Abstract
BACKGROUND
We previously identified a protein tumor signature of PTEN, SMAD4, SPP1 and CCND1 that, together with clinical features, was associated with lethal outcomes among prostate cancer patients. In the current study, we sought to validate the molecular model using time-dependent measures of AUC and predictive values for discriminating lethal from non-lethal prostate cancer.
METHODS
Using data from the initial study, we fit survival models for men with prostate cancer who were participants in the Physicians’ Health Study (PHS; n = 276). Based on these models, we generated prognostic risk scores in an independent population, the Health Professionals Follow-up Study (HPFS; n = 347) to evaluate external validity. In each cohort, men were followed prospectively from cancer diagnosis through 2011 for development of distant metastasis or cancer mortality. We measured protein tumor expression of PTEN, SMAD4, SPP1 and CCND1 on tissue microarrays.
RESULTS
During a median of 11.9 and 14.3 years follow-up in the PHS and HPFS cohorts, 24 and 32 men (9%) developed lethal disease. When used as a prognostic factor in a new population, addition of the four markers to clinical variables did not improve discriminatory accuracy through 15 years of follow-up.
CONCLUSIONS
Although the four markers have been identified as key biological mediators in metastatic progression, they do not provide independent, long-term prognostic information beyond clinical factors when measured at diagnosis. This finding may underscore the broad heterogeneity in aggressive prostate tumors and highlight the challenges that may result from overfitting in discovery-based research.
Keywords: Prognostic biomarkers, biomarker validation, prostate cancer
Introduction
A pressing challenge in the management of newly diagnosed localized prostate cancer is to distinguish men who will have an indolent course from those whose disease has lethal potential. The difficulty in making this distinction stems from both the considerable variability in prostate cancer’s biologic capacity to metastasize and the lack of tools to predict metastatic risk for an individual prostate cancer patient.
Our group previously identified a four-marker prostate tumor signature by focusing on molecular indicators of invasiveness and metastatic potential through the integration of genetically engineered mouse models of prostate cancer, mRNA expression profiling, and functional genomics[1]. This integrative genomic strategy established associations between aggressive prostate cancer and decreased tumor expression of SMAD4, a transforming growth factor-β (TGF-β) pathway signaling molecule, and PTEN, a regulator of the phosphoinositol-3 kinase pathway, as well as increased expression of both CCND1, a regulator of the G1/S cell cycle transition, and SPP1, a component of the TGF-β signaling cascade. Expression of the four markers in prostate cancer patient cohorts was significantly associated with risk of biochemical recurrence and cancer mortality, including in the Physicians’ Health Study (PHS) cohort in which the molecular signature in combination with tumor stage, Gleason score and age at diagnosis produced an AUC of 0.91 (95% confidence interval [CI] 0.86–0.96) for lethal prostate cancer[1].
A common barrier for the clinical adoption of prognostic biomarkers concerns successful validation of findings across multiple cohorts[2, 3]. One goal of our prior study was to assess the prognostic potential of a mouse model-derived molecular marker set within human populations. Models that included the four markers and Gleason score suggested strong discriminatory accuracy with respect to aggressive versus indolent disease trajectories. However, because the initial phase of marker discovery utilized the same data for model building and accuracy evaluation, it remains unclear whether the findings can be generalized to other populations or may result from overfitting[4].
The overarching aim of the current study is to assess whether the initial prediction model can improve prognostic accuracy in an independent population. Specifically, we will determine whether the four marker risk model developed in the PHS can improve discrimination of lethal prostate cancer over clinical factors alone in a new cohort, the Health Professionals Follow-up Study (HPFS). Time-dependent measures of AUC, positive predictive value (PPV), and negative predictive value (NPV), are used to evaluate the clinical utility of the four marker set.
