Abstract
Quasiracemic crystallography has been used to explore the significance of homochiral and heterochiral associations in a set of host-defense peptide derivatives. The previously reported racemic crystal structure of a magainin 2 derivative displayed a homochiral dimer association featuring a “phenylalanine zipper” notable for the dual roles of phenylalanines in mediating dimerization and formation of an exposed hydrophobic swath. This motif is seen as well in two new quasiracemate crystals that contain the D form of the magainin 2 derivative along with an L-peptide in which one Ala has been replaced by a β-amino acid residue. This structural trend supports the hypothesis that the Phe zipper motif has functional significance.
Host-defense peptides (HDPs) constitute a ubiquitous component of the eukaryotic innate immune response to bacterial invasion.1 Collectively, these peptides exert multiple antimicrobial mechanisms of action, some that cause damage to bacterial cells and others that modulate immunological signaling in the host. A large HDP subset comprises relatively short peptides that display little or no tendency to fold in aqueous solution, but that adopt a-helical structure upon encountering a bacterial surface.2 These HDPs typically contain many hydrophobic residues and many basic residues, and they seem to act by disrupting bacterial membranes, among other mechanisms.1–3 It has been difficult to develop detailed hypotheses regarding the biological function(s) of these HDPs, in part because their sequences are diverse, and only limited high-resolution structural characterization is available. In particular, crystal structures of amphiphilic peptides that are short and conformationally mobile are very rare.4
Here we report two quasiracemate crystal structures that feature polypeptides in the magainin 2 family. These structures, in conjunction with a related racemate structure previously reported,5 provide a basis for distinguishing a mode of peptide assembly that may be relevant to function from interactions that reflect crystal packing effects. These findings suggest that racemate/quasiracemate crystallography may represent a versatile strategy for gaining atomic-level insight on diverse polypeptide quaternary structures.
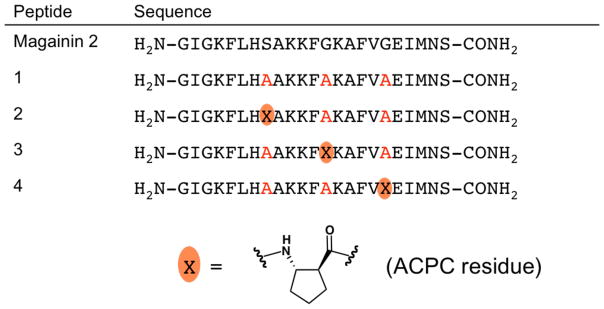
We recently determined the atomic-resolution structure of a potent derivative of the amphibian HDP magainin 2 by crystallizing the racemic form.5 Racemates are widely believed to be more susceptible to crystallization than are single enantiomers,6 and the application of racemic crystallography to polypeptides has grown in popularity since the first example was reported in 1993.7 The magainin 2 derivative we studied, 1 (Figure 1), contains three modifications relative to the natural HDP: Ser8→Ala, Gly13→Ala and Gly18→Ala.8 This variant is more active than magainin 2 itself against diverse bacteria.
Figure 1.
Peptides discussed in this paper. The structure of the β-amino acid residue designated ACPC is shown. Red A indicates a position at which the natural residue has been changed to alanine.
Magainin 2 is helical in the presence of vesicles,8 and the three residues altered in 1 are expected to enhance α-helical propensity.9 The crystal structure of 1 showed an α-helix that incorporated all residues except the C-terminal Ser, an observation consistent with previously described evidence of helicity in the presence of micelles.10 Of particular interest was an intimate association in the racemate crystal between pairs of peptides with the same absolute configuration. These homochiral dimers featured an antiparallel coiled-coil-like interaction (Figure 2A; helices of the same color have the same configuration). Phe residues, unusual within canonical coiled-coil interfaces,11 played a prominent role in the homochiral association observed for 1. The bulk of the aromatic side chain cannot be buried within a coiled-coil interface; thus, six phenyl rings, from Phe5, Phe12 and Phe16 of each peptide, were arrayed along one face of each homochiral dimer in the crystal of 1. This type of aromatic ring arrangement has previously been designated a “phenylalanine zipper.”12 Dimers of this type had been detected via NMR under micellar conditions for peptides derived from magainin 2,13 although not for magainin 2 itself or for variant 1. These NMR data led to antibacterial evaluation of covalently dimerized magainin 2 derivatives.14 Our structure provided the first crystallographic support for the Phe zipper proposal in the magainin context. We proposed that the extended hydrophobic surface projected by the Phe zipper motif mediates association of the coiled-coil dimer with bacterial membranes.
Figure 2.
