Abstract
Huntington’s disease (HD) is a progressive neurodegenerative disorder that profoundly impairs corticostriatal information processing. While late stage pathology includes cell death, the appearance of motor symptoms parallels more subtle changes in neuronal function and synaptic integration. Because of the difficulty in modeling the disease and the complexity of the corticostriatal network, understanding the mechanisms driving pathology has been slow to develop. In recent years, advances in animal models and network analysis tools have begun to shed light on the circuit-specific deficits. These studies have revealed a progressive impairment of corticostriatal synaptic signaling in sub-populations of striatal neurons, turning classical excitotoxicity models of HD upside down. Disrupted brain derived neurotrophic factor signaling appears to be a key factor in this decline.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by an expanded polyglutamine repeat in the huntingtin gene [1–4]. HD patients are plagued by progressive motor dysfunction. Initially, patients manifest uncontrolled, choreic “dance-like” movements [4]. This hyperkinetic phase is followed by a hypokinetic phase where purposeful movement is difficult [5]. Cognitive dysfunction parallels the declining motor control.
Consistent with the motor symptoms, postmortem studies of HD brains have found the basal ganglia to be a major site of pathology, in spite of the widespread expression of mutant huntingtin (mHtt) [2,6–8]. The earliest neuronal pathology in the basal ganglia is in the caudate and putamen (collectively referred to the striatum in rodents). Less profound pathology also is found in the cerebral cortex and thalamus, both structures that innervate the striatum [4].
The biphasic progression of symptoms in HD patients – and in many animal models – is consistent with the view that principal striatal GABAergic spiny projection neurons (SPNs) are not uniformly susceptible to mHtt [5,7]. Striatal SPNs can be divided into two roughly equally sized groups that differ along a number of dimensions, including peptide expression and axonal projection [9]. The first signs of pathology in HD patients are in one of these groups: indirect pathway SPNs (iSPNs) [7]; iSPNs anchor the basal ganglia network that suppresses contextually inappropriate movements. Later in the disease, direct pathway SPNs (dSPNs) that express substance P are affected [4]; dSPNs anchor the basal ganglia circuit that promotes contextually appropriate movements. Thus, HD neuropathology and symptoms align nicely with what is known about the functional circuitry of the striatum.
What is less clear is why SPNs should be particularly vulnerable to mHtt. A longstanding view posits that glutamate excitotoxicity is the culprit [10,11]. Support for this hypothesis comes primarily from the fact that intrastriatal injection of the glutamate receptor agonist quinolinic acid (an NMDAR agonist) mimics many characteristics of HD in rodents [12]. The proposition that NMDA receptors (NMDARs) drive neuronal loss in HD also is consistent with a large literature showing how this might happen [11,13]. However, more recent work has cast doubt on this theory. As outlined below, studies in animal models of HD suggest that there is a progressive loss of excitatory corticostriatal glutamatergic input to SPNs with advancing age, rather than a progressive growth of this input. In addition, there is evidence that the other major excitatory glutamatergic input to SPNs from the thalamus is also lost [14,15]. While these observations don’t unequivocally kill the excitotoxicity hypothesis, they make it less plausible.
As the excitotoxicity hypothesis has fallen in favor, another hypothesis has risen to prominence. Several lines of study suggest that something goes awry with cortical trophic support for the striatum. Brain derived neurotrophic factor (BDNF) is synthesized by cortical pyramidal neurons innervating the striatum, transported to the striatum and released [16]. BDNF activation of TrkB receptors on SPNs is necessary to maintain normal dendritic and synaptic function [17–19]. The expression of mHtt can impair corticostriatal BDNF signaling, suggesting that SPNs ‘wither’ in HD.
In what follows, we briefly outline recent developments that have led to our current opinion about striatal excitatory synaptic dysfunction in HD. The review is not all inclusive and readers are referred to a number of other recent reviews that cover similar or complementary aspects of the expansive HD literature [20–24].
A plethora of genetic mouse models of HD - which one is best?
Since the genetic locus of HD was identified in 1993 [1], a number of genetic models have been developed in mice. These models differ significantly in their genetic strategy, have different rates of progression and types of neuronal pathology (see reviews by [2,24,25]) (Fig. 1). The first models expressed an exon 1 fragment of mHtt with a large number (110–150) of CAG repeats (e.g., the R6/2 model) [26]. These mice are rapidly progressing, manifesting motor symptoms within a few weeks of birth and die prematurely. Subsequently, full length mHtt models were constructed with varying CAG repeat lengths (YAC72 (72 repeats), YAC128 (128 repeats) and BACHD (97 repeats). These models have slower disease progression with a clear dependence upon expansion length and gene dosage [27–29]. In the last few years, more biologically faithful knock-in models have been created (Q140, zQ175 lines). These models also manifest a slowly progressing pathology [30–32].
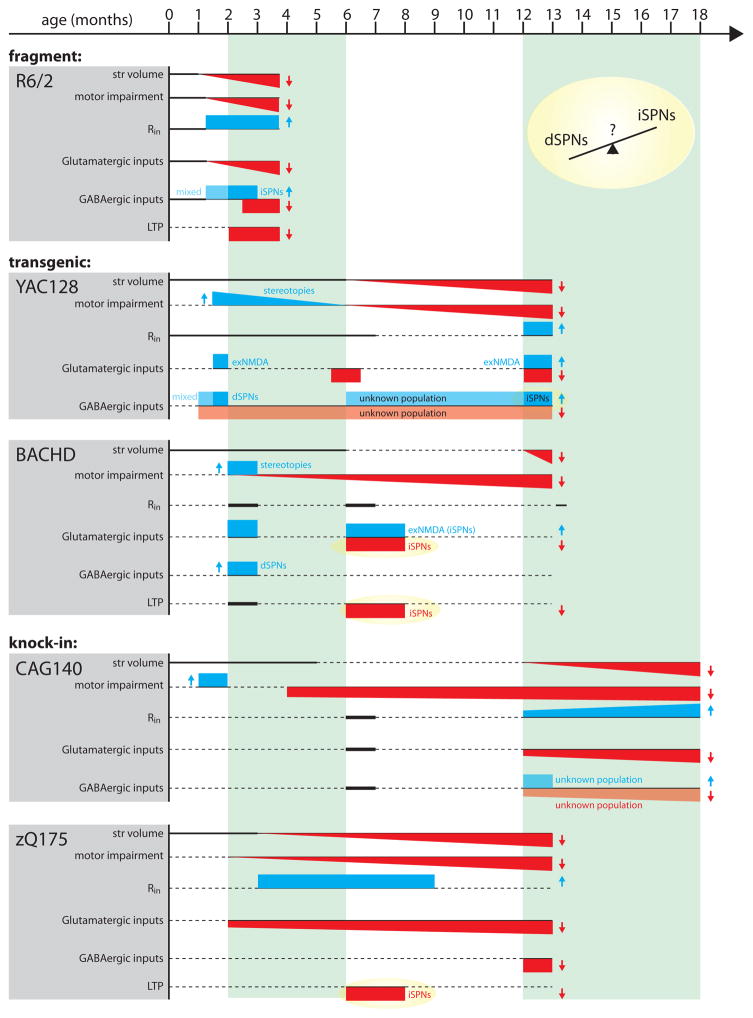
Figure 1.