Materials and methods
Cohorts
The study was based in the HPFS and PHS Prostate Tumor Cohorts[5–7], where the HPFS was used to assess validation performance following model-building in the PHS. HPFS is a prospective cohort study of 51,000 US male health professionals ongoing since 1986[8]. The PHS I and II were randomized trials of aspirin and nutritional supplements among US male physicians[9, 10] in the primary prevention of cardiovascular disease and cancer. We included a subset of men in the cohorts who were diagnosed with prostate cancer (HPFS, 1986–2002; PHS, 1983–2004) for whom we obtained archival formalin-fixed, paraffin-embedded tissue specimens, from radical prostatectomy (95%) or transurethral resection of prostate (5%). We used medical records to abstract data on pathologic tumor stage (TNM staging system), PSA, and age at diagnosis. All men were followed for the development of lethal disease, defined by metastases to bone or distant organs or prostate cancer-specific death through May 2011. A physician committee confirmed causes of death through medical record and death certificate review. This study was approved by the Institutional Review Boards of Harvard School of Public Health and Partners Healthcare.
Immunohistochemistry of the four markers
We measured protein expression of the four markers using immunohistochemistry on tumor tissue microarrays. Study pathologists (ML, RL) undertook a standardized histopathologic review which included Gleason score. Tissue microarrays were created using at least three tumor cores per case from areas representing the highest Gleason grade. Immunohistochemical staining was performed on 5-μm sections of the tissue microarrays to assess cytoplasmic PTEN (rabbit polyclonal, PN37, 18-0256, Zymed, South San Francisco, CA, USA), cytoplasmic SMAD4 (mouse monoclonal, clone B8, sc-7966, Santa Cruz, Sana Cruz, CA, USA), nuclear cyclin D1 (rabbit monoclonal, SP4, RM-9104-R7, Thermo Scientific, Waltham, MA, USA), and cytoplasmic SPP1 expression (rabbit polyclonal, O17, 18625, Immuno-Biological Laboratories, Minneapolis, MN, USA) after citrate-based antigen retrieval. The antibody optimization was performed using appropriate positive and negative controls for each marker and identical staining and interpretation techniques were used from the prior study[1].
Interpretation of the immunohistochemistry was undertaken using the Nuance v2.8 and inForm semiautomated image analysis system (Caliper, Hopkinton, MA, USA), blinded to the clinical data. A final score representing the percentage of cytoplasmic or nuclear area that was positively stained was determined for each core by the image analysis system.
Statistical analysis
The outcome was defined as lethal or non-lethal based on whether men experienced a lethal or metastatic event over the course of follow-up. Gleason score was modeled ordinally. We used the PHS as the training set and the HPFS cohort for validation.
Cox proportional hazards models were built in PHS, and the estimated regression coefficients were used to construct a prognostic score[11] in HPFS. We evaluated two models of interest: (1) a clinical factor model that included Gleason score and age at diagnosis, and (2) a model that included the four markers in addition to the two clinical factors. The proportional hazards assumption was tested globally and for each covariate using α-level 0.10 by assessing correlations between scaled Schoenfeld residuals and time.
Accuracy of the prognostic scores in the HPFS validation data was assessed via time-dependent measures of AUC, PPV, and NPV that are appropriate for censored time-to-event data[12–14]. AUC was estimated at each month of follow-up between 17 months and 180 months, and PPV and NPV were evaluated at 5, 10, and 15 years of follow-up. Confidence intervals were constructed using 500 bootstrap replicates.
Prognostic accuracy statistics were calculated with the survAccuracyMeasures[15] package. All analyses were conducted in R, version 3.1.1.
Results
Clinical characteristics
HPFS consisted of 347 men with available data on all four protein markers. During a median follow up of 14.3 years, 32 (9.2%) men developed lethal prostate cancer, defined as cancer death or metastatic disease. In PHS, there were 276 men followed a median of 11.9 years; 24 (8.7%) men developed lethal disease. The cohorts were generally similar across clinical and pathologic features (Table 1).
Table I.
