Abstract
Bovine respiratory disease (BRD) is the leading cause of morbidity and mortality in feedlot cattle, caused by multiple pathogens that become more virulent in response to stress. As clinical signs often go undetected and various preventive strategies failed, identification of genes affecting BRD is essential for selection for resistance. Selective DNA pooling (SDP) was applied in a genome wide association study (GWAS) to map BRD QTLs in Israeli Holstein male calves. Kosher scoring of lung adhesions was used to allocate 122 and 62 animals to High (Glatt Kosher) and Low (Non-Kosher) resistant groups, respectively. Genotyping was performed using the Illumina BovineHD BeadChip according to the Infinium protocol. Moving average of -logP was used to map QTLs and Log drop was used to define their boundaries (QTLRs). The combined procedure was efficient for high resolution mapping. Nineteen QTLRs distributed over 13 autosomes were found, some overlapping previous studies. The QTLRs contain polymorphic functional and expression candidate genes to affect kosher status, with putative immunological and wound healing activities. Kosher phenotyping was shown to be a reliable means to map QTLs affecting BRD morbidity.
Introduction
Bovine respiratory disease (BRD) complex is the leading world-wide cause of morbidity and mortality in feedlot cattle. It includes upper and lower respiratory tract infections, diphtheria and pneumonia [1,2]. Due to immature functionality of the respiratory system in young cattle [3], BRD occurs more frequently and severely at young age, regardless of immunological and management considerations [4,5]. BRD is the most costly feedlot disease due to prevention and treatment costs, morbidity, mortality, and production amortization that includes performance, carcass merit, meat tenderness and palatability [1,2,6,7].
BRD etiology is multifactorial, affected by a large number of stressors (e.g., weaning, transportation, commingling and others), and viral (infectious bovine rhinotracheitis; IBR, bovine virus diarrhea; BVD, bovine respiratory syncytial virus; BRSV) and bacterial pathogens (primarily, Pasteurella multocida, Haemophilus somnus and Mycoplasma hyponeumoniae). Many of these pathogens are normally present in the upper respiratory tract, but convert to pathogenic status as a consequence of stressful life history events [8]. Yet, individuals of the same age and environment, when exposed to BRD pathogens, vary greatly in whether they develop the disease [9], and the severity of clinical symptoms [10]. This variation suggests that genetic control is involved in susceptibility to BRD. Indeed, significant heritability for BRD resistance [11,12] and differences among breeds [11,13] have been documented.
During the years, various strategies have been implemented to prevent or minimize the prevalence of BRD. Among others, these include antibiotic treatment on a preventative or metaphylactic basis, non-antibiotic alternatives and vaccination [4]. Unfortunately, these strategies have collectively failed to reduce the prevalence of BRD (see citations in [14]). To date, methods for detecting morbid cattle involve subjective visual appraisal and depend on the stage and extent of the disease. However, clinical signs of BRD may often go undetected in feedlot calves [15], emphasizing the need of an objective and reliable early risk predictor [5]. Genomic-based approaches may thus serve as additional methods for control of BRD.
The importance of the genome in determining resistance and susceptibility to a wide variety of viral, bacterial and parasite-borne diseases is thoroughly documented [16,17]. Examples include mapping of quantitative trait loci (QTL) affecting trypanotolerance in the N’Dama cattle of West Africa [18], Marek’s Disease in layer chickens [19,20], and mastitis in Holstein dairy cattle [21]. With genome wide mapping and selection procedures based on high density SNP arrays [22], improving disease resistance through selection for resistance at the genome level has become realistic. Furthermore, high-resolution mapping of the QTLs responsible for genetic variation in resistance can serve as a platform to identify the causative genes themselves [23], opening further possibilities for disease control through greater understanding of the molecular mechanisms of resistance. For example, recent genome-wide association studies of Crohn’s Disease identified 71 loci associated with the disease [24] and new pathogenic mechanisms of the disease.
Reports on BRD QTL mapping are scarce. Using microsatellite markers, Neibergs et al. [25] identified QTLs affecting BRD on BTA 2 and 26 in a Brahman × Hereford sire half-sib family. Casas et al. [26] found association between BRD and SNPs in the ANKRA2 and CD180 genes on BTA 20 in a Brahman x Angus cross. Neibergs et al. [14] conducted genome-wide association study (GWAS) to map QTLs affecting BRD susceptibility in Holstein populations. Tizioto et al. [27] examined transcriptomes from bronchial lymph nodes of cattle challenged with three viral and three bacterial BRD pathogens, one at a time. Hundreds to thousands of genes changed expression in response to the pathogen challenge; 140 of which were located in previously mapped BRD QTLs.
To phenotypically distinguish between control and case individuals, Neibergs et al. [14] used common clinical signs of BRD, in combination with nasopharyngeal and pharyngeal recess swabs for the diagnosis of pathogens. An alternative well-accepted tool for retrospective diagnosis of BRD is lung lesions monitored at slaughter [28]. Among other categories, lung lesions include abscesses, parenchymal fibrosis, adhesions and emphysema [29]. Wittum et al. [15] found that damage resulting from BRD may leave persistent lesions in bovine lungs. Interestingly, lung lesions resulting from BRD are often found in animals never recorded for clinical BRD [2,6,15,30]. According to Gardner et al. [6], the high incidence of such cases reflected subclinical events, viral rather than bacterial infection, or disease occurrence at earlier stage, before calves were taken into feedlot. Still, pulmonary lesions at slaughter were indicative of BRD occurrence, which had a significant deleterious effect on production, independent of previous diagnosis of clinical illness [15]. In spite of the fact that it is the only practical way to detect subclinical BRD, only a few studies used lung lesions as an indicator of earlier life BRD episodes [4].
As mentioned above, Bryant et al. [29] classified lung adhesions in the cranioventral lobes as a specific example of lung lesions. Adhesions are formed as a normal part of the body's healing process and help to limit the spread of infection. They are fibrous bands of scar tissue that span the pleural space, between the parietal and visceral layers of the pleura and often between the lobes of the lungs, or between the lungs and the rib cage, and are caused by repeated episodes of inflammation of the lungs. The fibrin bands may eventually dissolve through fibrinolysis, and the traumatized site continues to heal, but there are cases in which fibrinolysis is inhibited [31]. Thus, uncontrolled fibrosis may render the repair process pathogenic, resulting in excess deposition of extracellular matrix (ECM) components, including collagen, and replacement of normal tissue by permanent scar tissue, which influences organ function [32]. In a recent report [33], calves free of pulmonary adhesions at slaughter were found biologically and economically more efficient than their affected peers. Interestingly, pleural adhesions found in ca. 40% of the calves accounted for reduced growth rate at early life stages [2].
Kosher slaughtering, a routine procedure in Israel and other countries [34], offers particular advantages for genetic analysis of BRD. Kosher slaughtering involves two steps: The actual slaughter, and then a close and detailed examination of the lungs of the slaughtered animals for adhesions. Cattle presenting lungs completely clear of adhesions, indicative of their having been BRD free, are classed as “Glatt” kosher. Cattle presenting severe pulmonary adhesions, indicative of one or more severe bouts of BRD, are classed as “non-kosher” or "treif". Animals presenting light adhesions are classed as standard-kosher [35].