Representation of disease progression in 5 genetic mouse models of HD. Parameters include striatal volume (a measure of frank cell loss), motor impairments (if not specified, based on rotorod performance and open field locomotor activity), input resistance (Rin), glutamatergic inputs (studies measuring both spontaneous and evoked glutamatergic events are included; exNMDA refers to extrasynaptic NMDA receptor engagement), GABAergic inputs (studies measuring both spontaneous and evoked GABAergic events are included) and LTP (long term potentiation). Unless otherwise noted, physiological changes are from mixed populations of iSPNs and dSPNs. Rectangles represent binned time points. Angled lines represent progressive alterations. Red-filled downward directed shapes and blue-filled upward directed shapes represent decreases and increases, respectively. Lightened colors represent unidentified SPN populations. Thick solid horizontal lines indicate no change was observed. Dashed horizontal lines indicate data at the corresponding time points were not reported in the referenced studies. Data compiled from [2,29,31–33,35,39,41,59,81–84].
Do these models mimic the human disease? As discussed above, humans transition through a hyperkinetic (choreic) and then a hypokinetic state [5]. Biphasic behavioral changes like this are seen in many HD models, but not all. In particular, only some slowly progressing models manifest a hyperkinetic phase, whereas all have a hypokinetic phase. The short repeat fragment model (R6/1) and full-length YAC72, YAC128, Q94 and Q140 models have an early hyperkinetic phase before ultimately becoming hypokinetic [25,30]. Why BACHD and zQ175 models do not have this early phase is not clear [29,32]. The absence of a hyperkinetic phase in the zQ175 model is puzzling, as it was derived from Q140 model.
One possible explanation for the discrepancy is that mHtt does not affect brain circuits controlling movement sequentially, but rather affects them in parallel. If the rate of disease progression within each of these circuits is independently affected by the genetic approach used to introduce mHtt, then the duration of each motor phase could vary. The early hyperkinetic phase should be the most sensitive to this kind of parallel process. For example, let’s suppose that the hyperkinetic phase is dependent upon reduced activity in the indirect pathway and the hypokinetic phase is dependent upon reduced activity in the direct pathway. A slower rate of progression in the direct pathway than the indirect pathway would yield the human pattern of staging. But if the rates of progression are similar is some models (e.g., BACHD), then the hyperkinetic phase would be absent.
Since no one mouse model perfectly recapitulates all aspects of the human HD condition, which model is the best? In principle, the heterozygous CAG140 and zQ175 models most accurately model the human condition as they place a single copy of full length mHtt in its native genomic locus; these models display a progressive behavioral phenotype and recapitulate many synaptic and anatomical pathologies present in more rapidly progressing models (Fig. 1). However, it must be acknowledged that the choice of the most appropriate model for a particular study will depend upon many factors.
Do SPNs get too much or too little excitatory input in HD?
For years, the pathology in HD was envisioned to be a consequence of glutamatergic excitotoxic damage to SPNs, as injection of NMDA receptor (NMDAR) agonists within the striatum produced a pattern of pathology resembling that found in HD brains [12]. But the ability of NMDAR agonists to phenocopy striatal changes does not mean that they mimic pathogenesis. Was there evidence that glutamatergic signaling in the striatum was altered in genetic models of HD? The first direct physiological evidence that something was changed came from the analysis of spontaneous excitatory events in ex vivo brain slices from the R6/2 HD mouse; in these studies, the frequency of spontaneous synaptic glutamate release onto SPNs increased and then decreased (Fig 1) [33]. Without knowing the identity of sampled SPNs (dSPNs or iSPNs) or the source of released glutamate, little can be inferred about why this happened or what it might mean for the striatal circuitry, but what these studies do show is that glutamatergic signaling is changing in HD models.
Does the glutamatergic input to both iSPNs and dSPNs change? Recall that in humans, iSPNs appear to be the most vulnerable to the disease process. The loss of iSPN functional integrity has long been thought to be responsible for the hyperkinetic features of early stage HD, as iSPNs anchor the indirect pathway responsible for movement suppression [9,34]. More recent studies have begun to address this question by crossing HD genetic models with BAC transgenic mice in which dSPNs or iSPNs express green fluorescent protein (GFP) [35–37]. Using this strategy Levine’s group suggested that the early increase in spontaneous glutamate EPSC frequency was specific to dSPNs, which would increase direct pathway excitability and promote hyperactivity. However, Andre et al. also showed that evoked glutamatergic responses were substantially larger in iSPNs in young HD models, but dSPN responses were normal [35]. These results suggest that early in the evolution of the disease, there is a complex set of pre- and post-synaptic changes taking place at SPN glutamatergic synapses, which might not map cleanly to the early and late motor symptoms of HD.