Clinical characteristics of men with prostate cancer who were participants in the Health Professionals Follow-up Study and Physicians’ Health Study. PSA was measured at diagnosis. Gleason score was rereviewed with the modern classification system and, as a result, there were only four Gleason 5 scores and no Gleason 2–4 across the cohorts.
| HPFS (n = 347) | PHS (n = 276) | |
|---|---|---|
| Age at diagnosis – median (Q1–Q3) | 66.0 (62.0–70.0) | 65.9 (62.2–69.8) |
| Lethal/metastatic events – n (%) | 32 (9) | 24 (9) |
| Years follow-up – median (Q1–Q3) | 14.3 (12.0–16.8) | 11.9 (9.3–15.6) |
| Gleason score – n(%) | ||
| 2–6 | 51 (15) | 74 (27) |
| 3+4 | 132 (38) | 97 (35) |
| 4+3 | 102 (29) | 57 (21) |
| 8–10 | 62 (18) | 48 (17) |
| Pathologic TNM stage – n(%) | ||
| T1/T2 | 224 (65) | 170 (62) |
| T3/T4/N1 | 109 (31) | 69 (25) |
| PSA ng/mL – n(%) | ||
| 0–4 | 42 (12) | 41 (15) |
| 4–10 | 162 (47) | 160 (58) |
| 10–20 | 57 (16) | 36 (13) |
| >20 | 40 (12) | 14 (5) |
Model development in PHS
Summaries of the Cox proportional hazards models fit in the PHS appear in Table 2. Tests for violation of the proportional hazards assumption were not significant. Effects of Gleason and age at diagnosis on hazard of lethality were relatively stable when modeled alone or when modeled alongside the four makers. In the multivariable model, coefficients for the effects SPP1 and SMAD4 were significant at α = 0.05, while those for PTEN and CCND1 were not. As found in the initial study[1], addition of the four markers significantly improved the overall model fit (p = 0.03 by likelihood ratio test). Directions of the marker associations with lethal prostate cancer were consistent with those previously reported.
Table II.
Estimates from the Cox model that included clinical factors (left) and the model that included clinical factors and the four markers (right), both fit in the PHS cohort. Age was coded in years, and Gleason score was modeled continuously with a score of 4 + 3 set to 7.5.
| HR (95% CI) | p | |
|---|---|---|
| Gleason score | 3.22 (2.04–5.07) | < 0.001 |
| Age at diagnosis | 1.09 (1.03–1.15) | 0.004 |
| HR (95% CI) | p | |
|---|---|---|
| Gleason score | 2.84 (1.70–4.74) | < 0.001 |
| Age at diagnosis | 1.09 (1.03–1.16) | 0.005 |
| SPP1 | 1.04 (1.00–1.07) | 0.02 |
| PTEN | 0.36 (0.02–7.66) | 0.51 |
| SMAD4 | 0.97 (0.94–1.00) | 0.04 |
| CCND1 | 1.00 (0.98–1.03) | 0.89 |
Performance of the prognostic scores in HPFS
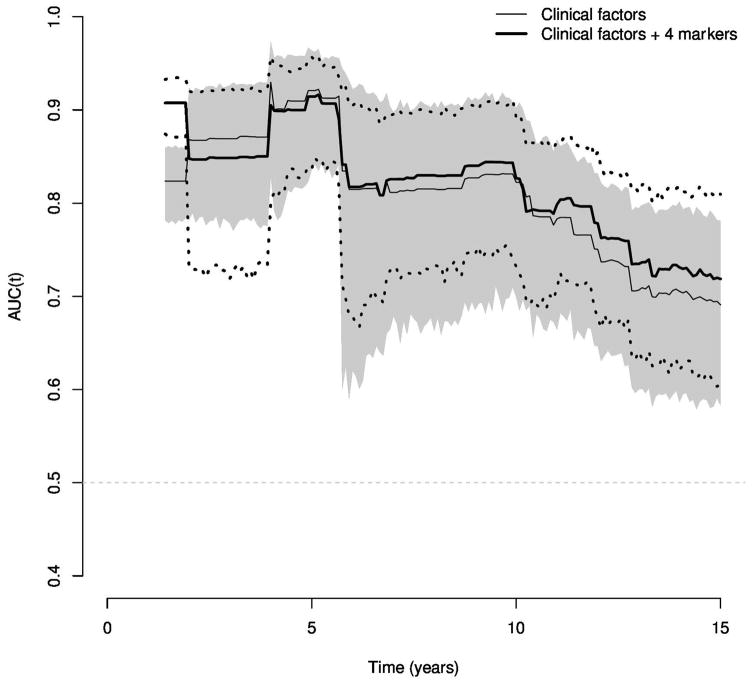
Figure 1 shows the discriminatory performance of the clinical and biomarker scores over time as measured by AUC. The AUC point estimates and 95% confidence intervals for the clinical factor model versus the clinical factors and four markers model are similar throughout 15 years of follow-up. Table 3 provides more detailed information concerning the prognostic value of the AUC estimates over time at 5, 10, and 15 years. Both the clinical factor model and the model with the four markers reveal the strongest prediction of lethal prostate cancer within the first 5 years of follow-up, and declines with greater time from diagnosis. The drop appears particularly pronounced at approximately 5 years of follow-up.