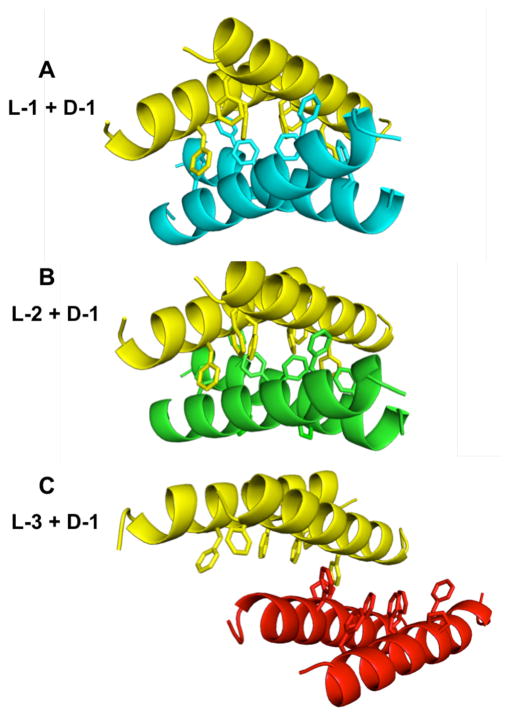
Pairs of homochiral dimers form distinct heterochiral crystal packing arrangements in the structures of racemic 1 (PDB 4MGP) and two quasiracemates. In each case, the yellow molecules are D-1. (A) Racemic 1; blue = L-1. (B) Quasiracemate [L-2 + D-1]; green = L-2. (C) Quasiracemate [L-3 + D-1]; red = L-3.
In addition to the homochiral pairing in crystalline 1, a heterochiral tetramer was observed in which the Phe zipper of an L-peptide dimer associates with the Phe zipper of a D-peptide dimer to create an aromatic-rich core (Figure 2A).5 We hypothesized that the heterochiral tetramer resulted from crystal packing effects, but this proposal raises the possibility that the homochiral dimer observed in the crystal of racemic 1 is also a consequence of crystallization rather than a native mode of peptide assembly with potential relevance to antibacterial function. The new crystal structures described here support the hypothesis that the homochiral dimer observed in the crystal structure of racemic 1 is functionally significant whereas the heterochiral associations serve merely as crystallization contacts.
We undertook crystallization of quasiracemic mixtures that contain D-1 and a variant of L-1 to assess the consistency of homochiral and heterochiral associations. Quasiracemates, in which each component is slightly different from the enantiomer of the other, often display the crystallization advantages associated with true racemates.15 In the variants of L-1 we examined, one Ala residue was replaced with the β-amino acid residue derived from (S,S)-trans-2-aminocyclopentanecarboxylic acid (ACPC; Figure 1). Previous work with oligomers that contain mixtures of α- and β-amino acid residues (“α/β-peptides”) has shown that the ACPC residue supports formation of a helical conformation strongly resembling the α-helix.16 Peptides L-2, L-3 and L-4 contain a single ACPC residue in place of Ala8, Ala13 or Ala18, respectively. Two quasiracemates, [L-2 + D-1] and [L-3 + D-1], were crystallized under the conditions that were successful for racemic 1: 0.1 M sodium citrate tribasic, pH 5.6, 35% v/v tert-butanol. Structure determination by molecular replacement and refinement using a new library of D-amino acid residues with canonical peptide bond restraints are described in the SI.
The [L-2 + D-1] quasiracemate, refined at 2.2 Å resolution in space group P21212, contains a pseudo-four-fold rotoinversion that forms a tightly packed tetrameric quaternary arrangement (Figure 2B). (Were this a true rotoinversion the space group would be .) Each asymmetric unit contains two of these L-2 dimer + D-1 dimer “tetramers”, which differ slightly in the curvature of their D-1 peptides. As observed for [L-1 + D-1], the C-terminal Ser residues of L-2 and D-1 could not be reliably modeled.
The asymmetric unit of the [L-3 + D-1] quasiracemate contains four polypeptides, one L-3 dimer and one D-1 dimer that are related by a pseudo-inversion center (Figure 2C). This structure, refined at 1.50 Å, can be viewed as pseudo-P1; however, the chemical distinction between the quasienantiomers required refinement in space group P1. The relationship between the dimers of the quasienantiomers in the [L-3 + D-1] structure is pseudo-centrosymmetric. (Packing representations and electron density maps for [L-2 + D-1] and [L-3 + D-1] are found in Figures S3 and S4.)