Nevertheless, these studies show that synaptic glutamate receptor function is not changing in a way that is consistent with an excitotoxicity model of HD. The ‘out’ here for the excitotoxicity model is that it does not specify that synaptic glutamate receptors are driving pathology. In the last decade it has become clear that while synaptic NMDARs promote neuronal viability, extrasynaptic NMDARs are coupled to signaling cascades that promote degeneration and death [11,38]. In a landmark paper, Milnerwood and Raymond demonstrated that the abundance of extrasynaptic GluN2B-containing NMDARs rises in HD SPNs [39]. Subsequently, they have shown that the insertion of extrasynaptic NMDARs is regulated by calpain- and striatal enriched protein tyrosine phosphatase (STEP) [40] in aged YAC128 mice (Fig. 2). Moreover, the elevation in extrasynaptic NMDARs appears to occur first in iSPNs [37], then ultimately spreads into both SPN types [41], establishing a parallel with the evolution of pathology in humans.
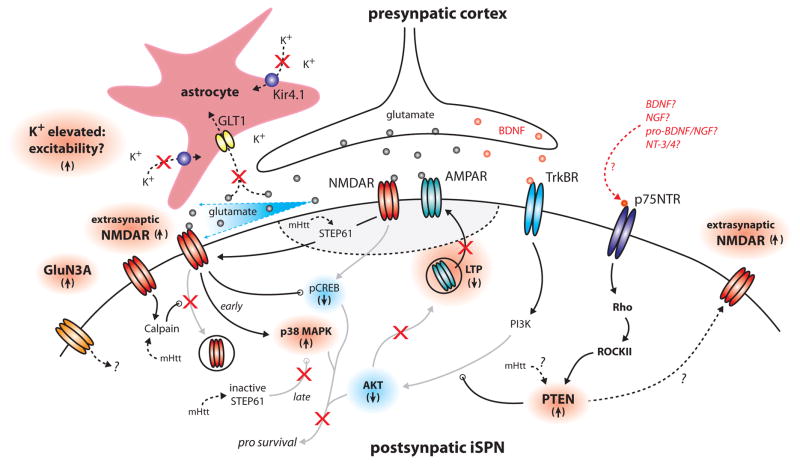
Figure 2.
Schematic diagram of alterations in iSPNs leading to attenuated LTP, NMDA receptor signaling and cell death. STEP61, striatal enriched protein tyrosine phosphatase 61; pCREB, phosphorylated cyclic adenosine monophoshate response element; Rho, Rho A small GTPase; ROCKII, Rho-associated, coiled-coil containing protein kinase II; NGF, nerve growth factor; pro-NGF, pro-nerve growth factor; pro-BDNF, pro-brain derived neurotrophic factor; NT-3/4, neurotropic 3/neurotrophic 4 [37,39,40,43–46].
The proposition that there is a progressive engagement of extrasynaptic NMDARs driving pathology is also consistent with changes in astrocytic function seen in the HD models. Clearance of glutamate from the synaptic cleft is controlled by astrocytic uptake through transporters [42]. In symptomatic HD mice, astrocytic expression of glutamate transporter 1 (GLT1) is down-regulated [43]. In principle, this should lead to more robust engagement of extrasynaptic NMDARs (Fig. 2). In addition, recent work in aged R6/2 HD mice has shown that astrocytes down-regulate the expression of a key K+ channel (Kir4.1), resulting in an elevation in extracellular K+ concentration [44]. Elevating extracellular K+ should depolarize SPNs and make it easier to remove the Mg2+ block of NMDARs. Thus, astrocytic dysfunction could potentiate the negative impact of rising extrasynaptic NMDARs (Fig. 2).
As plausible as this scenario seems, work in the last year has cast doubt on the proposition that extrasynaptic NMDARs are driving late stage striatal pathology in HD. Gladding et al (2014) found that while intrastriatal injection of quinolinate (an NMDAR agonist) induces neuronal death in young presymptomatic (6 week) YAC128 mice, the same injection into aged (1 year) YAC128 mice had little effect [45]. The resistance to NMDAR excitotoxicity in the older HD mice appeared to be due to an attenuation of the ability of extrasynaptic NMDA receptors to activate pro-apoptotic p38 mitogen-activated protein kinase (p38 MAPK). In young mice, extrasynaptic NMDARs robustly engaged p38 MAPK, whereas in older HD mice, this linkage seemed to be attenuated. The change appeared to be dependent upon processing of STEP (Fig. 2).
Although this work suggests that NMDARs are not driving HD pathology, the issue is not resolved. Work by Perez-Otano et al. argues that striatal expression of GluN3A in very old (16 month) YAC128 mice is critical to late stage synaptic dysfunction and neurodegeneration [46]. Sorting out these seemingly discrepant findings is one of the challenges facing the field.
Is BDNF at the center of striatal dysfunction in HD?
There is a significant body of literature suggesting that mHtt impairs the ability of cortical pyramidal neurons to provide the striatum with needed BDNF, leading to striatal ‘withering’ [20,47,48]. There is no question that cortical BDNF expression and axonal transport can be impaired by mHtt. However, these studies have relied largely upon over-expression of mHtt in cell culture models, not in vivo or ex vivo approaches in mouse HD models. Those studies that have been done in mouse models that support this conclusion have focused upon BDNF mRNA abundance assays that have used less than optimal strategies for quantitation.
Another prickly issue with this model is that both dSPNs and iSPNs express the receptor for BDNF – TrkB receptors [49] – and receive overlapping cortical inputs [50]. How then, if non-cell autonomous factors are driving striatal HD pathology, does selective vulnerability come about?
Recent work using very different approaches argues that there are corticostriatal BDNF signaling deficits in HD models, but that these deficits are in the postsynaptic response to BDNF, not its delivery, release or binding to TrkBRs [37,51]. In 6 month old BACHD and heterozygous zQ175 knock-in mice (before striatal cell loss) neither cortical nor striatal BDNF mRNA or protein was abnormal. Neither was striatal TrkBR expression or activation of TrkBRs by stimulation of corticostriatal axons. However, TrkBR activation of Akt – a key signaling kinase in prosurvival pathways – was impaired [37].
To pinpoint why this was the case, a functional assay for TrkBR signaling was developed using the fact that corticostriatal long term potentiation (LTP) requires SPN TrkBR signaling (Fig 2) [18]. This led to the conclusion that TrkBR signaling through phosphoinositide-3 kinase (PI3K) was blunted specifically in HD iSPNs due to up-regulation of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [37]. It also was found that p75 neurotrophic factor receptor (p75NTR) signaling was necessary for PTEN-mediated attenuation of TrkBR signaling. Why PTEN is up-regulated initially in iSPNs remains to be determined (Fig 2) [37], but one intriguing possibility is that D2 receptors, which are expressed by iSPNs and not dSPNs, are involved, as D2 receptors have been shown to elevate the engagement of several downstream targets of p75NTRs [52]. Whether this turns out to be the mechanism or not, these studies support the proposition that neuronal phenotype is a determinant of the staging of HD pathology.
