
GATCCCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA |
| Long-Strand Adaptor |
GAAGAGTAACCGTTGCTAGGAGAGACCGTGGCTGAATGAGACTGGT GTCGACACTAGTGG |
| Splinkerette PCR | |
| Splink1 | GAAGAGTAACCGTTGCTAGGAGAGACC |
| U3LTR#5 | GCGTTACTTAAGCTAGCTTGCCAAACCTAC |
| Splink2 | GTGGCTGAATGAGACTGGTGTCGAC |
| U3LTR#1 | CCAAACCTACAGGTGGGGTCTTTC |
| qRT-PCR | |
| ATP8B2-F | GGGTGCTAACCGGAGAC |
| ATP8B2-R | AGTATGGCCAGTGACTATGAA |
| DCAF12-F | CTTGCCATCTATCGACTACCT |
| DCAF12-R | CCATAGTGTCGCTGATCCA |
| FNBP4-F | TGGCAGGAGTCGGAATTGAG |
| FNBP4-R | ACCTGTGTGGCAAGATATTGG |
| LAT2-F | GACCAACAGAGCTTTACGG |
| LAT2-R | CTGGGGTAGAATTGC |
| BIK-F | GGTTCTTGGCATGACTGA |
| BIK-R | GGCCAATGCGTCACT |
| AGPAT6-F | GGCAGGGAGGATTGGTAG |
| AGPAT6-F | TCATGGTTTCCGACTATCAT |
| AIG-F | CGCACCATCAGTATCCC |
| AIG-R | ACACCCACATGCCAGTTACA |
| PTC7-F | AAACCTGGGCGATTC |
| PTC7-R | ATCCGGACTGTCGCTCAAGA |
| GAPDH-F | ACATCGCTCAGACACCATG |
| GAPDH-R | TGTAGTTGAGGTCAATGAAGGG |
Table 2.
Adaptor ligation.
| Component | Quantity/tube (µl) |
|---|---|
| Sau3A1 digested DNA | 4.5 |
| Adaptor mix (25 µM) | 1 |
| T4 DNA ligase (20 U/µl) | 1 |
| 10× T4 DNA ligase buffer | 4 |
| Double-distilled water | 29.5 |
| Final volume | 40 |
Note: Ligation reaction setup, utilizing a distinct adaptor mix with T4DNA ligase. Required quantities per 40 µl total reaction volume.
III. Amplification of genome segment disrupted by virus insertion
A second round of digestion was performed in order to remove the adaptors ligated onto the viral end of the Sau3A1 digested DNA fragments using restriction enzyme Nhe1, preventing internal viral sequence amplification (Table 3). The Nhe1 digest was cleaned using the QIAquick® PCR Purification kit (Qiagen, Valencia, CA), eluting the DNA with 40 µl of double-distilled water instead of elution buffer. Purified DNA samples were stored at −20 °C. A series of PCR rounds were then performed to amplify the genome segment disrupted by viral insertion. The primary round of PCR utilized primers Splink1 and U3LTR#5, followed by a second round using primers Splink2 and U3LTR#1 (Table 4). The primary PCR mix contained Nhe1 digest as the template, Splink1 and (10 µM), U3LTR#5 (10 µM), 10 mM dNTP mix, 10× Pfu buffer, Pfu Turbo HotStart DNA Polymerase, and double-distilled water to bring the total reaction volume up to 50 µl. The primary PCR protocol was initialized heating to 94 °C for 3 min followed by 29 cycles of 94 °C for 15 s, 68 °C for 30 s, and 72 °C for 5 min, with a final extension of 72 °C for 5 min. The secondary PCR then used the primary PCR product as template and was initialized by heating to 94 °C for 15 min followed by 25 cycles of 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 5 min, with a final extension of 72 °C for 5 min. The secondary PCR products were visualized on an 2% agarose gel, and then extracted and purified from the gel using the QIAquick® Gel Extraction kit (Qiagen, Valencia, CA). The purified and concentrated PCR was then sequenced, and the obtained sequences of each PCR product were then run through a basic local alignment search tool (BLAST) and compared to the human genome database to determine the genomic segments disrupted by virus insertion.
Table 3.