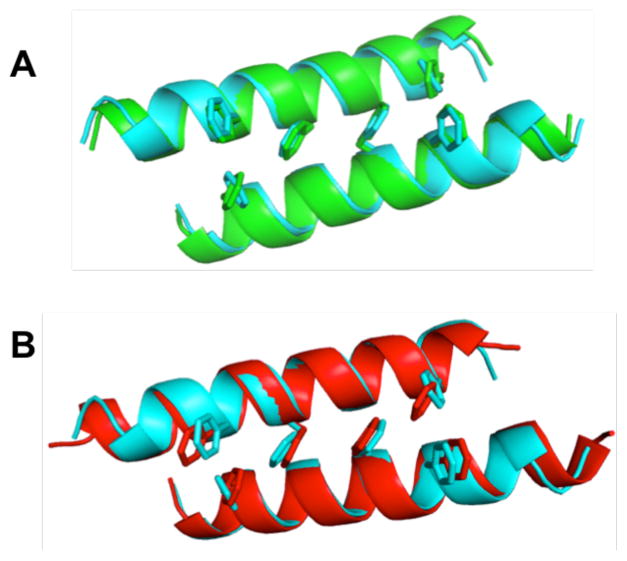
In both of the quasiracemate structures, each component forms the type of homochiral coiled-coil dimer that was observed for the constituents of the racemic crystal of 1. The Phe zipper motif is evident in each case (Figure 3). Backbone atom overlay of a D-1 dimer from the [L-2 + D-1] quasiracemate or from the [L-3 + D-1] quasiracemate with the D-1 dimer from the racemate gives RMSD 1.1 Å or 1.4 Å, respectively. Comparable similarities are observed for the peptides containing L residues. (Discussion of RMSD calculations may be found in the SI.)
Figure 3.
Overlay comparisons on peptide chains containing L-amino-acid residues. Only the backbones (ribbon) and Phe side chains are shown. (A) L-1 (blue) from racemic 1 (PDB 4MGP) vs. L-2 (green) from the [L-2 + D-1] quasiracemate. (B) L-1 (blue) from racemic 1 (PDB 4MGP) vs. L-3 (red) from the [L-3 + D-1] quasiracemate.
In contrast to the similarities among homochiral dimers in the three crystals, significant variations are observed among the heterochiral quaternary associations. In the [L-2 + D-1] quasiracemate, the L and D homodimers display a tetrameric assembly that is very similar to the assembly seen in the crystal of racemic 1 (Figure 2B). However, the analogous tetrameric association in the [L-3 + D-1] quasiracemate is unique in that there is not a close packing of the phenylalanine zippers from each dimer (Figure 2C). The fact that the homochiral coiled-coil assembly, featuring the Phe zipper motif, is universal among the three data sets, involving five independent dimer structures, despite variations in the heterochiral associations, strongly supports the conclusion that the homochiral dimerization mode is relevant to a native interaction of magainin-family peptides, while the heterochiral associations reflect crystal packing forces that differ among the systems.
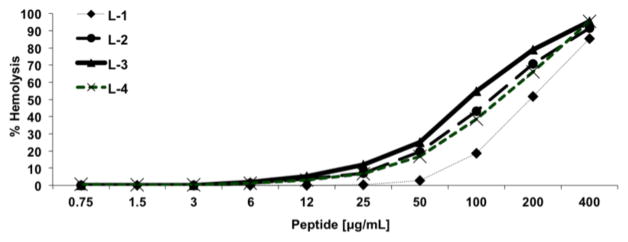
The two new ACPC-containing magainin 2 variants, L-2 and L-3, show similar or slightly superior levels of growth-inhibitory activity relative to L-1 against a small panel of bacteria (Table 1). However, both variants are somewhat more prone to cause lysis of human red blood cells (“hemolysis”) than is L-1 (Figure 4). (The quasiracematic mixtures containing D-1 and either L-2 or L-3 show very similar biological activities to those of pure L-2 or pure L-3, respectively.) These trends presumably arise because the ACPC residue in L-2 or L-3 is more hydrophobic than the alanine residue that has been replaced from L-1. Among α-peptides, it is known that increasing hydrophobicity can increase both antibacterial and hemolytic activities.2a For example, L-1 is more hydrophobic than magainin 2 itself because one Ser and two Gly residues have been replaced by Ala, and L-1 is more potent against bacteria and more hemolytic than is magainin 2.
Table 1.
Minimum inhibitory concentration (MIC) for L-peptides toward four bacterial species.
| Peptide | MIC μg/mL | |||
|---|---|---|---|---|
| E. coli | B. subtilis | S. aureus | E. faecium | |
| L-1 | 12 | 6 | 50 | 50 |
| L-2 | 12 | 6 | 12 | 12 |
| L-3 | 12 | 6 | 12 | 25 |
| L-4 | 12 | 6 | 12 | 12 |
Figure 4.