A deficit in iSPN TrkBR signaling and LTP induction is consistent with the progressive attenuation of corticostriatal glutamatergic signaling described in HD models (Fig. 1). While the earliest deficits in striatal TrkBR signaling have been found in iSPNs, it is likely that as the disease progresses, this deficit will spread to dSPNs. It is of some note that HD patients have a lower incidence of cancer and PTEN (the proximal culprit in impaired TrkBR signaling) is a tumor suppressor [53]; this suggests that mHtt might ultimately induce PTEN up-regulation broadly. Not only might this lead to impaired TrkBR signaling in dSPNs, but the deficit should appear in cortical pyramidal neurons and neurons in other regions of the brain.
An important implication of these findings is that delivery of small molecule TrkBR agonists is unlikely to be effective in HD. Rather, targeting p75NTR signaling should be more effective as well as less burdened by side-effects, as this receptor is developmentally downregulated and has a restricted tissue distribution.
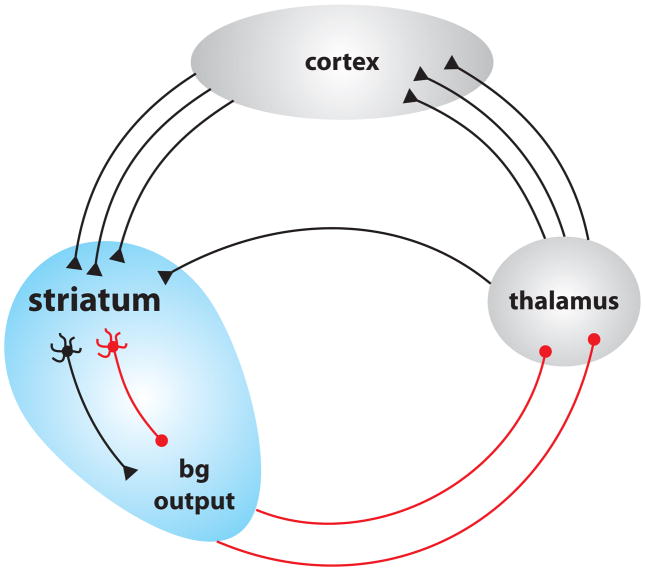
It also should be remembered that the cerebral cortex and striatum form part of a richly connected network involved in movement and thought control [9]. It is impossible to affect one component of this network without affecting other components (Fig 3) [54]. For example, expression of mHtt in a discrete cortical neuron populations induces pathology in neighboring neurons [55]. Since BDNF production and release are activity dependent [56,57], deficits in corticostriatal signaling are very likely to ultimately lead to deficits in BDNF signaling within other components of the circuit, most importantly the cerebral cortex [47,58].
Figure 3.
Schematic representation of the basal ganglia-thalamus-cortex feedback loop.
Does GABAergic input to SPNs change?
For some time it has been thought that GABAergic input to SPNs increased in HD models, nominally compensating for their increased intrinsic excitability [59]. This conclusion was based upon a reported increase in the frequency of spontaneous inhibitory postsynaptic currents (sIPSCs) in a random sample of SPNs in symptomatic R6/2, YAC128 and CAG140 HD models [60]. Subsequent work in BAC-eGFP transgenic mice found that the increase occurred first in dSPNs and then later in iSPNs [61].
The trouble with measurements of sIPSCs is that they are heterogeneous. This is particularly problematic in the striatum where the GABAergic circuitry is very complex. SPNs have GABAergic synapses arising from other SPNs and a heterogeneous group of interneurons (there are at least 3 classes). Because spontaneous IPSCs are a mixture of miniature IPSCs (whose frequency depends upon terminal release probability) and spike-drivien IPSCs, there is no way of knowing which circuit elements are driving changes in sIPSC frequency or why.
Recent work using another approach has challenged the general notion that GABAergic inhibition of SPNs is increased in HD models. Dvorzhak et al. found that in both R6/2 and zQ175 models, GABAergic responses evoked by minimal local stimulation were reduced in SPNs [62]. The reduction was attributable to an up-regulation in mGluR5 mediated endocannabinoid (eCb) production, resulting in presynaptic CB1 receptor activation and suppression of GABA release. Although the identity of the presynaptic axons was not determined, it is not unreasonable to make the conjecture that they were collaterals from neighboring GABAergic SPNs given the strong CB1R mediated suppression of GABA release [63]. Consistent with this interpretation, Cepeda et al. [36] found that collateral connectivity between SPNs was attenuated in the R6/2 model. In this same study, the authors found that optogenetic activation of fast-spiking interneurons (FSIs) evoked larger responses in HD SPNs. Stimulation of another major interneuron input to SPNs – from PLTSIs – yielded similar amplitude responses, but the spontaneous activity of these interneurons was elevated in HD brain slices. Taken together, these studies suggest that GABAergic input to SPNs from interneurons is elevated in the HD striatum, whereas that arising from collaterals is diminished.
As both FSIs and PLTSIs appear to be part of a corticostriatal feedforward inhibitory circuit [64], the changes described could serve to limit the duration of the striatal response to cortical glutamatergic excitation. In contrast, collateral feedback between SPNs would be expected to limit the spatial dimensions of the cortically evoked striatal activity. That said, there are some fundamental unknowns in this equation. One is that it is far from clear whether it is appropriate to characterize GABAA receptors as inhibitory. At rest, Kir2 K+ channels dominate the SPN conductance profile (though there is evidence their somatic density may be reduced in HD SPNs [65]), holding the membrane potential in a ‘down’ state near the K+ equilibrium potential (~−90 mV). Although it has not been determined rigorously in adult SPNs, the Cl− reversal potential is sure to be more depolarized than this, probably in the range of −65 to −70 mV [66]. Thus, GABAergic input is very likely to be depolarizing to quiescent SPNs. The shunting effect of open GABAA receptors could counter-balance this effect to some extent when there is temporally coincident excitatory input [67]. But this effect is short-lived and because the local depolarizing effect of GABA should globally collapse the electrotonic structure of SPNs, glutamatergic inputs that trail the GABAergic input should be amplified. This could bring NMDA receptors into a voltage range where Mg2+ block is less potent. Sorting how these interactions occur in SPN dendrites still faces technical hurdles but advances like multi-color opsins [68] should make profitable study feasible soon.