Nhe1 enzyme digestion.
| Component | Quantity/tube (µl) |
|---|---|
| Adaptor ligated DNA | 40 |
| 10× NEBuffer 2 | 10 |
| 100× BSA | 1 |
| Nhe1 (10 U/µl) | 2 |
| Double-distilled water | 47 |
| Final volume | 100 |
Note: Nhe1 enzyme digestion reaction setup. Required quantities per 100 µl total reaction volume.
2.5. Quantitative real-time PCR
Total RNA was extracted using TRIzol Reagent (Life Tech., Carlsbad CA). mRNAs were reverse-transcribed using a SuperScript III cDNA synthesis kit (Life Tech., Carlsbad CA). Gene expression of ATP8B2, DCAF12, LAT2, BIK, FNBP4, AGPAT6, AIG1, and PPTC7 in individual selected CPF resistant cells and passage matched parental cells was measured using quantitative real-time reverse transcription (RT)-PCR using primers listed in Table 4. Real-Time PCR was performed using Bio-Rad CFX96 Touch™ Real-Time PCR Detection System and a SYBR Green Supermix Kit (Bio-Rad Laboratories, Hercules, CA). The PCR efficiency was examined by serially diluting template cDNA and the melting curve data were collected to check the PCR specificity. Results were calculated using the delta, delta Ct method normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression for each sample.
2.6. Statistical analysis
Results were shown as mean ± standard deviation (SD), and comparisons were made by one-way ANOVA with Tukey post hoc analysis. p-value < 0.05 was considered statistically significant.
3. Results
3.1. Identification of CPF toxicity resistant genes
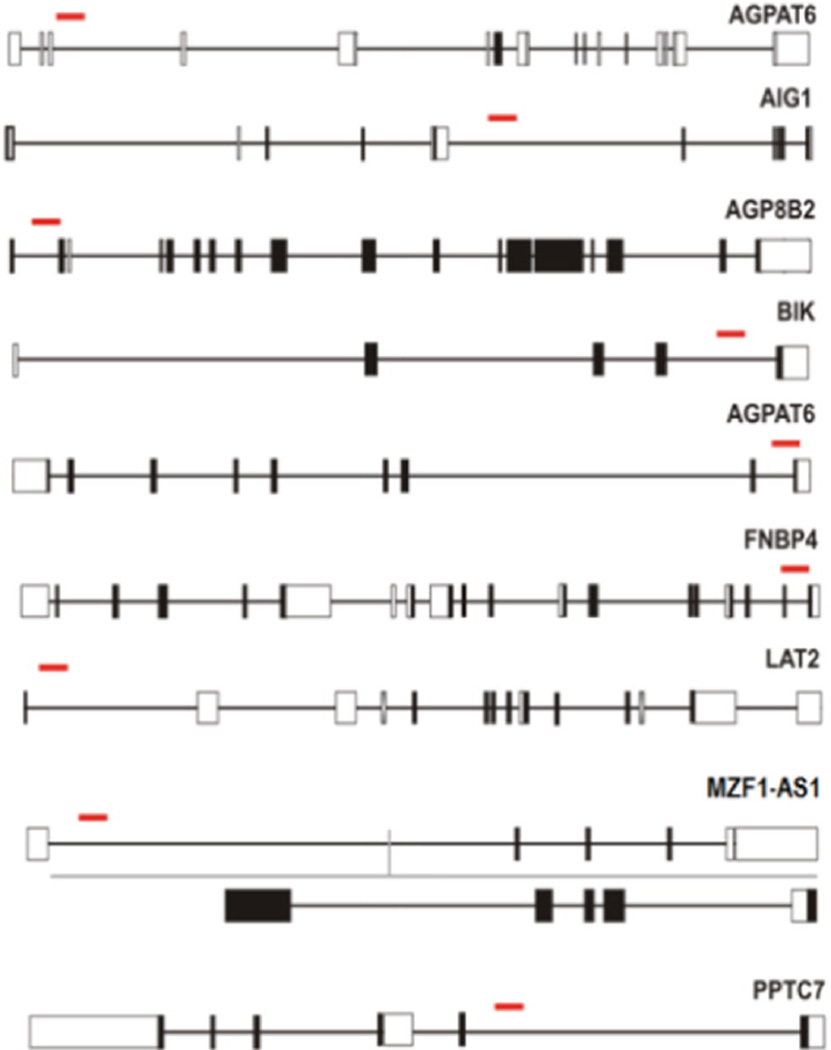
Following the protocol of cell treatment and recovery, each CPF resistant single cell colony recovered from 96-well plate was labeled using a three digit system, the plate number (1, 2, or 3) followed by the letter row and number column in which it was located. CFP resistant cells collected from well 3D4 had a gene trap insertion site in AGPAT6 (1-acylglycerol-3-phosphate O-acyltransferase 6). Cells collected from well 2D10 had a gene trap insertion site in AIG1 (Androgen-induced 1). Wells 1G7, 2D7, 2E7, 3D2 and 3C2 contained cells which had a gene trap insertion site in ATP8B2 (ATPase, aminophospholipid transporter, class I, type 8B, member 2). Resistant cells collected from well 3D8 had a gene trap insertion site in BIK (BCL-2 interacting killer). Wells 1C9, 1C11, 2F8, and 2D3 contained