Lysis of human red blood cells by L-peptides as a function of peptide concentration.
Conclusions regarding the relationship between a polypeptide’s function in solution and an association mode found in a single crystal structure are tenuous; trends manifested in multiple crystal structures obtained for related molecules engender stronger conclusions. The study of a series that includes a racemate and related quasiracemates is particularly interesting in this regard because one component can be held constant (D-1 in our system) while the other is varied. Collectively, three crystal structures based on 1 and related peptides, including the two new quasiracemate structures reported here, support the hypothesis, initially proposed based on NMR data acquired in solution,13 that a coiled-coil-like dimerization mode plays a functional role in the antibacterial activities of peptides in the magainin 2 family. Our crystallographic data offer insights beyond those available via NMR, because the crystal structures reveal a mode of helix association that involves pairing of nonpolar surfaces but nevertheless leaves considerable nonpolar surface area exposed for interaction with a bacterial membrane. These observations are important in light of a proposal that helix dimerization would prevent hydrophobic surface exposure.17
Our findings raise the possibility that quasiracemate crystallography will prove to be a general strategy for assessing polypeptide quaternary structure preferences. Specifically, exploration of multiple quasiracemates based on a given racemate structure appears to be a logical and efficient approach to obtaining sets of atomic-resolution data that collectively provide insights on noncovalent association preferences. This approach might be useful for other types of molecules as well.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 GM061238 (to S.H.G.) and GM100346 (to Michael G. Thomas and K.T.F.). Partial support was provided to K.T.F. by IMéRA, to Z.H. by a Fulbright Fellowship the Nanoscale Science and Engineering Center at UW-Madison (NSF DMR-0832760), and to D.E.M. by a Molecular Biophysics Training Grant (T32 GM08293). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Footnotes
The authors declare no competing financial interest.
Experimental details for peptide synthesis and X-ray crystallography. This material is available free of charge via the Internet at http://pubs.acs.org. Model coordinates and structure factors have been deposited in the Protein Data Bank as entries 5CGN and 5CGO.
References
- 1.(a) Diamond G, Beckloff N, Weinberg A, Kisich KO. Curr Pharm Des. 2009;15:2377. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zasloff M. Nature. 2002;415:389. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]; (c) Rathinakumar R, Walkenhorst WF, Wimley WC. J Am Chem Soc. 2009;131:7609. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Boman HG. J Int Med. 2003;254:197. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]; (e) Hancock RE. Lancet Infect Dis. 2001;1:156. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 2.(a) Tossi A, Sandri L, Giangaspero A. Biopolymers. 2000;55:4. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]; (b) Shai Y. Biochim Biophys Acta. 1999;1462:55. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]; (c) Wang Z, Wang GS. Nucleic Acids Res. 2004;32:D590. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wimley WC. ACS Chem Biol. 2010;5:905. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Song C, Weichbrodt C, Salnikov ES, Dynowski M, Forsberg BO, Bechinger B, Steinem C, de Groot BL, Zachariae U, Zeth K. Proc Natl Acad Sci USA. 2013;110:4586. doi: 10.1073/pnas.1214739110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Terwilliger TC, Eisenberg D. J Biol Chem. 1982;257:6016. [PubMed] [Google Scholar]