Another unresolved issue is the impact of tonic, extrasynaptic GABAA receptor currents in the HD models. Recent data suggest they may be decreased in R6/2 iSPNs [36]. Tonic currents are prominent in SPNs [69,70] and help set the membrane potential and input resistance of dendrites – key determinants of synaptic integration. Neurogliaform GABAergic interneurons, which have not been examined in HD models to date, appear to exert a strong influence on these receptors [71]. Also, if there is astrocytic dysfunction in HD, GABA could easily ‘overflow’ from synaptic sites to these receptors.
Are there other plastic changes in HD?
Although recent studies provide a rich working framework for understanding how mHtt alters the moment-to-moment activity in the corticostriatal network, there are fundamental questions about long-term plasticity of synapses that remain unanswered. For example, is corticostriatal LTP the only form of synaptic plasticity altered in the HD striatum? This seems unlikely. The best understood form of synaptic depression at both cortical glutamatergic and intrastriatal GABAergic synapses requires postsynaptic group I metabotropic glutamate receptor (mGluR) mediated production of endocannabinoids (eCBs), which then diffuse to the presynaptic terminal, bind to presynaptic CB1 receptors (CB1Rs), and reduce neurotransmitter release probability [72–74]. One of the earliest molecular changes in HD patients is a decrease in CB1R expression in both the striatum and cortex [75,76]. It is unknown if corticostriatal synaptic depression is altered in HD, but recent work showed that genetically correcting CB1R down-regulation in SPNs rescues axospinous (largely corticostriatal) synapse loss in HD mice [77]. Why would elevating SPN CB1R expression increase glutamatergic axospinous synapses? Perhaps the effect is indirect. Much of the intrastriatal GABAergic input to SPNs comes from recurrent collateral synapses [64]. Suppression of SPN collateral GABA release might engage homeostatic mechanisms to increase glutamatergic innervation (leading to more spines). This makes sense if GABAergic synapses were excitatory. As mentioned above, this is not so far-fetched since SPNs rest about 20 mV more hyperpolarized than the Cl− reversal potential (GABAA receptors are Cl− permeable channels) [66]. In this scenario, suppression of GABAergic signaling could lead to SPN hypoexcitability, triggering homeostatic up-regulation of glutamatergic synapses [78].
Finally, though striatal glutamatergic hypoexcitability is a common theme among late stage HD models, it is far from certain that this hypoexcitability is entirely synaptic in origin. For example, dendritic morphology and ion channel composition exquisitely shape how SPNs integrate glutamatergic inputs [79,80]. Alterations in either of these parameters might diminish the response to activation of glutamatergic synapses. In fact, the near universal increase in SPN input resistance (Fig. 1) [21] might reflect an intrinsic homeostatic adaptation of this sort. Although SPN dendrites are too small to directly record, two photon laser scanning microscopy, optogenetics and two photon uncaging techniques make these regions accessible to investigation [79,80]. These approaches should make the role of intrinsic mechanisms in HD pathogenesis clearer in the near future.
Concluding Remarks
The development of new mouse models and tools for analyzing brain networks and synapses has led to fundamental new insights into the mechanisms driving HD pathogenesis. This is most evident in our understanding of how mHtt affects the corticostriatal network. In the last few years, the field has moved from thinking that striatal pathology in HD was driven by excitotoxic mechanisms to the view that, if anything, it is a hypoexcitability disorder driven by impaired corticostriatal signaling. Although there remain fundamental gaps in our understanding of synaptic dysfunction in HD, the tools at our disposal should accelerate progress toward filling these gaps. With that understanding should come the first generation of effective therapies for HD.
Highlights.
Advances in genetic mouse models and tools to probe neural circuits have led to new insights about pathogenic mechanisms in Huntington’s disease (HD).
Recent work suggests that there is a progressive decline in corticostriatal glutamatergic signaling in HD.
The progressive decline in corticostriatal glutamatergic signaling appears to be driven by both cell-autonomous and network mechanisms.
Deficits in brain derived neurotrophic factor signaling have emerged as a major factor in synaptic pathogenesis and the death of striatal neurons in HD.