cells that had a gene trap insertion site in DCAF12 (DDB1 and CUL4 associated factor 12). Cells collected from wells 1F3, 1C5, 3E8, and 2E6 had a gene trap insertion site in FNBP4 (Formin binding protein 4). Wells 2C6 and 3D4 contained cells that had a gene trap insertion site in LAT2 (Linker for activation of T cells family, member 2). Resistant cells in well 1F3 had a gene trap insertion site in MZF1-AS1 (MZF1 antisense RNA 1). Cells collected from wells 2G10 and 3B6 had a gene trap insertion site in PPTC7 (PTC7 protein phosphatase homolog). In summary, the identified genes, which were responsible for a CPF resistant phenotype following disruption by virus insertion, were AGPAT6, AIG1, ATP8B2, BIK, DCAF12, FNBP4, LAT2, MZF1-AS1, and PPTC7 (Fig. 2). The detailed information of the above genes was summarized in (Table 5).
Fig. 2.
Schematic outline of the gene-trap insertion sites (red lines) in mutant cells response to CPF. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 5.
Information of genes trapped by retrovirus insertion in CPF resistant KBM-7 clones.
| Gene-trapped | Chr | Gene ID | Protein ID | Description | Function |
|---|---|---|---|---|---|
| ATP8B2 | 1 | 57198 | NP_001005855 | ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
Transport phosphatidylserine and phosphatidylethanolamine between bilayer |
| DCAF12 | 9 | 25853 | NP_056212 | DDB1 and CUL4 associated factor 12 | Interact with the COP9 signalosome to regulate cullin-RING E3 ligases activity |
| FNBP4 | 11 | 23360 | NP_056123 | Formin binding protein 4 | Associated with anophthalmos disease |
| LAT2 | 7 | 7462 | NP_054865 | Linker for activation of T cells family, member 2 | Commonly deleted in Williams syndrome |
| BIK | 22 | 638 | NP_001188 | BCL-2 interacting killer | Interaction with anti-apoptotic members of the BCL2 family, and viral survival-promoting proteins |
| AGPAT6 | 8 | 137964 | NP_848934 | 1-acylglycerol-3-phosphate O-acyltransferase 6 | Active against both saturated and unsaturated long-chain fatty acyl-CoAs. |
| AIG1 | 6 | 51390 | NP_001273516 | Androgen-induced 1 | Associated with carcinogenesis |
| PPTC7 | 12 | 160760 | NP_644812 | PTC7 protein phosphatase homolog | Up-regulated during lymphocyte activation |
| MZF1-AS1 | 19 | 100131691 | N/A | MZF1 antisense RNA 1 | N/A |
N/A: non available.