- 5.Hayouka Z, Mortenson DE, Kreitler DF, Weisblum B, Forest KT, Gellman SH. J Am Chem Soc. 2013;135:15738. doi: 10.1021/ja409082w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Brock CP, Schweizer WB, Dunitz JD. J Am Chem Soc. 1991;113:9811. [Google Scholar]; (b) Zawadzke LE, Berg JM. J Am Chem Soc. 1992;114:4002. [Google Scholar]; (c) Wukovitz SW, Yeates TO. Nat Struct Biol. 1995;2:1062. doi: 10.1038/nsb1295-1062. [DOI] [PubMed] [Google Scholar]; (d) Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SBH. J Am Chem Soc. 2008;130:9695. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Zawadzke LE, Berg JM. Proteins. 1993;16:301. doi: 10.1002/prot.340160308. [DOI] [PubMed] [Google Scholar]; (b) Toniolo C, Peggion C, Crisma M, Formaggio F, Shui X, Eggleston DS. Nat Struct Biol. 1994;1:908. doi: 10.1038/nsb1294-908. [DOI] [PubMed] [Google Scholar]; (c) Yeates TO, Kent SBH. Annu Rev Biophys. 2012;41:41. doi: 10.1146/annurev-biophys-050511-102333. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki K, Harada M, Funakoshi S, Fujii N, Miyajima K. Biochim Biophys Acta. 1991;1063:162. doi: 10.1016/0005-2736(91)90366-g. [DOI] [PubMed] [Google Scholar]
- 9.Chen HC, Brown JH, Morell JL, Huang CM. FEBS Lett. 1988;236:462. doi: 10.1016/0014-5793(88)80077-2. [DOI] [PubMed] [Google Scholar]
- 10.Hicks RP, Mones E, Kim H, Koser BW, Nichols DA, Bhattacharjee AK. Biopolymers. 2003;68:459. doi: 10.1002/bip.10325. [DOI] [PubMed] [Google Scholar]
- 11.Woolfson DN. Adv Protein Chem. 2005;70:79. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 12.(a) Yoder NC, Kumar K. J Am Chem Soc. 2006;128:188. doi: 10.1021/ja055494k. [DOI] [PubMed] [Google Scholar]; (b) Javadpour MM, Barkley MD. Biochemistry. 1997;36:9540. doi: 10.1021/bi961644f. [DOI] [PubMed] [Google Scholar]; (c) Liu J, Zheng Q, Deng Y, Kallenbach NR, Lu M. J Mol Biol. 2006;361:168. doi: 10.1016/j.jmb.2006.05.063. [DOI] [PubMed] [Google Scholar]; (d) Dhe-Paganon S, Werner ED, Nishi M, Hansen L, Chi YI, Shoelson SE. Nat Struct Mol Biol. 2004;11:968. doi: 10.1038/nsmb829. [DOI] [PubMed] [Google Scholar]
- 13.(a) Wakamatsu K, Takeda A, Tachi T, Matsuzaki K. Biopolymers. 2002;64:314. doi: 10.1002/bip.10198. [DOI] [PubMed] [Google Scholar]; (b) Porcelli F, Buck-Koehntop BA, Thennarasu S, Ramamoorthy A, Veglia G. Biochemistry. 2006;45:5798. doi: 10.1021/bi0601813. [DOI] [PubMed] [Google Scholar]; (c) Matsuzaki K, Murase O, Tokuda H, Funakoshi S, Fujii N, Miyajima K. Biochemistry. 1994;33:3342. doi: 10.1021/bi00177a027. [DOI] [PubMed] [Google Scholar]; (d) Bechinger B, Zasloff M, Opella SJ. Protein Sci. 1993;2:2077. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hicks RP, Mones E, Kim H, Koser BW, Nichols DA, Bhattacharjee AK. Biopolymers. 2003;68:459. doi: 10.1002/bip.10325. [DOI] [PubMed] [Google Scholar]
- 14.(a) Hara T, Kodama H, Kondo M, Wakamatsu K, Takeda A, Tachi T, Matsuzaki K. Biopolymers. 2001;58:437. doi: 10.1002/1097-0282(20010405)58:4<437::AID-BIP1019>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (b) Mukai Y, Matsushita Y, Niidome T, Hatekeyama T, Aoyagi H. J Peptide Sci. 2002;8:570. doi: 10.1002/psc.416. [DOI] [PubMed] [Google Scholar]
- 15.(a) Pasteur L. Ann Chim Phys. 1853;28:437. [Google Scholar]; (b) Wheeler KA, Grove RC, Davis RE, Kassel WS. Angew Chem Int Ed. 2008;47:78. doi: 10.1002/anie.200704007. [DOI] [PubMed] [Google Scholar]; (c) Karle IL, Karle J. J Am Chem Soc. 1966;88:24. [Google Scholar]; (d) Grove RC, Malehorn SH, Breen ME, Wheeler KA. Chem Commun. 2010;46:7322. doi: 10.1039/c0cc02775h. [DOI] [PubMed] [Google Scholar]; (e) Kelley SP, Fabian L, Brock CP. Acta Crystallogr. 2011;B67:79. doi: 10.1107/S0108768110048135. [DOI] [PubMed] [Google Scholar]; (f) Mandal K, Pentelute BL, Bang D, Gates ZP, Torbeev VY, Kent SBH. Angew Chem Int Ed. 2012;51:1481. doi: 10.1002/anie.201107846. [DOI] [PubMed] [Google Scholar]; (g) Mortenson DE, Satyshur KA, Guzei IA, Forest KT, Gellman SH. J Am Chem Soc. 2012;134:2473. doi: 10.1021/ja210045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Horne WS, Gellman SH. Acc Chem Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Johnson LM, Gellman SH. Methods Enzymol. 2013;523:407. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory SM, Pkorny A, Almeida PFF. Biophys J. 2009;96:116. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.