Acknowledgments
This work was supported by CHDI and NS34696.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
• Special Interest
•• Outstanding Interest
- 1.Group HsDCR. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Menalled LB, Chesselet MF. Mouse models of Huntington’s disease. Trends Pharmacol Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 3.Cepeda C, Wu N, Andre VM, Cummings DM, Levine MS. The corticostriatal pathway in Huntington’s disease. Prog Neurobiol. 2007;81:253–271. doi: 10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 5.Berardelli A, Noth J, Thompson PD, Bollen EL, Curra A, Deuschl G, van Dijk JG, Topper R, Schwarz M, Roos RA. Pathophysiology of chorea and bradykinesia in Huntington’s disease. Mov Disord. 1999;14:398–403. doi: 10.1002/1531-8257(199905)14:3<398::aid-mds1003>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 6.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Reiner A, Albin RL, Anderson KD, D’Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vonsattel JP, Keller C, Cortes Ramirez EP. Huntington’s disease - neuropathology. Handb Clin Neurol. 2011;100:83–100. doi: 10.1016/B978-0-444-52014-2.00004-5. [DOI] [PubMed] [Google Scholar]
- 9.Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones L, Hughes A. Pathogenic mechanisms in Huntington’s disease. Int Rev Neurobiol. 2011;98:373–418. doi: 10.1016/B978-0-12-381328-2.00015-8. [DOI] [PubMed] [Google Scholar]
- 11.Parsons MP, Raymond LA. Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron. 2014;82:279–293. doi: 10.1016/j.neuron.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Beal MF, Kowall NW, Ellison DW, Mazurek MF, Swartz KJ, Martin JB. Replication of the neurochemical characteristics of Huntington’s disease by quinolinic acid. Nature. 1986;321:168–171. doi: 10.1038/321168a0. [DOI] [PubMed] [Google Scholar]
- 13.Koh JY, Peters S, Choi DW. Neurons containing NADPH-diaphorase are selectively resistant to quinolinate toxicity. Science. 1986;234:73–76. doi: 10.1126/science.2875522. [DOI] [PubMed] [Google Scholar]
- 14.Deng YP, Wong T, Bricker-Anthony C, Deng B, Reiner A. Loss of corticostriatal and thalamostriatal synaptic terminals precedes striatal projection neuron pathology in heterozygous Q140 Huntington’s disease mice. Neurobiol Dis. 2013;60:89–107. doi: 10.1016/j.nbd.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng YP, Wong T, Wan JY, Reiner A. Differential loss of thalamostriatal and corticostriatal input to striatal projection neuron types prior to overt motor symptoms in the Q140 knock-in mouse model of Huntington’s disease. Front Syst Neurosci. 2014;8:198. doi: 10.3389/fnsys.2014.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altar CA, Cai N, Bliven T, Juhasz M, Conner JM, Acheson AL, Lindsay RM, Wiegand SJ. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 17.Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 18.Jia Y, Gall CM, Lynch G. Presynaptic BDNF promotes postsynaptic long-term potentiation in the dorsal striatum. J Neurosci. 2010;30:14440–14445. doi: 10.1523/JNEUROSCI.3310-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nithianantharajah J, Hannan AJ. Dysregulation of synaptic proteins, dendritic spine abnormalities and pathological plasticity of synapses as experience-dependent mediators of cognitive and psychiatric symptoms in Huntington’s disease. Neuroscience. 2013;251:66–74. doi: 10.1016/j.neuroscience.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 20.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Raymond LA, Andre VM, Cepeda C, Gladding CM, Milnerwood AJ, Levine MS. Pathophysiology of Huntington’s disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 2011;198:252–273. doi: 10.1016/j.neuroscience.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JY, Wang EA, Cepeda C, Levine MS. Dopamine imbalance in Huntington’s disease: a mechanism for the lack of behavioral flexibility. Front Neurosci. 2013;7:114. doi: 10.3389/fnins.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrada-Sanchez AM, Rebec GV. Role of cerebral cortex in the neuropathology of Huntington’s disease. Front Neural Circuits. 2013;7:19. doi: 10.3389/fncir.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat Rev Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 25.Levine MS, Cepeda C, Hickey MA, Fleming SM, Chesselet MF. Genetic mouse models of Huntington’s and Parkinson’s diseases: illuminating but imperfect. Trends Neurosci. 2004;27:691–697. doi: 10.1016/j.tins.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 27.Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 28.Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 29.Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 31.Heikkinen T, Lehtimaki K, Vartiainen N, Puolivali J, Hendricks SJ, Glaser JR, Bradaia A, Wadel K, Touller C, Kontkanen O, et al. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington’s disease. PLoS One. 2012;7:e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menalled LB, Kudwa AE, Miller S, Fitzpatrick J, Watson-Johnson J, Keating N, Ruiz M, Mushlin R, Alosio W, McConnell K, et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PLoS One. 2012;7:e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cepeda C, Hurst RS, Calvert CR, Hernandez-Echeagaray E, Nguyen OK, Jocoy E, Christian LJ, Ariano MA, Levine MS. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23:961–969. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••35.Andre VM, Cepeda C, Fisher YE, Huynh M, Bardakjian N, Singh S, Yang XW, Levine MS. Differential electrophysiological changes in striatal output neurons in Huntington’s disease. J Neurosci. 2011;31:1170–1182. doi: 10.1523/JNEUROSCI.3539-10.2011. This study represents a technical leap forward in the electrophysiological examination of the HD striatum. The authors crossed multiple HD mouse lines with BAC transgenics expressing fluorescent SPN specific reporters, allowing changes in spontaneous and evoked glutamatergic transmission to be interrogated in defined SPN populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Cepeda C, Galvan L, Holley SM, Rao SP, Andre VM, Botelho EP, Chen JY, Watson JB, Deisseroth K, Levine MS. Multiple sources of striatal inhibition are differentially affected in Huntington’s disease mouse models. J Neurosci. 2013;33:7393–7406. doi: 10.1523/JNEUROSCI.2137-12.2013. The authors used a combination of paired electrophysiological recordings and affarent specific ontogenetic stimulation to probe GABAergic inputs to defined SPN populations. This study suggested, for the first time, that alterations in GABAergic inputs to SPNs may arise from specific presynaptic sources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••37.Plotkin JL, Day M, Peterson JD, Xie Z, Kress GJ, Rafalovich I, Kondapalli J, Gertler TS, Flajolet M, Greengard P, et al. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington’s disease. Neuron. 