3.2. Expression levels of identified genes
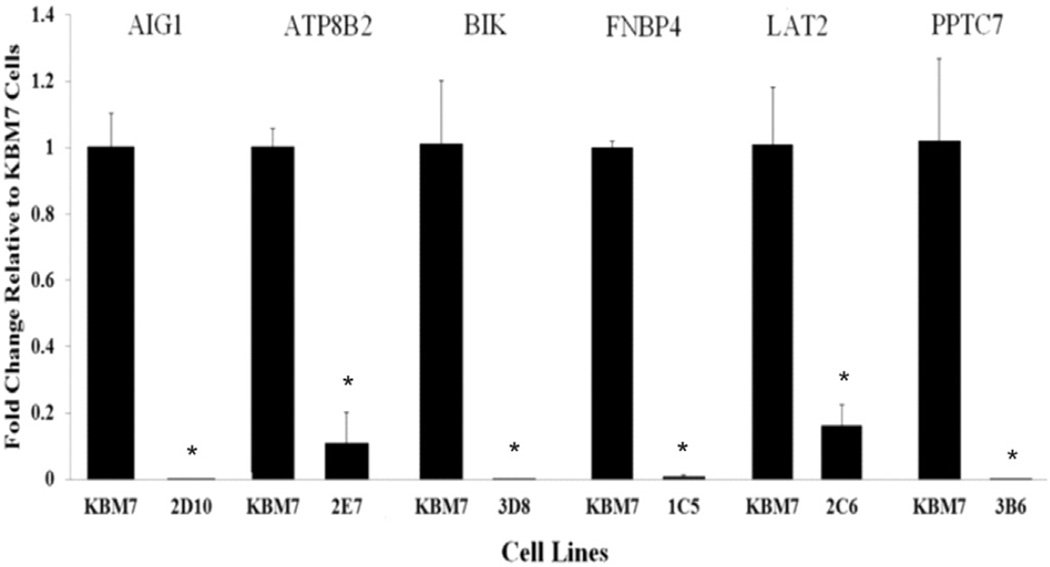
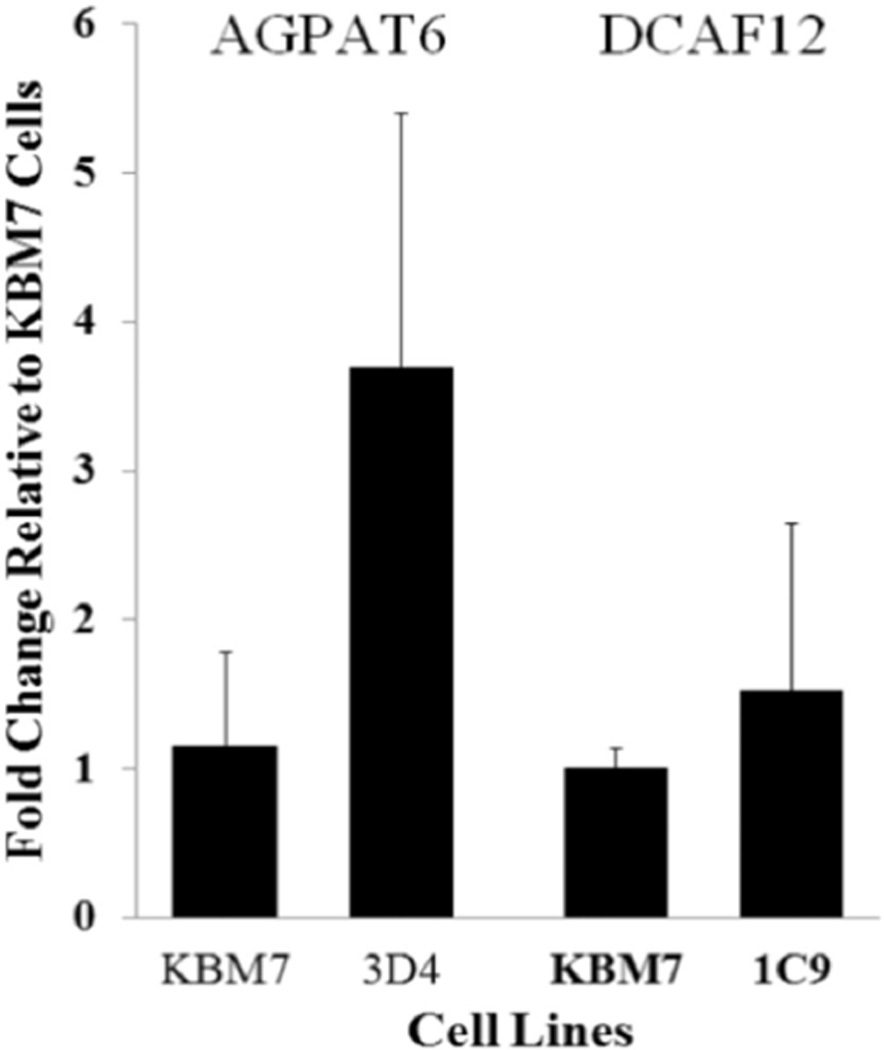
qRT-PCR was performed to determine the effects of virus insertion within specific genomic regions on the expression of genes located in those regions (Fig. 3). mRNA expression levels of genes AIG1, ATP8B2, BIK, FNBP4, LAT2, and PPTC7 were significantly decreased. Whereas no significant change was detected in genes of AGPAT6 and DCAF12 (Fig. 4).