Figure 1.
Time-dependent AUC estimates through 15 years of follow-up when applying prognostic models built in the PHS cohort to the HPFS cohort. The shaded region provides a 95% confidence interval for the clinical factor model, while the dotted lines give a 95% interval for the model with clinical factors and the four markers.
Table III.
Time-dependent AUC estimates at 5, 10, and 15 years of follow-up when applying prognostic models built in the PHS cohort to the HPFS cohort.
| Clinical factorsAUCt (95% CI) | Clinical factors + 4 markersAUCt (95% CI) | N events | At risk | |
|---|---|---|---|---|
| 5 years | 0.92 (0.83–0.96) | 0.91 (0.85–0.95) | 7 | 325 |
| 10 years | 0.82 (0.69–0.91) | 0.83 (0.73–0.89) | 17 | 289 |
| 15 years | 0.69 (0.58–0.79) | 0.72 (0.61–0.81) | 28 | 148 |
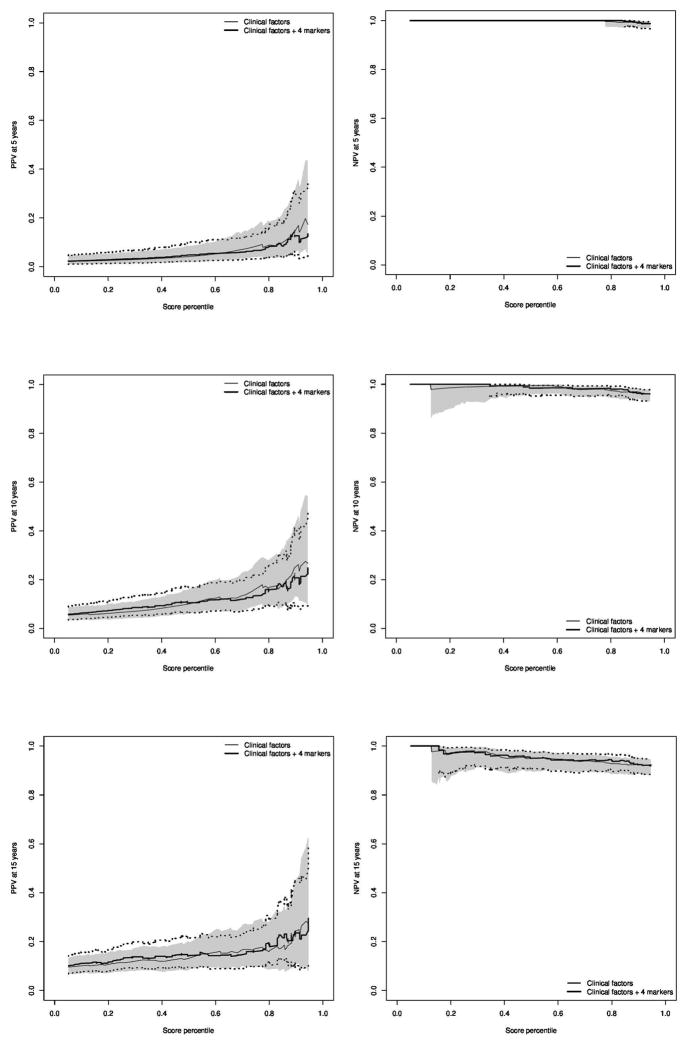
Figure 2 contains positive and negative predictive values at 5, 10, and 15 years of follow-up. We observe that neither PPV nor NPV appear improved by inclusion of the four protein markers with the clinical factors. Confidence intervals for certain areas of the NPV curves were not estimable, as the point estimate for NPV was 1.0 over these regions. The consistent pattern of low PPV and high NPV is indicative of prognostic scores that have adequate sensitivity, yet lack specificity for the lethal prostate cancer event that is relatively uncommon.