In the current study, we mapped BRD QTLs in Israeli Holstein male calves by means of selective DNA pooling (SDP), using the kosher slaughtering three-level classification system to distinguish between High (Glatt) and Low (Treif) resistant phenotypes, Numerous QTLs were found and their surrounding regions (QTLRs) searched for candidate genes and for amino acid (AA) polymorphisms within these genes. The candidate genes involved immunological response and wound healing activities that include cell adhesion, extra cellular matrix (ECM) remodeling, epithelial-to-mesenchyme transition, and profibrotic responses. The QTLRs and their genes partially overlap previous QTLs mapping studies, supporting the use of kosher phenotype for BRD QTL mapping.
Results
Markers
Animals were allocated to High and Low resistant groups by a kosher inspection of lung adhesions. Frequencies of allele B in the pools were obtained by Illumina software. Frequency differences between High and Low groups (Di) and an empirical estimate of the standard error of Di, were used to obtain Comparison-wise-Type I error rate (P-values). The P-values over all autosomes are presented in S1 Fig. Table 1 shows critical marker P-values required to achieve significance at the given PFP thresholds. A total of 749 markers were in the range of PFP ≤ 0.2, distributed over all autosomes. Of course, the number of QTLs is much less than the numbers of significant markers, as many significant markers are associated with the same QTL. The estimated number of falsified and true null hypotheses, n1 and n2 [36,37] were 2,114 and 568,449 respectively. Thus, effective power at PFP ≤ 0.2 was 0.35.
Table 1. Critical P-values and number of significant SNPs, by PFP level.
| PFP | Critical P | Sig SNPs |
|---|---|---|
| < 0.01 | 1.63E-06 | 95 |
| > 0.01–0.05 | 2.06E-05 | 148 |
| > 0.05–0.10 | 6.91E-05 | 152 |
| > 0.10–0.20 | 2.64E-04 | 354 |
| > 0.20–0.40 | 1.02E-03 | 712 |
QTLRs detection
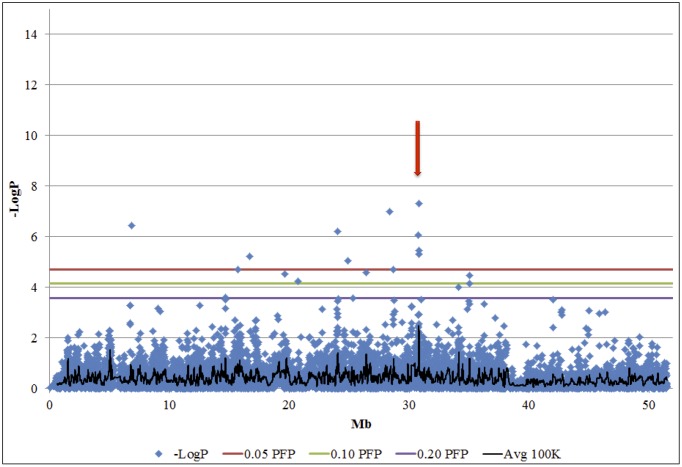
Visual inspection of chromosomal scatter charts (S1 Fig) revealed clusters of significant marker -LogP values (e.g., Fig 1). We considered such clusters to represent QTLRs. Detailed examination of the makers in the QTLR showed that they were a mixture of significant markers and non-significant markers, making it difficult to set boundaries for the QTLR. To circumvent this, we used a moving average of marker -LogP values with a window size of 23 markers (about 100 kb), and a Log drop of 1 to define QTLRs and their boundaries (details in Methods). This worked well and clear peaks with monotonic shoulders were obtained. A moving average of -LogP ≥ 2, corresponding to P = 0.01, was set as the threshold value for declaration of a window as a QTLR, irrespective of whether the window contained a significant marker or not. Fig 2 presents a detailed example of defining a QTLR based on the cluster on BTA 29 (Fig 1).
Fig 1. A cluster of significant -LogP values on BTA 29 at about 30 Mb (red arrow; Fig 2 and Table 2).
Blue diamonds, -LogP values of the markers; Avg 100K, moving average -LogP values of windows of 23 markers (≈ 100Kb; see text). Note that for this cluster the peak value of the moving average exceeds the -LogP = 2.0 threshold chosen to declare significance.
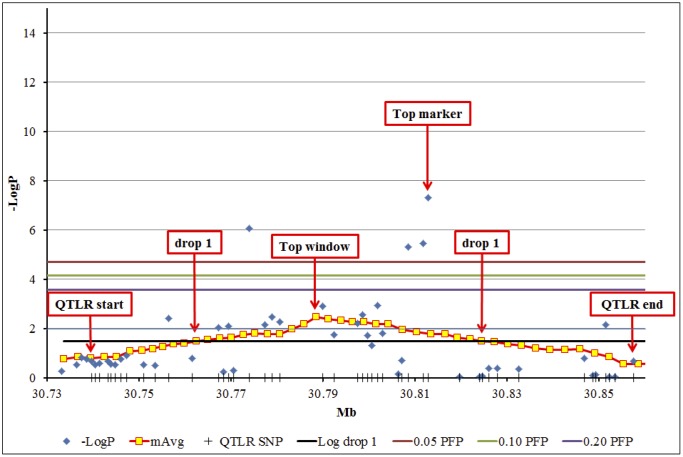
Fig 2. Expanded view of the QTLR at 30 Mb on BTA 29 (Fig 1 and Table 2).
Vertical bars on the X-axis, QTLR marker locations; Blue diamonds, -LogP values of the markers; Yellow squares: X-axis, mean location of the markers in the window; Y-axis, mean -LogP of the window. QTLR start and end, up- and down-stream boundary markers of the QTLR; Drop 1, up and down-stream Log drop 1 boundary windows; Top window, the window with highest average -LogP; Top marker, the most significant marker of the cluster. Three uppermost horizontal bars from top down: significance thresholds for individual markers at PFP = 0.05, 0.10 and 0.20, respectively. Two lowest horizontal bars, from top down: significance threshold for moving average of -LogP = 2.0; Log drop 1 threshold (from Peak window), respectively.