Fig. 3.
mRNA levels of ATP8B2, LAT2, BIK, FNBP4, AIG1, and PPTC7 genes. The expression of ATP8B2, LAT2, BIK, FNBP4, AIG1, and PPTC7 mRNAs were either significantly decreased or completely lost in recovered KBM7-mu cells compared to the control KBM7 cells. Data are expressed as means ± standard deviations (n = 3), *P < 0.05. (Recovered KBM7-mu cell was labeled using a three digit system, the 96-well plate number (1, 2, or 3) followed by the letter row and number column in which it was locatEd.)
Fig. 4.
mRNA levels of DCAF12 and AGPAT6 genes. No significant changes of the expression of DCAF12 and AGPAT6 mRNAs were observed in recovered KBM7-mu cells compared to the control KBM7 cells. Data are expressed as means ± standard deviations (n = 3). (Recovered KBM7-mu cell was labeled using a three digit system, the 96-well plate number (1, 2, or 3) followed by the letter row and number column in which it was locatEd.)
4. Discussion
Genome-scale loss-of-function screens have provided a wealth of information in diverse model systems (Berns et al., 2004; Carette et al., 2009; Rad et al., 2010; Shalem et al., 2014). The yeast based genetic screening model is the most commonly available and widely used model system due to its haploid status, which makes recessive mutations relatively easy to produce. However, the yeast genome is not directly relevant to humans when screening for susceptibility genes to specific environmental toxicants. Although RNA interference (RNAi) based screening methods in mammalian cells have shown potentials, it is greatly limited by the inability to fully reduce and block gene expression in genome scales (Berns et al., 2004; Boutros et al., 2004). In this study we are able to demonstrate that a newly developed human haploid cell (KBM7)-based insertional mutagenic screening model can be modified and used for screening and identifying genes whose absence are associated with the resistance to specific environmental toxicants.
The screening procedure used in this proposed study has been modified. The major modification of the procedure compared to the previous study (Carette et al., 2009) is that we applied the newly developed Splinkerette PCR method (Uren et al., 2009) to amplify and identify the targeted insertional genomic regions. Results showed that this modified screening procedure greatly improved the flow of the experiment, and more importantly, with better reproducibility and success rate of the experiment. In this pilot study, we recovered 48 single cell colonies, and were able to identify virus insertion position in the genome from about half of these recovered colonies. The virus insertion in genomic regions are linked to 9 genes, AGPAT6, AIG1, ATP8B2, BIK, DCAF12, FNBP4, LAT2, MZF1-AS1, and PPTC7. Among them, 6 genes, ATP8B2, LAT2, BIK, FNBP4, AIG1, and PPTC7 were demonstrated to be significantly down-regulated in the cells carrying the disrupted genes. However, the expression of two genes, DCAF12 and AGPAT6, did not change, indicating that other mechanisms may be involved and lead to the observed resistance to CPF exposure.