2014;83:178–188. doi: 10.1016/j.neuron.2014.05.032. This study used a novel LTP induction technique to assay BDNF and TrkB receptor function with cell specific resolution in several mouse models of HD. What the study showed is that contrary to popular dogma, BDNF signaling is impaired first in iSPNs due to postsynaptic shunting of TrkB receptor signaling, not an impairment in in BDNF production or delivery to the striatum. This work suggests that targeting p75NTR signaling, and not BDNF replacement, is a promising direction for future therapies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••39.Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RW, Vasuta OC, Graham RK, Hayden MR, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. This seminal study used electrophysiological techniques to show that while synaptic glutamate receptor composition may not be altered in SPNs of HD mice, there is a progressive enhancement of extrasynaptic NMDA receptors. Because of the historic association of extrasynaptic NMDA receptors with cell-death related cascades, this study was one of the first to rationalize how excitotoxicity may be possible in the absence of elevated synaptic glutamatertic responses. [DOI] [PubMed] [Google Scholar]
- 40.Gladding CM, Sepers MD, Xu J, Zhang LY, Milnerwood AJ, Lombroso PJ, Raymond LA. Calpain and STriatal-Enriched protein tyrosine phosphatase (STEP) activation contribute to extrasynaptic NMDA receptor localization in a Huntington’s disease mouse model. Hum Mol Genet. 2012;21:3739–3752. doi: 10.1093/hmg/dds154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Botelho EP, Wang E, Chen JY, Holley S, Andre V, Cepeda C, Levine MS. Differential Synaptic and Extrasynaptic Glutamate-Receptor Alterations in Striatal Medium-Sized Spiny Neurons of Aged YAC128 Huntington’s Disease Mice. PLoS Curr. 2014;6 doi: 10.1371/currents.hd.34957c4f8bd7cb1f5ec47381dfc811c3. The study used used SPN-specific transgenic mice crossed with YAC128 HD mice to show that in the late stage HD striatum inflated extrasynaptic NMDA receptor responses spread to both direct and indirect pathway SPNs. Combined with the observation that extrasynaptic NMDA receptor responses are initially seen only in iSPNs in BACHD mice (37), this study offers a potential example of how impairments originating in iSPNs may ultimately spread in the striatum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergles DE, Diamond JS, Jahr CE. Clearance of glutamate inside the synapse and beyond. Curr Opin Neurobiol. 1999;9:293–298. doi: 10.1016/s0959-4388(99)80043-9. [DOI] [PubMed] [Google Scholar]
- 43.Miller BR, Dorner JL, Shou M, Sari Y, Barton SJ, Sengelaub DR, Kennedy RT, Rebec GV. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •44.Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, Anderson MA, Mody I, Olsen ML, Sofroniew MV, et al. Astrocyte Kir4. 1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat Neurosci. 2014;17:694–703. doi: 10.1038/nn.3691. This study showed that in R6/2 and zQ175 HD mice there is a deficit in astrocytic Kir4.1 K+ channels. The authors demonstrate that this leads to elevated extracellular K+ concentration, which may in turn lead to increased neuronal excitability in the striatum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••45.Gladding CM, Fan J, Zhang LY, Wang L, Xu J, Li EH, Lombroso PJ, Raymond LA. Alterations in STriatal-Enriched protein tyrosine Phosphatase expression, activation, and downstream signaling in early and late stages of the YAC128 Huntington’s disease mouse model. J Neurochem. 2014;130:145–159. doi: 10.1111/jnc.12700. In this study the authors elegantly dissected how extrasynaptic NMDA receptors, calpain and STEP converge or act separately to regulate the phosphorylation state of the pro-apoptotic signal p38 MAPK. The authors showed that extrasynaptic NMDA receptors only directly elevate p38 MAPK activity in young YAC128 mice, but not 1 year old mice. This leads to excitotoxic resistance in the aged HD striatum, arguing against NMDA-induced excitotoxicity as a cause of late striatal pathology, consistent with the aged HD striatum being defined by a hypoglutamatergic state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marco S, Giralt A, Petrovic MM, Pouladi MA, Martinez-Turrillas R, Martinez-Hernandez J, Kaltenbach LS, Torres-Peraza J, Graham RK, Watanabe M, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med. 2013;19:1030–1038. doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 48.Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 49.Freeman AY, Soghomonian JJ, Pierce RC. Tyrosine kinase B and C receptors in the neostriatum and nucleus accumbens are co-localized in enkephalin-positive and enkephalin-negative neuronal profiles and their expression is influenced by cocaine. Neuroscience. 2003;117:147–156. doi: 10.1016/s0306-4522(02)00802-3. [DOI] [PubMed] [Google Scholar]
- 50.Kress GJ, Yamawaki N, Wokosin DL, Wickersham IR, Shepherd GM, Surmeier DJ. Convergent cortical innervation of striatal projection neurons. Nat Neurosci. 2013;16:665–667. doi: 10.1038/nn.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brito V, Puigdellivol M, Giralt A, del Toro D, Alberch J, Gines S. Imbalance of p75(NTR)/TrkB protein expression in Huntington’s disease: implication for neuroprotective therapies. Cell Death Dis. 2013;4:e595. doi: 10.1038/cddis.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deyts C, Galan-Rodriguez B, Martin E, Bouveyron N, Roze E, Charvin D, Caboche J, Betuing S. Dopamine D2 receptor stimulation potentiates PolyQ-Huntingtin-induced mouse striatal neuron dysfunctions via Rho/ROCK-II activation. PLoS One. 2009;4:e8287. doi: 10.1371/journal.pone.0008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji J, Sundquist K, Sundquist J. Cancer incidence in patients with polyglutamine diseases: a population-based study in Sweden. Lancet Oncol. 2012;13:642–648. doi: 10.1016/S1470-2045(12)70132-8. [DOI] [PubMed] [Google Scholar]
- 54.Kozorovitskiy Y, Saunders A, Johnson CA, Lowell BB, Sabatini BL. Recurrent network activity drives striatal synaptogenesis. Nature. 2012;485:646–650. doi: 10.1038/nature11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu X, Li C, Wei W, Lo V, Gong S, Li SH, Iwasato T, Itohara S, Li XJ, Mody I, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 56.Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29:12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park H, Popescu A, Poo MM. Essential Role of Presynaptic NMDA Receptors in Activity-Dependent BDNF Secretion and Corticostriatal LTP. Neuron. 2014;84:1009–1022. doi: 10.1016/j.neuron.2014.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynch G, Kramar EA, Rex CS, Jia Y, Chappas D, Gall CM, Simmons DA. Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington’s disease. J Neurosci. 2007;27:4424–4434. doi: 10.1523/JNEUROSCI.5113-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cepeda C, Starling AJ, Wu N, Nguyen OK, Uzgil B, Soda T, Andre VM, Ariano MA, Levine MS. Increased GABAergic function in mouse models of Huntington’s disease: reversal by BDNF. J Neurosci Res. 2004;78:855–867. doi: 10.1002/jnr.20344. [DOI] [PubMed] [Google Scholar]