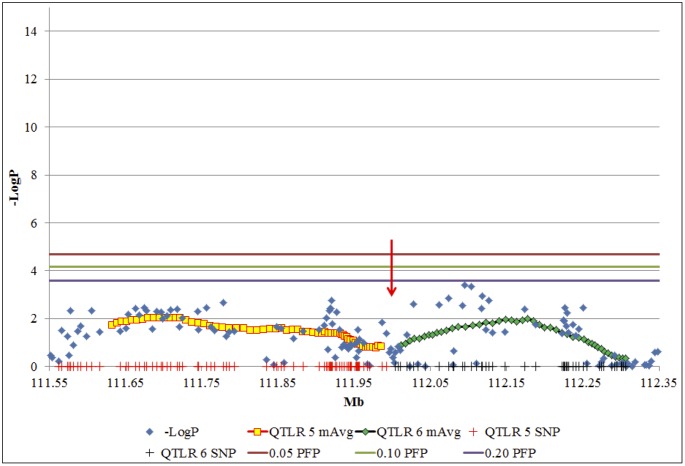
Based on our criteria for declaring a QTLR, a total of 19 QTLs were found, distributed over 13 chromosomes (Table 2). Of these, 16 represent clearly distinguished clusters of significant markers, while three represent windows that reached the -LogP < 2 criterion, but did not contain significant markers. They were, however, characterized by a stretch of high value -LogP values, without admixture of low -LogP value markers. Although declared as individual QTLs on our Log drop 1 criterion, the two closely linked QTLRs 5 and 6 on BTA 2 may possibly represent only a single QTLR (Fig 3). The two QTLR cover the region of 111.5–112.3 Mb on BTA 2. Close inspection reveals an intermediate cluster of low -LogP values, around 112.0 Mb (red arrow in Fig 3). It was this low cluster that dropped the -LogP of the intermediate windows below the Log drop 1 of both sides, and thus split this region into two QTLR.
Table 2. Autosomal QTLRs.
| QTLRs | Avg of top windowd | Top SNPe | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| QTLR | BTA | Starta | Endb | Length | Distancec | SNPs | bp | P | Name | bp | P |
| 1 | 1 | 32,762,621 | 33,000,982 | 238,362 | 49 | 32,922,068 | 7.2x10-2 | rs43229554 | 32,892,280 | 2.7x10-15 | |
| 2 | 1 | 92,104,370 | 92,235,747 | 131,378 | 59,103,388 | 46 | 92,172,725 | 2.8x10-1 | rs42857937 | 92,194,011 | 9.8x10-10 |
| 3 | 1 | 136,141,849 | 136,617,538 | 475,690 | 43,906,102 | 101 | 136,234,581 | 8.3x10-2 | rs109727900 | 136,269,109 | 2.6x10-5 |
| 4 | 2 | 103,578,039 | 103,858,856 | 280,818 | 71 | 103,604,186 | 7.4x10-2 | rs110744763 | 103,622,473 | 7.4x10-6 | |
| 5 | 2 | 111,562,742 | 111,992,526 | 429,785 | 7,703,886 | 73 | 111,702,216 | 1.3x10-2 | rs41718804 | 111,920,205 | 1.7x10-3 |
| 6 | 2 | 112,003,692 | 112,306,444 | 302,753 | 11,166 | 46 | 112,177,753 | 4.7x10-2 | rs109625954 | 112,094,944 | 3.9x10-4 |
| 7 | 2 | 114,545,480 | 114,830,722 | 285,243 | 2,239,036 | 50 | 114,692,948 | 1.1x10-1 | rs42619825 | 114,679,293 | 1.4x10-7 |
| 8 | 8 | 41,810,125 | 42,014,357 | 204,233 | 47 | 41,951,028 | 2.5x10-1 | rs109097634 | 41,958,832 | 1.6x10-6 | |
| 9 | 9 | 103,591,751 | 103,738,680 | 146,930 | 64 | 103,644,910 | 6.1x10-3 | rs109603023 | 103,658,874 | 7.2x10-8 | |
| 10 | 10 | 55,539,090 | 55,719,332 | 180,243 | 57 | 55,606,200 | 1.7x10-2 | rs43633836 | 55,591,993 | 2.3x10-3 | |
| 11 | 12 | 7,221,507 | 7,469,740 | 248,234 | 53 | 7,357,534 | 1.3x10-2 | rs134347273 | 7,306,510 | 8.9x10-5 | |
| 12 | 15 | 2,491,081 | 2,679,720 | 188,640 | 45 | 2,593,214 | 4.5x10-3 | rs132966783 | 2,560,990 | 8.1x10-5 | |
| 13 | 15 | 35,995,220 | 36,429,977 | 434,758 | 33,315,500 | 72 | 36,183,585 | 2.1x10-2 | rs110068780 | 36,198,691 | 3.7x10-5 |
| 14 | 16 | 38,146,214 | 38,575,149 | 428,936 | 97 | 38,370,212 | 2.0x10-2 | rs136111126 | 38,382,048 | 2.0x10-5 | |
| 15 | 18 | 58,445,044 | 58,667,459 | 222,416 | 44 | 58,614,523 | 1.4x10-1 | rs43073607 | 58,634,645 | 2.1x10-5 | |
| 16 | 22 | 55,739,008 | 55,889,024 | 150,017 | 47 | 55,804,668 | 3.9x10-2 | rs135721055 | 55,798,814 | 5.2x10-5 | |
| 17 | 24 | 26,944,578 | 27,174,025 | 229,448 | 56 | 27,068,708 | 1.2x10-1 | rs134945287 | 27,080,758 | 2.8x10-5 | |
| 18 | 26 | 43,067,146 | 43,222,533 | 155,388 | 46 | 43,172,648 | 8.4x10-2 | rs132928018 | 43,178,710 | 1.6x10-5 | |
| 19 | 29 | 30,739,666 | 30,860,467 | 120,802 | 47 | 30,788,390 | 1.0x10-1 | rs134937987 | 30,812,830 | 5.0x10-8 | |
aFirst marker of the first significant window.
bLast (23rd) marker of the last significant window.
cThe distance between the present and the previous QTLR on the same chromosome = the length between the end and the start of the up- and down- stream QTLRs.
dThe window with highest -LogP value.
eMost significant SNP in the QTLR.
Fig 3. Example of a region with two mapped QTLRs that are possibly only one putative QTLR.
QTLRs 5 and 6 on BTA 2. Red and black vertical bars on the X-axis, QTLRs 5 and 6 marker locations; Blue diamonds, -LogP values of the markers; Yellow squares (QTLR 5) and green diamonds (QTLR 6): X-axis, mean location of the markers in the window; Y-axis, mean -LogP of the window; red arrow, inter QTLR cluster. Three uppermost horizontal bars from top down: significance thresholds for individual markers at PFP = 0.05, 0.10 and 0.20, respectively.