Figure 2.
Discussion
In this prospective study of prostate cancer patients with long-term follow-up, we found that protein tumor expression of PTEN, SMAD4, CCND1 and SPP1 did not significantly enhance prediction of lethal prostate cancer beyond age and Gleason score in the HPFS when using a prognostic scoring system built in the PHS. In addition to a standard AUC measure of discrimination, we evaluated whether the four markers could improve positive- or negative-predictive values and also evaluated how potential prognostication changed over time. Importantly, PPV and NPV incorporate information on the rate of lethal prostate cancer relative to indolent disease and, hence, can provide a clearer picture with respect to potential clinical impact. A complete assessment of prognosis may be missed by evaluation of the summary AUC measure alone[16].
Interestingly, our analysis of the clinical factor model suggests that Gleason score has high prognostic accuracy during the initial 5 years of follow-up, with a decrease in prognostic value with longer follow-up time. To our knowledge, the longitudinal trajectory of Gleason as a prognostic indicator has not been previously evaluated in this manner. Future biomarker discovery efforts may benefit from a focus on discriminatory performance at 10 or 15 years of follow-up in order to seek strong prognostic complements to Gleason scoring depending on the clinical question.
Our finding that the four markers could improve overall model fit in the PHS was consistent with the initial study[1]. Several factors could underlie the lacking transportability of the four-marker model to the HPFS. SMAD4 and SPP1 maintained the closest correlations of expression to one another across cohorts (data not shown). It is possible that the other two markers introduced additional variability in the validation step due to greater within-tumor heterogeneity. On the technical level, quantification of PTEN and CCND1 may not be sufficiently consistent by immunohistochemistry suggesting the need for more quantitative methods of assessing the strength and subcellular location of these signals in prostate cancer cells. More broadly, the identification of molecular signatures to predict metastatic and lethal prostate cancer that can be externally validated continues to pose a considerable challenge[3], and our findings are in line with similar investigations[17, 18].
We note that the inability of the four markers to improve upon established clinical factors in a prognostic context does not imply that they have no biological role or that they may not be potential candidates for treatment strategies. Indeed, the four markers examined in this study were identified through an integrated, cross-species oncogenomic, genetic and functional approach, and have been shown in model systems to be direct drivers of invasion and metastasis. This iterative, biologically-based approach may have enhanced the likelihood that our model reflects drivers of prostate cancer biology. Nonetheless, it is possible to observe minimal correspondence between relative hazards which reflect biology and measures of prognostic success which reflect clinical utility[13, 19].
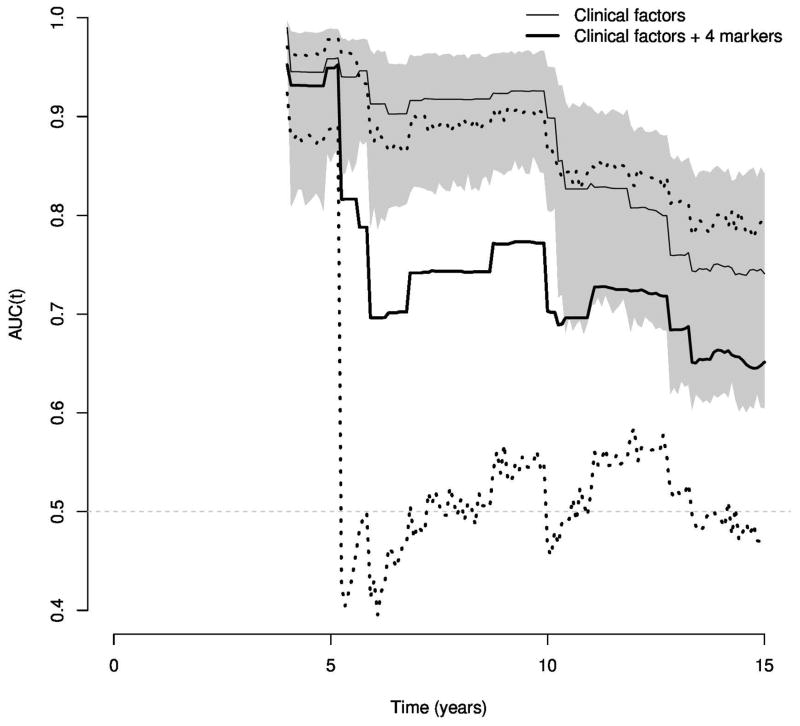