Other than the well-known neurotoxicity of CPF, prenatal exposure to CPF, even at low levels, may negatively alter the neurodevelopment in the fetus or newborn (Potera, 2012). Chronic CPF exposure could affect liver and heart development (Meyer, Seidler, & Slotkin, 2004; Song et al., 1997). Although some mechanisms through which CPF mediates deficits in development of brain and other organs have been proposed, the modes of action overall remains elusive and apparently is not related with cholinesterase inhibition (Kamel & Hoppin, 2004; Kamel et al., 2005; Moser et al., 2005; Ray & Richards, 2001). In the present study, genes identified in screening cells that exhibit a resistant phenotype to CPF exposure are involved in many different biological processes. AIG1, androgen-induced 1, whose expression is induced by androgen (Chen et al., 2008), with high AIG1 mRNA level found in the testis, ovary, kidney, liver, and heart, whereas low levels found in small intestine, colon, pancreas, spleen, prostate, skeletal muscle, and brain (Seo, Kim, & Kim, 2001). In a previous study, Wu G. et al. found 63% of human hepatocellular carcinomas (HCCs) had reductions in expression of AIG1, suggesting that AIG1 could serve as a new diagnostic and prognostic biomarker for HCCs (Wu, Sun, Zhang, & Huo, 2011). ATP8B2, the ATPase aminophospholipid transporter, class I, type 8B, member 2, encodes a protein belonging to the catalytic component of a P4-ATPase flippase complex, and also to the aminophospholipid-transporting ATPases subfamily. Inactivation of murine P4-ATPases could lead to fertility-related disorders, insulin resistance and obesity (Folmer, Elferink, & Paulusma, 2009). BIK encodes the BCL-2 interacting killer, which is thought to be part of pro-apoptotic protein family referred to as BCL-2 homology domain 3. BCL-2 family-regulated apoptosis is believed to be involved in the pathogenesis of several human diseases, including cancer, neurodegenerative, asthma, cardiovascular disease and autoimmune disease (Kluck, 2010). Thus, BIK has been proposed to be an eminent target for anti-cancer drug design and discovery (Huang, 2000). FNBP4 plays a role in Bone morphogenetic proteins (BMP) signaling and a recent study identified a homozygous mutation in FNBP4, which results in a microphthalmia, an eye abnormality, with a limb anomalies-like condition (Olma et al., 2009). As for PPTC7, also known as PTC7 protein phosphatase homolog (S. cerevisiae), no gene function data were reported in the literature. However, it appears to have several serine/threonine phosphatase conserved domains, suggesting a possible function involved in cell signaling (Mumby & Walter, 1993).
In summary, we demonstrated that the KBM7-mu genetic screening system can be modified and suitable for studying gene-environmental interactions, specifically to identify genes that contribute to the resistance to specific environmental toxicants. Additionally, our study results suggest that multiple genetic factors may impact the CPF-induced toxicity, and further studies are warranted to address mechanisms behind these involvements.
Acknowledgments
This work was supported by a start-up fund (to X.R.) provided by University at Buffalo. Support was also provided by NIEHS grant R21ES022329 and R01ES022629 to X.R.
Abbreviations
- AChE
acetylcholinesterase
- AGPAT6
1-acylglycerol-3-phosphate O-acyltransferase 6
- AIG1
androgen-induced 1
- ATP8B2
ATPase, aminophospholipid transporter, class I, type 8B, member 2
- BChE
butyrylcholinesterase
- BIK
BCL2-interacting killer
- Cat. No
Catalog Number
- CPF
Chlorpyrifos
- CPF-O
Chlorpyrifos-Oxon
- DCAF12
DDB1 and CUL4 associated factor 12
- DMSO
dimethyl sulfoxide
- FBS
Fetal Bovine Serum
- FNBP4
formin binding protein 4
- GFP
green fluorescent protein
- KBM7-mu-s
KBM7-mutated cells sorted
- LAT2
Linker for activation of T cells family member 2
- MTT
Thiazolyl Blue Tetrazolium Bromide
- PBS
Phosphate Buffered Saline
- PON1
Paraoxonase 1
- PPTC7
PTC7 protein phosphatase homolog
- P/S
Pencillin/Streptomycin
- TCPy
3, 5, 6-trichloro-2-pyridinol.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Reference