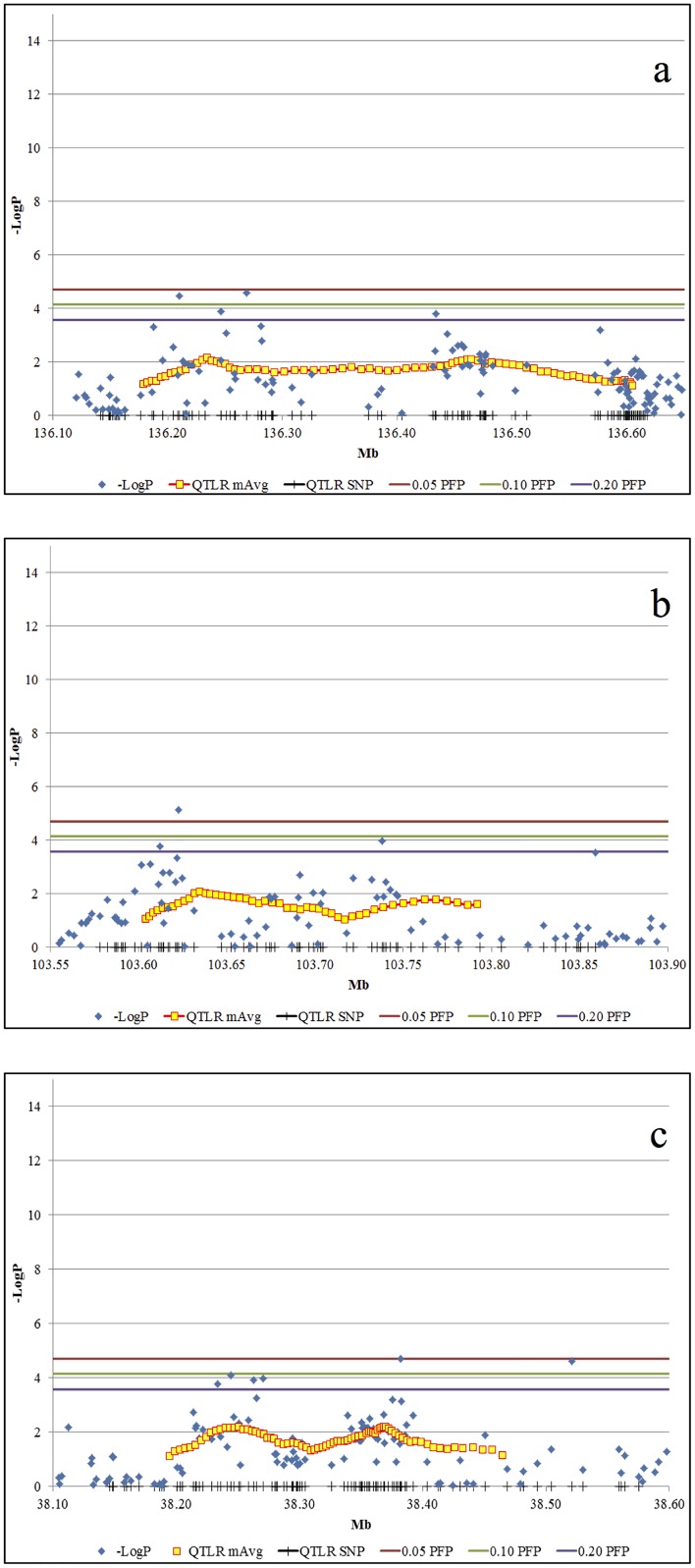
Conversely, QTLR 3, 4, and 14, each present two separated peaks but the intermediate regions did not meet our Log drop criteria for declaration as separate QTLRs. Thus, they may possibly represent a closely linked pair of QTLR each (Fig 4).
Fig 4. Examples of chromosomal regions with one QTLR but possibly including two putative QTLRs.
a. QTLR 3 on BTA 1. b. QTLR 4 on BTA 2. c. QTLR 14 on BTA 16. Vertical bars on the X-axis, QTLR markers locations; Blue diamonds, -LogP values of the markers; Yellow squares: X-axis, mean location of the markers in the window; Y-axis, mean -LogP of the window. Three uppermost horizontal bars from top down: significance thresholds for individual markers at PFP = 0.05, 0.10 and 0.20, respectively.
The QTLRs averaged 58 markers (range 44–101 markers), 255,478 bp (range 120,802–475,690 bp), covering a total of 4,854,074 bp, 0.19% of total of the 2.6 Gb bovine genome.
Genes in the QTLR
The 19 QTLRs (Table 2) were composed of a total of 1,111 array markers (S1 Table, SNP report sheet). Based on public databases, all except one of these array marker SNPs were non-coding. These regions and markers were used for detailed bioinformatics analyses. Among the 1,111 markers, 545 mapped within 35 genes (Table 3). KCNE4 in QTLR 5 on BTA 2 is also presented in Table 3, even though no SNP was mapped to it. Thus, Table 3 presents a total of 36 genes. Among these, annotation data were available in David Database for only 20 genes.
Table 3. Complete list of all genes within the QTLRs or within 0.5 Mb upstream or downstream of the QTLR boundaries.
| QTLR | Gene | Intergenic SNPs No.c | |||
|---|---|---|---|---|---|
| No. | BTA | Name/Flank | SNPs | ||
| No.a | P<0.01b | ||||
| 1 | 1 | Upstream | CADM2 | ||
| CADM2 | 49 | 10 | 0 | ||
| Downstream | CADM2 | ||||
| 2 | 1 | Upstream | NAALADL2 | ||
| NAALADL2 | 46 | 8 | 0 | ||
| Downstream | NAALADL2 | ||||
| 3 | 1 | Upstream | EPHB1, KY, CEP63, ANAPC13, AMOTL2 | ||
| RYK | 26 | 11 | 25 | ||
| SLCO2A1 | 21 | 9 | |||
| RAB6B | 29 | 2 | |||
| Downstream | RAB6B, SRPRB, TF, LOC525947, TOPBP1, CDV3, BFSP2, TMEM108 | ||||
| 4 | 2 | Upstream | VWC2L, BARD1, ABCA12 | ||
| ABCA12 | 46 | 14 | 20 | ||
| ATIC | 5 | 1 | |||
| Downstream | ATIC, FN1, MIR2285L | ||||
| 5 | 2 | Upstream | PAX3, SGPP2 | ||
| FARSB | 5 | 1 | 58 | ||
| LOC538702 | 1 | 0 | |||
| MOGAT1 | 2 | 1 | |||
| ACSL3 | 7 | 0 | |||
| KCNE4 | 0 | ||||
| Downstream | - | ||||
| 6 | 2 | Upstream | FARSB, LOC538702, MOGAT1, ACSL3, KCNE4 | ||
| - | 0 | 46 | |||
| Downstream | AP1S3, SCG2, WDFY1 | ||||
| 7 | 2 | Upstream | - | ||
| NYAP2 | 16 | 0 | 34 | ||
| Downstream | LOC104969998 | ||||
| 8 | 8 | Upstream | RFX3, LOC101905621, LOC101905770 | ||
| KIAA0020 | 18 | 9 | 29 | ||
| Downstream | KCNV2, VLDLR | ||||
| 9 | 9 | Upstream | RPS6KA2, RNASET2, FGFR1OP, CCR6, GPR31, TTLL2, UNC93A | ||
| LOC101905262 | 5 | 0 | 59 | ||
| Downstream | MLLT4, KIF25, FRMD1, DACT2, SMOC2 | ||||
| 10 | 10 | Upstream | PIGB, RAB27A, RSL24D1, SNORA25 | ||
| UNC13C | 1 | 0 | 56 | ||
| Downstream | - | ||||
| 11 | 12 | Upstream | - | ||
| - | 0 | 53 | |||
| Downstream | - | ||||
| 12 | 15 | Upstream | MSANTD4 | ||
| GRIA4 | 45 | 20 | 0 | ||
| Downstream | - | ||||
| 13 | 15 | Upstream | OTOG, USH1C, ABCC8, KCNJ11, NCR3LG1, NUCB2, PI3KC2a, RPS13 | ||
| PLEKHA7 | 36 | 14 | 23 | ||
| C15H11orf58 | 5 | 1 | |||
| SOX6 | 8 | 0 | |||
| Downstream | - | ||||
| 14 | 16 | Upstream | NME7, BLZF1, CCDC181, SLC19A2, F5, SELP, SELL | ||
| SELL | 6 | 0 | 24 | ||
| SELE | 16 | 5 | |||
| C16H1orf112 | 16 | 4 | |||
| SCYL3 | 5 | 0 | |||
| KIFAP3 | 29 | 9 | |||
| METTL11B | 1 | 0 | |||
| Downstream | GORAB, PRRX1 | ||||
| 15 | 18 | Upstream | SIGLEC5, MIR99B, MIRLET7E, MIR125A, HAS1, VN2R408P, ZNF613, ZNF615, ZNF614, ZNF432, ZNF350 | ||
| PPP2R1A | 1 | 0 | 33 | ||
| BOSTAUV1R416 | 1 | 0 | |||
| BOSTAUV1R417 | 4 | 0 | |||
| ZNF415 | 5 | 0 | |||
| Downstream | LOC539675, LOC787057, LOC100848895, LOC506495, LOC785630 | ||||
| 16 | 22 | Upstream | SLC6A11, SLC6A1 | ||
| HRH1 | 13 | 0 | 34 | ||
| Downstream | ATG7, VGLL4, TAMM41 | ||||
| 17 | 24 | Upstream | DSC3 | ||
| - | 0 | 0 | 56 | ||
| Downstream | - | ||||
| 18 | 26 | Upstream | PLEKHA1, HTRA1, DMBT1, SPADH1, SPADH2 | ||
| C26H10orf88 | 1 | 0 | 16 | ||
| PSTK | 1 | 0 | |||
| IKZF5 | 5 | 0 | |||
| ACADSB | 23 | 15 | |||
| Downstream | HMX3, HMX2, BUB3 | ||||
| 19 | 29 | Upstream | KIRREL3 | ||
| KIRREL3 | 47 | 15 | 0 | ||
| Downstream | KIRREL3, LOC104976256 | ||||
aNumber of SNPs within the gene;
bNumber of SNPs with P ≤ 0.01 within the gene.