A considerable strength of the present study is the long and complete clinical follow-up from two independent, prospective cohorts focusing on the development of lethal and metastatic disease rather than the surrogate endpoint of biochemical failure. Given the long-term follow-up needed to study lethal prostate cancer, the cohorts included patients with somewhat higher-risk disease than is common today. Thus, the absolute rate of lethal prostate cancer may be somewhat higher in the cohort, although the absolute change in predictive probabilities is unlikely to differ. Moreover, a proportion of the cases were diagnosed before the introduction of PSA screening, and thus the pre-PSA cases will not have lead-time that is introduced with screening. A sensitivity analysis evaluated time-trends for the markers in models that included PSA at diagnosis when available. Though confidence intervals were wider due to reduced sample sizes, patterns were generally similar (Figure 3). Finally, the study was undertaken in a surgical cohort, and a proportion of the men with nonlethal prostate cancer may have had an aggressive disease that was cured by surgery. However, as shown by data from the US and Scandinavian randomized trials of watchful waiting versus prostatectomy, this proportion of patients is small (2–11%)[20, 21].
Figure 3.
Time-dependent AUC estimates through 15 years of follow-up when applying prognostic models built in the PHS cohort to the HPFS cohort among those men with non-missing PSA at diagnosis. PSA was included as a factor in both the clinical and marker models. The shaded region provides a 95% confidence interval for the clinical factor model, while the dotted lines give a 95% interval for the model with clinical factors and the four markers.
In summary, we found that the tumor expression of four markers shown to be central to the development of lethal disease in model systems did not significantly improve prediction of lethal disease in surgically treated men with prostate cancer. This finding highlights the challenges of finding robustly translatable biomarkers for aggressive prostate cancers that exhibit broad biological heterogeneity.
Acknowledgments
We would like to thank the participants in the Health Professionals Follow-up Study and Physicians’ Health Study for their dedicated participation over the past 25 years. We would like to acknowledge the Health Professionals Follow-up Study staff for their valuable contributions, and the following state cancer registries for their help in ascertaining cancer diagnosis: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MN, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data. We are grateful to Chungdak Li for tumor tissue microarray construction at the Dana Farber/Harvard Cancer Center, Li Moy and Lingxin Xu for technical assistance.
Funding
National Institutes of Health research grants UM1CA16755201 (PI Willett), P50CA090381 (PI Kantoff), R01CA133891, R01CA136578, R01CA141298, U01MMHCC, Metamark Genetics, Inc, the Belfer Institute for Applied Cancer Science and the Prostate Cancer Foundation. National Research Service Award T32 CA009001 NCI (Gerke)
Footnotes
Disclosure statement:
The authors of this paper have nothing to disclose.