- Albers JW, Garabrant DH, Berent S, Richardson RJ. Paraoxonase status and plasma butyrylcholinesterase activity in chlorpyrifos manufacturing workers. Journal of Exposure Science & Environmental Epidemiology. 2010;20:79–89. doi: 10.1038/jes.2009.9. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Chen YQ, Kuo MS, Li S, Bui HH, Peake DA, Sanders PE, et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. The Journal of Biological Chemistry. 2008;283:10048–10057. doi: 10.1074/jbc.M708151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer DE, Elferink RP, Paulusma CC. P4 ATPases—Lipid flippases and their role in disease. Biochimica et Biophysica Acta. 2009;1791:628–635. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Holland N, Furlong C, Bastaki M, Richter R, Bradman A, Huen K, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environmental Health Perspectives. 2006;114:985–991. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Bcl-2 family proteins as targets for anticancer drug design. Oncogene. 2000;19:6627–6631. doi: 10.1038/sj.onc.1204087. [DOI] [PubMed] [Google Scholar]
- Kamel F, Engel LS, Gladen BC, Hoppin JA, Alavanja MC, Sandler DP. Neurologic symptoms in licensed private pesticide applicators in the agricultural health study. Environmental Health Perspectives. 2005;113:877–882. doi: 10.1289/ehp.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel F, Hoppin JA. Association of pesticide exposure with neurologic dysfunction and disease. Environmental Health Perspectives. 2004;112:950–958. doi: 10.1289/ehp.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluck GDaRM. Bcl-2 family-regulated apoptosis in health and disease. Cell Health and Cytoskeleton. 2010;9–22 [Google Scholar]
- Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Experimental Cell Research. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- Meyer A, Seidler FJ, Slotkin TA. Developmental effects of chlorpyrifos extend beyond neurotoxicity: critical periods for immediate and delayed-onset effects on cardiac and hepatic cell signaling. Environmental Health Perspectives. 2004;112:170–178. doi: 10.1289/ehp.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser VC, Phillips PM, McDaniel KL, Marshall RS, Hunter DL, Padilla S. Neurobehavioral effects of chronic dietary and repeated high-level spike exposure to chlorpyrifos in rats. Toxicological Sciences. 2005;86:375–386. doi: 10.1093/toxsci/kfi199. [DOI] [PubMed] [Google Scholar]
- Mumby MC, Walter G. Protein serine/threonine phosphatases: Structure, regulation, and functions in cell growth. Physiological Reviews. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- Olma MH, Roy M, Le Bihan T, Sumara I, Maerki S, Larsen B, et al. An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. Journal of Cell Science. 2009;122:1035–1044. doi: 10.1242/jcs.043539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potera C. Newly discovered mechanism for chlorpyrifos effects on neurodevelopment. Environmental Health Perspectives. 2012;120:a270–a271. doi: 10.1289/ehp.120-a270a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DE, Richards PG. The potential for toxic effects of chronic, low-dose exposure to organophosphates. Toxicology Letters. 2001;120:343–351. doi: 10.1016/s0378-4274(01)00266-1. [DOI] [PubMed] [Google Scholar]
- Rusyniak DE, Nanagas KA. Organophosphate poisoning. Seminars in Neurology. 2004;24:197–204. doi: 10.1055/s-2004-830907. [DOI] [PubMed] [Google Scholar]
- Searles Nielsen S, Mueller BA, De Roos AJ, Viernes HM, Farin FM, Checkoway H. Risk of brain tumors in children and susceptibility to organophosphorus insecticides: The potential role of paraoxonase (PON1) Environmental Health Perspectives. 2005;113:909–913. doi: 10.1289/ehp.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Kim J, Kim M. Cloning of androgen-inducible gene 1 (AIG1) from human dermal papilla cells. Molecules and Cells. 2001;11:35–40. [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Seidler FJ, Saleh JL, Zhang J, Padilla S, Slotkin TA. Cellular mechanisms for developmental toxicity of chlorpyrifos: Targeting the adenylyl cyclase signaling cascade. Toxicology and Applied Pharmacology. 1997;145:158–174. doi: 10.1006/taap.1997.8171. [DOI] [PubMed] [Google Scholar]
- Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nature Protocols. 2009;4:789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA (U.S. Environmental Protection Agency) Chlorpyrifos facts. Washington, D.C: 2002. [accessed January 13, 2013]. Report 738-F-01-006 http://www.epa.gov/oppsrrd1/REDs/factsheets/chlorpyrifos_fs/htm. [Google Scholar]
- USEPA (U.S. Environmental Protection Agency) Chlorpyrifos; receipt of requests for amendments, cancellations, and notifications of tolerance revocation and modifications. Washington, D.C: 2000. [accessed January 13, 2013]. Report 65 FR 56886 http://www.gpo.gov/fdsys/granule/FR-2000-09-20/00-24211. [Google Scholar]
- Wu G, Sun M, Zhang W, Huo K. AIG1 is a novel Pirh2-interacting protein that activates the NFAT signaling pathway. Frontiers in Bioscience (Elite Edition) 2011;3:834–842. doi: 10.2741/E291. [DOI] [PubMed] [Google Scholar]