cNumber of SNPs in the QTLR in the regions between genes.
Considering all genes included within or in the 0.5 Mb flanks of the QTLRs, a total of 130 genes were found, of which annotation data were available for 98. Their biological processes (BP), cellular components (CC), molecular function (MF) and metabolic pathways (KEGG) are detailed in S1 Table (‘gene not clustered’ and ‘Kegg pathway’ sheets, respectively). Among these 98 genes, we focused on 18 candidates, based on biological and statistical considerations as detailed in the Discussion. We searched NCBI dbSNP for non-synonymous (AA substitution) polymorphisms, candidates to be the causative mutation of the found BRD effect. The NCBI dbSNP present a substantial polymorphism in all 18 genes (Table 4). These genes averaged 664.5 AA, 1,993.5 bp. All genes had SNPs in the coding regions (average 7.8% of all SNPs); all had AA substitutions (average 78.4% of the coding SNPs) and all had AA substitutions involving change of properties (average 60.4% of all AA substitutions). Although these are published polymorphisms, not polymorphisms found in the study population, they present the potential functional polymorphisms of the QTLR genes.
Table 4. Non-synonymous polymorphisms in the selected 18 candidate genes as reported in NCBI dbSNP.
| SNPsc | Non-synon.d | Properties changee | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QTLR | BTA | Gene | AAa | Bpb | No. | Prop. | No. | Prop. | No. | Prop. |
| 1 | 1 | CADM2 | 444 | 1,332 | 52 | 0.039 | 39 | 0.750 | 19 | 0.487 |
| 2 | 1 | NAALADL2 | 882 | 2,646 | 156 | 0.059 | 130 | 0.833 | 78 | 0.600 |
| 3 | 1 | SLCO2A1 | 644 | 1,932 | 98 | 0.051 | 72 | 0.735 | 37 | 0.514 |
| 3 | 1 | TF | 703 | 2,109 | 113 | 0.054 | 88 | 0.779 | 60 | 0.682 |
| 4 | 2 | ABCA12 | 2,593 | 7,779 | 297 | 0.038 | 215 | 0.724 | 132 | 0.614 |
| 6 | 2 | AP1S3 | 154 | 462 | 15 | 0.032 | 12 | 0.800 | 8 | 0.667 |
| 8 | 8 | KIAA0020 | 647 | 1,941 | 136 | 0.070 | 105 | 0.772 | 68 | 0.648 |
| 9 | 9 | SMOC2 | 445 | 1,335 | 214 | 0.160 | 196 | 0.916 | 115 | 0.587 |
| 9 | 9 | CCR6 | 375 | 1,125 | 75 | 0.067 | 53 | 0.707 | 32 | 0.604 |
| 10 | 10 | RAB27A | 221 | 663 | 59 | 0.089 | 43 | 0.729 | 23 | 0.535 |
| 13 | 15 | PLEKHA7 | 1,224 | 3,672 | 605 | 0.165 | 505 | 0.835 | 307 | 0.608 |
| 14 | 16 | SELE | 485 | 1,455 | 46 | 0.032 | 37 | 0.804 | 22 | 0.595 |
| 14 | 16 | SELP | 646 | 1,938 | 43 | 0.022 | 31 | 0.721 | 20 | 0.645 |
| 14 | 16 | SELL | 370 | 1,110 | 30 | 0.027 | 24 | 0.800 | 17 | 0.708 |
| 15 | 18 | PPP2R1A | 589 | 1,767 | 443 | 0.251 | 383 | 0.865 | 207 | 0.540 |
| 16 | 22 | HRH1 | 491 | 1,473 | 191 | 0.130 | 158 | 0.827 | 94 | 0.595 |
| 16 | 22 | ATG7 | 629 | 1,887 | 151 | 0.080 | 117 | 0.775 | 53 | 0.453 |
| 18 | 26 | IKZF5 | 419 | 1,257 | 60 | 0.048 | 45 | 0.750 | 36 | 0.800 |
| Avg | 664.5 | 1,993.5 | 154.7 | 0.078 | 125.2 | 0.784 | 73.8 | 0.604 | ||
| Min | 154 | 462 | 15 | 0.022 | 12 | 0.707 | 8 | 0.453 | ||
| Max | 2,593 | 7,779 | 605 | 0.251 | 505 | 0.916 | 307 | 0.800 | ||
aNumber of amino acid.
bNumber of coding nucleotides in the gene.
cNo. and Prop.: number of coding SNPs and their proportion out of all coding nucleotides.
dNo. and Prop.: number of non- synonymous SNPs and their proportion out of all SNPs.
eNo. and Prop.: number of AA substitution involving change of AA physical properties, and their proportion out of all non-synonymous SNPs).
Discussion
BRD is the most prevalent disease in the cattle industry in many parts of the world, with symptoms that often go undetected. Using the kosher phenotypic classification we performed a genome-wide association study (GWAS) to map QTL affecting BRD morbidity in Israeli Holstein calves. SDP and Illumina BovineHD BeadChip were used to scan the genome.