References
- 1.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470(7333):269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman DG, Royston P. What do we mean by validating a prognostic model? Stat Med. 2000;19(4):453–73. doi: 10.1002/(sici)1097-0258(20000229)19:4<453::aid-sim350>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Shariat SF, Kattan MW, Vickers AJ, Karakiewicz PI, Scardino PT. Critical review of prostate cancer predictive tools. Future Oncol. 2009;5(10):1555–84. doi: 10.2217/fon.09.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4(4):309–14. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 5.Fiorentino M, Judson G, Penney K, Flavin R, Stark J, Fiore C, Fall K, Martin N, Ma J, Sinnott J, Giovannucci E, Stampfer M, Sesso HD, Kantoff PW, Finn S, Loda M, Mucci L. Immunohistochemical expression of BRCA1 and lethal prostate cancer. Cancer Res. 2010;70(8):3136–9. doi: 10.1158/0008-5472.CAN-09-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickson WK, Flavin R, Kasperzyk JL, Fiorentino M, Fang F, Lis R, Fiore C, Penney KL, Ma J, Kantoff PW, Stampfer MJ, Loda M, Mucci LA, Giovannucci E. Vitamin D receptor protein expression in tumor tissue and prostate cancer progression. J Clin Oncol. 2011;29(17):2378–85. doi: 10.1200/JCO.2010.30.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, Suppan C, Flavin R, Sesso HD, Rider JR, Sweeney C, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci EL, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21(9):1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, Stampfer MJ. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338(8765):464–8. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 9.Final report on the aspirin component of the ongoing Physicians’ Health Study. Steering Committee of the Physicians’ Health Study Research Group. N Engl J Med. 1989;321(3):129–35. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 10.Christen WG, Gaziano JM, Hennekens CH. Design of Physicians’ Health Study II–a randomized trial of beta-carotene, vitamins E and C, and multivitamins, in prevention of cancer, cardiovascular disease, and eye disease, and review of results of completed trials. Ann Epidemiol. 2000;10(2):125–34. doi: 10.1016/s1047-2797(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 11.Royston P, Altman DG. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol. 2013;13:33. doi: 10.1186/1471-2288-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Cai T, Pepe MS, Levy WC. Time-dependent Predictive Values of Prognostic Biomarkers with Failure Time Outcome. J Am Stat Assoc. 2008;103(481):362–8. doi: 10.1198/016214507000001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepe MS, Zheng Y, Jin Y, Huang Y, Parikh CR, Levy WC. Evaluating the ROC performance of markers for future events. Lifetime Data Anal. 2008;14(1):86–113. doi: 10.1007/s10985-007-9073-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Cai T, Zheng Y. Evaluating the predictive value of biomarkers with stratified case-cohort design. Biometrics. 2012;68(4):1219–27. doi: 10.1111/j.1541-0420.2012.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown M. survAccuracyMeasures. 2014 Apr 1; https://github.com/mdbrown/survAccuracyMeasures. commit 594f71d4d7.
- 16.Dodd LE, Pepe MS. Partial AUC estimation and regression. Biometrics. 2003;59(3):614–23. doi: 10.1111/1541-0420.00071. [DOI] [PubMed] [Google Scholar]
- 17.Mucci LA, Pawitan Y, Demichelis F, Fall K, Stark JR, Adami HO, Andersson SO, Andren O, Eisenstein AS, Holmberg L, Huang W, Kantoff PW, Perner S, Stampfer MJ, Johansson JE, Rubin MA. Nine-gene molecular signature is not associated with prostate cancer death in a watchful waiting cohort. Cancer Epidemiol Biomarkers Prev. 2008;17(1):249–51. doi: 10.1158/1055-9965.EPI-07-0722. [DOI] [PubMed] [Google Scholar]
- 18.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, Hoshida Y, Perner S, Adami HO, Fall K, Mucci LA, Kantoff PW, Stampfer M, Andersson SO, Varenhorst E, Johansson JE, Gerstein MB, Golub TR, Rubin MA, Andren O. Molecular sampling of prostate cancer: A dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emir B, Wieand S, Su JQ, Cha S. Analysis of repeated markers used to predict progression of cancer. Stat Med. 1998;17(22):2563–78. doi: 10.1002/(sici)1097-0258(19981130)17:22<2563::aid-sim952>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 20.Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, Nsouli I, Iyer P, Cartagena R, Snider G, Roehrborn C, Sharifi R, Blank W, Pandya P, Andriole GL, Culkin D, Wheeler T. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367(3):203–13. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bill-Axelson A, Holmberg L, Garmo H, Rider JR, Taari K, Busch C, Nordling S, Haggman M, Andersson SO, Spangberg A, Andren O, Palmgren J, Steineck G, Adami HO, Johansson JE. Radical prostatectomy or watchful waiting in early prostate cancer. N Engl J Med. 2014;370(10):932–42. doi: 10.1056/NEJMoa1311593. [DOI] [PMC free article] [PubMed] [Google Scholar]