- 60.Cummings DM, Cepeda C, Levine MS. Alterations in striatal synaptic transmission are consistent across genetic mouse models of Huntington’s disease. ASN Neuro. 2010;2:e00036. doi: 10.1042/AN20100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andre VM, Fisher YE, Levine MS. Altered Balance of Activity in the Striatal Direct and Indirect Pathways in Mouse Models of Huntington’s Disease. Front Syst Neurosci. 2011;5:46. doi: 10.3389/fnsys.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •62.Dvorzhak A, Semtner M, Faber DS, Grantyn R. Tonic mGluR5/CB1-dependent suppression of inhibition as a pathophysiological hallmark in the striatum of mice carrying a mutant form of huntingtin. J Physiol. 2013;591:1145–1166. doi: 10.1113/jphysiol.2012.241018. This study shows that mGluR5 signaling is elevated in aged HD SPNs. This leads to an excessive eCB-mediated reduction in presynaptic GABA release probability in the striatum. When interpreted in light of (36), these data suggest that specific GABAergic sources to SPNs may be differentially impaired in HD, and imply that there may be a shift towards diminished feedback and enhanced feedforward inhibition of SPNs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adermark L, Talani G, Lovinger DM. Endocannabinoid-dependent plasticity at GABAergic and glutamatergic synapses in the striatum is regulated by synaptic activity. Eur J Neurosci. 2009;29:32–41. doi: 10.1111/j.1460-9568.2008.06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tepper JM, Koos T, Wilson CJ. GABAergic microcircuits in the neostriatum. Trends Neurosci. 2004;27:662–669. doi: 10.1016/j.tins.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Ariano MA, Cepeda C, Calvert CR, Flores-Hernandez J, Hernandez-Echeagaray E, Klapstein GJ, Chandler SH, Aronin N, DiFiglia M, Levine MS. Striatal potassium channel dysfunction in Huntington’s disease transgenic mice. J Neurophysiol. 2005;93:2565–2574. doi: 10.1152/jn.00791.2004. [DOI] [PubMed] [Google Scholar]
- 66.Stein V, Nicoll RA. GABA generates excitement. Neuron. 2003;37:375–378. doi: 10.1016/s0896-6273(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 67.Gulledge AT, Stuart GJ. Excitatory actions of GABA in the cortex. Neuron. 2003;37:299–309. doi: 10.1016/s0896-6273(02)01146-7. [DOI] [PubMed] [Google Scholar]
- 68.Prigge M, Schneider F, Tsunoda SP, Shilyansky C, Wietek J, Deisseroth K, Hegemann P. Color-tuned channelrhodopsins for multiwavelength optogenetics. J Biol Chem. 2012;287:31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ade KK, Janssen MJ, Ortinski PI, Vicini S. Differential tonic GABA conductances in striatal medium spiny neurons. J Neurosci. 2008;28:1185–1197. doi: 10.1523/JNEUROSCI.3908-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santhakumar V, Jones RT, Mody I. Developmental regulation and neuroprotective effects of striatal tonic GABAA currents. Neuroscience. 2010;167:644–655. doi: 10.1016/j.neuroscience.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo R, Janssen MJ, Partridge JG, Vicini S. Direct and GABA-mediated indirect effects of nicotinic ACh receptor agonists on striatal neurones. J Physiol. 2013;591:203–217. doi: 10.1113/jphysiol.2012.241786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kreitzer AC, Malenka RC. Striatal plasticity and basal ganglia circuit function. Neuron. 2008;60:543–554. doi: 10.1016/j.neuron.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Surmeier DJ, Plotkin J, Shen W. Dopamine and synaptic plasticity in dorsal striatal circuits controlling action selection. Curr Opin Neurobiol. 2009;19:621–628. doi: 10.1016/j.conb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lovinger DM. Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology. 2010;58:951–961. doi: 10.1016/j.neuropharm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glass M, Dragunow M, Faull RL. The pattern of neurodegeneration in Huntington’s disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington’s disease. Neuroscience. 2000;97:505–519. doi: 10.1016/s0306-4522(00)00008-7. [DOI] [PubMed] [Google Scholar]
- 76.Van Laere K, Casteels C, Dhollander I, Goffin K, Grachev I, Bormans G, Vandenberghe W. Widespread decrease of type 1 cannabinoid receptor availability in Huntington disease in vivo. J Nucl Med. 2010;51:1413–1417. doi: 10.2967/jnumed.110.077156. [DOI] [PubMed] [Google Scholar]
- ••77.Naydenov AV, Sepers MD, Swinney K, Raymond LA, Palmiter RD, Stella N. Genetic rescue of CB1 receptors on medium spiny neurons prevents loss of excitatory striatal synapses but not motor impairment in HD mice. Neurobiol Dis. 2014;71:140–150. doi: 10.1016/j.nbd.2014.08.009. This study demonstrated that rescuing pathologically low CB1 receptor expression specifically in SPNs can rescue dendritic spine deficits in the R6/2 striatum. This implies that impaired eCB signaling in the basal ganglia-thalamus-cortex loop may be primarily at GABAergic synapses, and glutamatergic corticostriatal synapse loss may be a response to this impairment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fieblinger T, Graves SM, Sebel LE, Alcacer C, Plotkin JL, Gertler TS, Chan CS, Heiman M, Greengard P, Cenci MA, et al. Cell type-specific plasticity of striatal projection neurons in parkinsonism and L-DOPA-induced dyskinesia. Nat Commun. 2014;5:5316. doi: 10.1038/ncomms6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Day M, Wokosin D, Plotkin JL, Tian X, Surmeier DJ. Differential excitability and modulation of striatal medium spiny neuron dendrites. J Neurosci. 2008;28:11603–11614. doi: 10.1523/JNEUROSCI.1840-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plotkin JL, Day M, Surmeier DJ. Synaptically driven state transitions in distal dendrites of striatal spiny neurons. Nat Neurosci. 2011;14:881–888. doi: 10.1038/nn.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klapstein GJ, Fisher RS, Zanjani H, Cepeda C, Jokel ES, Chesselet MF, Levine MS. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington’s disease transgenic mice. J Neurophysiol. 2001;86:2667–2677. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- 82.Heng MY, Detloff PJ, Albin RL. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Cepeda C, Cummings DM, Andre VM, Holley SM, Levine MS. Genetic mouse models of Huntington’s disease: focus on electrophysiological mechanisms. ASN Neuro. 2010;2:e00033. doi: 10.1042/AN20090058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lerner RP, del Trejo Martinez LC, Zhu C, Chesselet MF, Hickey MA. Striatal atrophy and dendritic alterations in a knock-in mouse model of Huntington’s disease. Brain Res Bull. 2012;87:571–578. doi: 10.1016/j.brainresbull.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]