Typically, significant markers found by a GWAS are intermingled with non-significant markers (S1 Fig). The lack of a monotonic relation between marker location and marker significance makes it impossible to apply the widely used LOD drop method to set boundaries for the QTLR. To circumvent this we used a moving average of -LogP. This worked well, and clear peaks with monotonic shoulders were obtained (Fig 2). Using a threshold of moving average of -LogP = 2 (corresponding to P = 0.01) to identify QTLs yielded 19 QTLRs distributed over 13 autosomes (Table 2).
The commonly used Log drop 1 method was effective in setting clear boundaries for the QTLR, in one case distinguishing between QTLR as close as 11 Kb (between QTLRs 5 and 6; Table 2 and Fig 2). While distinguishing between two very close QTL on BTA 2 (Fig 3), it nevertheless merged possibly distinct QTL on BTAs 1, 2 and 16 (Fig 4).
Among the highly significant markers (with P < 0.001) in the QTLRs, 48.1% were generated by markers with MAF < 0.15 (S1 Table). This high proportion could be a result of an underestimation of allele frequency variance for markers with low MAF. However, in this study P-values were obtained by SD adjusted to allele B frequency (see Materials and Methods). The distribution of the SD values against allele B frequency showed no indication of a secondary mode at the high SD levels that would indicate presence of an appreciable group of unstable markers (data not shown). We are not aware of any report on the distribution to compare with of MAF in QTLRs. The high proportion of low MAFs among highly significant QTLRs markers could be a result of recent new mutations, selection, or both. This, however, is beyond the scope of the present study.
Combining moving average with Log drop resulted in high resolution mapping, with average QTLR size of 0.26 Mb, allowing thorough bioinformatic analysis to identify candidate genes in the proximity of those QTLR. Numerous databases and analyses were used to locate and categorize genes in and around the 19 QTLRs. Among the genes within or in the 0.5 Mb flanks of the QTLRs (Table 3), we focused on 18 candidate gene, based on biological and statistical considerations (Table 4). These included involvement of the genes in immunity, wound repair, cells adhesion and pulmonary function; the number of significant SNPs within the gene; and proximity of previously reported relevant QTLRs.
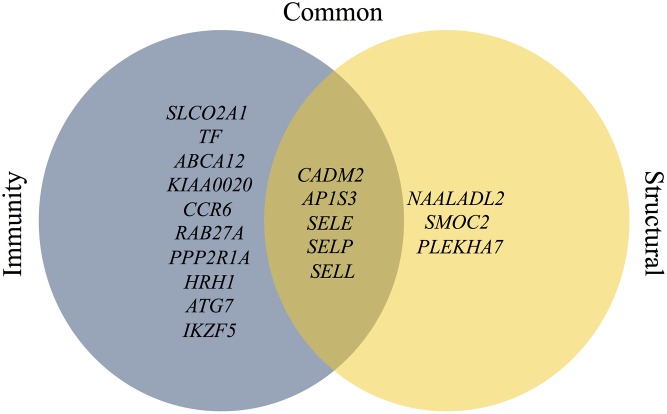
Within the 15 candidates that possess various immune functions, 5 genes, CADM2 [38] AP1S3 [39], SELE [40,41], SELP [41] and SELL [5,41,42], are associated with adhesive activity, ECM remodeling, epithelial-to-mesenchyme transition (EMT) and profibrotic activities. All are part of the repair/wound healing process that might lead to the generation of adhesions (Fig 5). Ten genes, SLCO2A1 [43], TF [44], ABCA12 [45], KIAA0020 [46], CCR6 [47], RAB27A [48], PPP2R1A [49], HRH1 (GeneCards, http://www.genecards.org/), ATG7 [50,51] and IKZF5 [52] are known for their immunological activity. Three genes, NAALADL2 [53,54], SMOC2 [55,56] (GeneCards), PLEKHA7 [57,58], possess the above scar formation activities without apparent immune function (Fig 5). Thus, although they may respond to immunological cues [32], it is tempting to refer to these three candidates as potential exclusive markers for kosher status while the other 15 may serve as markers for both BRD and kosher status. Interestingly, Tizioto et al. [27] showed that one of these three genes, SMOC2, was differentially expressed in lymph nodes in response to BVD infection. It belongs to a protein family that mainly presents in tissues undergoing repair or remodeling [59]. Possibly, in response to BVD infection, SMOC2 plays a role as an early wound repair gene.
Fig 5. Genes and systems.
Structural, genes involved in repair/wound healing processes without apparent immune function, including: adhesive activity, extracellular matrix (ECM) remodeling, epithelial-to-mesenchyme transition (EMT) and profibrotic activities; Immunity, genes involved in immunological activity; Common, genes involved in both.
The definition of the non-Kosher status as a phenotype characterized by pulmonary adhesions makes the eight structural and common genes, namely, structural and immune functions, interesting candidates. In line with the above, functional polymorphism in genes reported herein may alter the molecular pathways involved in the wound healing process, rendering individuals susceptible/resistant to the lung adhesion phenotype. If this would occur in the three structural genes, it might exclusively affect the kosher phenotype. However, if it would take place also in the common and/or immunity genes, the pulmonary adhesion phenotype may be representative of BRD susceptibility/resistance as well.
A total of 2,253 AA substitutions were found in the NCBI dbSNP in 18 of the QTLR genes (Table 4). This polymorphism constitutes a potential source of structural changes of the proteins, and hence potentially a large number of quantitative alleles. It is more than enough to be a source of protein conformation alleles that differ from one another in their molecular efficiency. Thus, all QTLRs found in the present study harbor a plethora of candidate causative mutations potentially responsible for the quantitative effects found.
Such a polymorphism in protein coding genes is in accord with the AA polymorphism found in the chicken OCX gene by [60]. It suggests that a substantial portion of the AA may not be important to the protein function.
Another evidence that a gene may be a QTG is changing the level of expression in response to intrinsic and extrinsic cues. Tizioto et al. [27] searched for Differentially Expressed (DE) genes in cattle bronchial lymph node in response to a separate challenge by each of 3 viral and 3 bacterial BRD pathogens. Forty-three of the DE genes were located in the QTLRs of the present study (Table 5). On average 2.9 genes in a QTLR responded to a challenge by one or more pathogens. Fifteen of the QTLRs included genes that changed expression in response to at least one pathogen. QTLRs 3, 4, 8 and 18, contained genes that among them responded to all pathogens. All pathogens changed expression of genes in at least 4 QTLRs, averaging 9.3 QTLRs. The 43 genes found by Tizioto et al. [27] located in the QTLR of the present study responded to an average of 2.5 pathogens (Table 6), and 32 of these genes (74.4%) responded to more than 2 pathogens (Table 5). The six pathogens changed expression of an average of 27.7 of our 43 genes (5 to 30; Table 6).
Table 5. Tizioto et al. [27] differentially expressed genes found in the QTLRs of the present study.
| Gene | QTLR | BTA | BRSVc | BVDVd | IBRe | M. bovisf | M. haemolyticag | P. multocidah | Pathogensa |
|---|---|---|---|---|---|---|---|---|---|
| TF | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| SLCO2A1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| BARD1 | 4 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| FARSB | 5 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| KCNE4 | 5 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| VLDLR | 8 | 8 | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| RFX3 | 8 | 8 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| RNASET2 | 9 | 9 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| MLLT4 | 9 | 9 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| RAB27A | 10 | 10 | 1 | 1 | 1 | 0 | 1 | 0 | 4 |
| OTOG | 13 | 15 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| SELP | 14 | 16 | 1 | 0 | 1 | 0 | 1 | 0 | 3 |
| SELL | 14 | 16 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| BLZF1 | 14 | 16 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| LOC787057 | 15 | 18 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| ZNF614 | 15 | 18 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| LOC785630 | 15 | 18 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| ZNF432 | 15 | 18 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| VGLL4 | 16 | 22 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| DSC3 | 17 | 24 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| DMBT1 | 18 | 26 | 1 | 1 | 1 | 0 | 0 | 1 | 4 |
| IKZF5 | 18 | 26 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| PLEKHA1 | 18 | 26 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| KIRREL3 | 19 | 29 | 1 | 0 | 0 | 0 | 1 | 0 | 2 |
| EPHB1 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| AMOTL2 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| TOPBP1 | 3 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| FN1 | 4 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 5 |
| SCG2 | 6 | 2 | 0 | 1 | 1 | 0 | 1 | 0 | 3 |
| UNC93A | 9 | 9 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| SMOC2 | 9 | 9 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| USH1C | 13 | 15 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| NUCB2 | 13 | 15 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| SELE | 14 | 16 | 0 | 1 | 1 | 0 | 1 | 0 | 3 |
| SLC19A2 | 14 | 16 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| F5 | 14 | 16 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| HRH1 | 16 | 22 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| HTRA1 | 18 | 26 | 0 | 1 | 1 | 1 | 1 | 1 | 5 |
| BFSP2 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| GRIA4 | 12 | 15 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ZNF613 | 15 | 18 | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| RAB6B | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| PRRX1 | 14 | 16 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Genesb | 24 | 29 | 30 | 5 | 14 | 7 | 109 | ||
aNumber of pathogens affected the expression of the gene.
bNumber of genes affected by the pathogen.
cBovine respiratory syncytial virus.
dBovine viral diarrhea virus Infectious.
eBovine rhinotracheitis.
fMycoplasma bovis.
gMannheimia haemolytica.
hPasteurella multocida.
Table 6. Effects of QTLR’s genes and pathogens.
| Pathogens/genea | Genes/pathogenb | |||
|---|---|---|---|---|
| All | Virus | Bacteria | ||
| Avg | 2.5 | 18.2 | 27.7 | 8.7 |
| Min | 1 | 5 | 24 | 5 |
| Max | 6 | 30 | 30 | 14 |
aNumber of pathogens which changed the expression of a gene.
bNumber of genes whose expression was changes by a pathogen.
Thus, most of the QTLRs found in this study based on the kosher phenotype, harbor an abundance of genes responding to BRD pathogens and thus are candidates for containing QTG.
Some of the QTLRs found in this study are in the vicinity of related QTLs mapped previously. QTL 2 on BTA 1 (Table 2) is within a QTLR affecting "Veterinary treatments" located at 91.9–97.3 Mb [61] and near a QTL located at 91.6–91.7 Mb affecting heat tolerance in beef cattle [62]. QTLR 5 on BTA 2 is near a BRD QTL [25]; QTLR 11 on BTA 12 and QTLR 18 on BTA 26 overlap BRD QTLs [14] and a BVD QTL region associated with the bovine viral diarrhea persistent infection, a virus frequently identified as a causative pathogen for BRD outbreaks [63]. QTL associated with carcass, production, reproduction and behavior traits were reported in different cattle breeds in the region of QRLR 19 on BTA 20 [64].
Being a part of the regular routine in kosher slaughterhouses, obtaining the kosher phenotype is free of charge, allowing large scale studies. The function and expression of tens of candidate genes, ampleness of candidate AA polymorphisms in the QTLR genes, and proximity of previously reported associated QTL, support the QTLRs found in this study, and thus support kosher phenotyping as an efficient means of mapping BRD QTL, QTG and causative mutations.
Materials and Methods
Samples and genotyping
All samples were collected as they came from male Holstein calves slaughtered in a commercial slaughterhouse (Adom Adom abattoir, Israel) under stringent kosher meat inspection requirements. Genetically, the Israel Holstein cattle population is very homogeneous. There is a single artificial insemination center that serves the entire country, so all female replacements are produced from the same pool of sires. Thus, the dams are a homogenous group and there is no reason to suspect population subdivision or stratification. The calves in the study population were produced over the course of a year, and hence represent a set of sire half-sib progeny groups. Examination of the pedigree of the study animals did not uncover any differential allocation of herds, sires, or maternal grandsires among the Glatt kosher and non-kosher groups. Hence, we believe that population structure was not a factor in our findings.
The inspector of the internal organs of the animal for assigning kosher status is trained to look for lung adhesions in the animal both before and after its lungs are removed. To test a lung, the inspector first removes all adhesions. If the lung is still intact it is classified as "Kosher"; Torn adhesions that cause perforations in the lung render it "Non- Kosher" (NK); Finally, lungs that are adhesion-free are referred to as “Glatt Kosher” (GK), referring to the fact that the animal’s lungs do not have any adhesions. In the current study GK individuals were taken as the High resistant group, while NK individuals were taken as the Low resistant group, as also recently appeared in Hayes et al. [65].
Blood was sampled immediately after slaughter, using evacuated tubes (Greiner Bio-One GmbH, Kremsmunster, Austria) containing EDTA as anticoagulant. DNA was isolated from the whole blood using Sigma DNA extraction kit, according to the manufacturer's instructions. DNA was quantified using NanoDrop (Wilmington, DE) spectrophotometry and purity was estimated using the 260/280 ratio. The quality control was performed on each sample to verify the DNA integrity on Invitrogen E-Gel 1% Agarose Gel.
Pools were constituted as reported by Strillacci et al. [66]. Five GK and two NK pools were made of 21 − 31 Holstein male calves each (Table 7).
Table 7. Number of individuals in the pools.
aOrdinal number of the pool.
bNumber of calves in the pool.
cGlatt kosher.
dNon-kosher.
The pooled DNA samples were each genotyped in two duplicates, on two independent microarrays for a total of 14 microarray positions. Genotyping was performed at the University of Milan using the Illumina BovineHD BeadChip (777,962 SNPs). Each analysis was based on the genomic position of SNPs according to the bovine UMD3.1 genome assembly. Proportion in the pools of the Allele defined by Illumina as B (pB) was obtained by Illumina software.
Quality Control (QC)
Quality control filters were: SNP mapped to specific autosome location; no more than 50% or 25% missing genotypes for pools or markers, respectively; no more than a difference of 0.10 between the pB values of the two duplicates of the same pool-marker combination; and a minimum of 0.05 average marker pB. A total of 570,563 autosomal markers were retained after the QC and used to map QTLs.
Statistical Methods
QTL Mapping: Testing for significance of marker-trait association
Let pBijk be the mean frequency of the B-allele across both duplicate arrays of the ith marker in the jth pool of the kth tail.
i = 1 − M, where M is the total number of markers analyzed;
j = 1 − 5 for the GK pools, j = 1 − 2 for the NK pools;
k = 1 − 2, where 1 = GK pools, and 2 = NK pools.
We used a single-marker test for marker-trait association, where CWER P-value for the ith marker was calculated as
Pi = 2 x the area of the standard normal curve to the right of
| (1) |
where,
Di = pBi.1 − pBi.2;
pBi.1 = mean pBij1 across the 5 GK pools;
pBi.2 = mean pBij2 across the 2 NK pools.
For SE(Di) we used an empirical estimate of the standard erroof Di under the null hypothesis of absence of marker-QTL association. This estimate is based on the variance among individual replicate pools within the same tail [67]. The basic assumption is that under the null hypothesis, the sampling variance among individual pools across tails is the same as the sampling variance among individual pools within tails. On this assumption, it follows from elementary principles, that
| (2) |
| (3) |
where,
Var(pBi.1) = the variance among the pBij1 of the 5 replicate pools in the GK tail;
Var(pBi.2) = the variance among the pBij2 of the 2 replicate pools in the NK tail.
We expect Var(pBi.1) = (VarpBi.2) to be the same, hence denoted Var(pBi). Then the best estimate of Var(pBi) will be the mean of Var(pBi.1) and Var(pBi.2) weighted by the number of degrees of freedom in each variance.
| (4) |
and
| (5) |
| (6) |
Because the estimate of Var(pBi) is based on a small number of pools, we used a global estimate of Var(pBi), averaged across markers and denoted Var(pB), as our estimate of the sampling variance of pBi across pools. Because pB is a binomial variate, Var(pB) is a function of pB(1-pB), which maximizes at pB = 0.5 and drops off to either side. Thus, each marker requires an appropriate SE2(D), depending on its frequency. Consequently, we binned the markers in bins of width 0.1 from 0.0 to 1.0, according to their average pB across all pools (Avg pBi), and calculated average Var(pB) across the markers in each bin. Table 8 shows that indeed Var(pB) maximized in the range 0.4 to 0.6, dropping off to either side, thus justifying the use of Var(pB) according the marker mean pB. As a result, SE2D differed somewhat for the different markers, according to marker frequency in the pools.
Table 8. Weighted average of variances (Var(pB) of marker frequencies among replicates within GK and NK tails, pooled across all markers within bins.
| Avg pBi | Var(pB) |
|---|---|
| ≤ 0.1 | 0.00091 |
| > 0.1–0.2 | 0.00562 |
| > 0.2–0.3 | 0.00718 |
| > 0.3–0.4 | 0.00796 |
| > 0.4–0.5 | 0.00833 |
| > 0.5–0.6 | 0.00847 |
| > 0.6–0.7 | 0.00827 |
| > 0.7–0.8 | 0.00716 |
| > 0.8–0.9 | 0.00492 |
| > 0.9–1.0 | 0.00109 |
Binning is by average marker frequency across all pools (Avg pBi).
The proportion of false positives (PFP; [37]) was used to correct for multiple tests.
QTL regions (QTLRs): Declaration of a QTLR
While searching for regions containing QTLs (QTLRs), significance of a single marker may not be enough to declare a QTLR, as a singleton significant P, without any support from surrounding markers, is prone to be false positive. On the other hand, visual inspection of chromosomal scatter charts reveals clusters of significant marker -LogP values (e.g., Fig 1). We interpret these clusters as putative QTLR. Note, however, that high -LogP values are interspersed with very low values across the cluster region. This behavior is related to complex LD patterns observed across small chromosomal regions [68]. Nevertheless, average -LogP value across a cluster region will be greater than in the adjoining regions lacking a concentration of high -LogP markers. Thus, to identify the clusters quantitatively, assign them an overall -LogP value, and determine their boundaries, we used a moving average of -LogP in 1 nucleotide steps taken across a window of markers. QTLRs were declared on the basis of windows having average -LogP above some chosen threshold. For the present study, average spacing between markers was 4,394 bp. Hence, windows of size 23 markers, equivalent to an average window size of 100 Kb were used. The size of a window was chosen to give a reasonable physical length yet not be overly influenced by the -LogP value of any single marker. A moving average of -LogP = 2, corresponding to P = 0.01, was set as the threshold value for declaration of a window as a QTLR. This threshold was set to give a reasonable number of ranked QTLR for further study in depth.
The windows worked well, and clear peaks with monotonic shoulders were obtained. See Fig 2 for a detailed example of the cluster of Fig 1. For this cluster, a run of 8 consecutive windows was found, all with a moving average above the chosen threshold of -LogP = 2. The top window among these had average -LogP = 2.470. The windows are located on the chromosome by the average location of their markers. The locations of the markers in the top window averaged 30,788,390, and this was taken as the point location of the QTL. The marker in this cluster with highest -LogP value (SNP BovineHD2900009163 located at 30,812,830 bp, -LogP = 7.304) was contained within this window, at a remove of 20,440 bp from the window average marker location.
Setting the boundaries of the QTLR
The popular LOD drop method [69] works well when applied to family-based linkage mapping and the low-density microsatellite maps of the previous generation of QTL mapping. In such studies, there is usually a monotonic inverse relation between marker significance and marker location relative to the most significant marker; with marker significance ascending monotonically marker by marker to the most significant marker and descending monotonically from that point. With GWAS and high-density marker maps, however, we are faced with the above-mentioned phenomenon where highly significant and non-significant markers are interspersed across the cluster. Consequently, there is no longer a monotonic relation between marker location and marker significance, making it impossible to apply the LOD drop method. That is, considering -LogP values of individual markers of the cluster, going out from the putative point location of the QTL (the most significant marker of the cluster), one will meet one or more non-significant markers, ostensibly setting a LOD drop boundary, but then just beyond these, are a new series of significant markers, clearly part of the same cluster and QTLR (e. g., Fig 2).
We found that the moving windows present a monotonic inverse relation of window location and window average -LogP values relative to the top window, forming a clear peak. This allowed the use of the LOD drop method for setting QTLR boundaries. For the present study, since we did not have LOD scores but -LogP, we used instead a -LogP drop of 1 (denoted "Log drop"). As noted above, for the cluster on BTA 29, the top window had average -LogP = 2.470. Accordingly, the Log drop 1 boundary of the QTLR were at -LogP = 1.470. The upstream and downstream windows with means closest to this value (-LogP = 1.470 and 1.492, respectively; Fig 2) were taken as the boundary windows. Since the windows are located by the average location of their markers, the actual boundaries of the QTLR were from the first marker of the upstream boundary window, to the final marker of the downstream boundary window. These markers were taken to define the final boundaries of the QTLR.
Distinguishing adjoining clusters
When two clusters and consequent runs of windows above the threshold were close to each other, two top windows and two peaks are seen (Figs 3 and 4). In this case, declaring the region as consisting of one or two QTLRs was based on the -LogP values of the region between the two top windows, relative to the lower of the Log drop 1 boundary thresholds of the two peaks. If the entire region between the peaks was above the higher Log drop 1 threshold, they were taken as one QTLR; otherwise, they were taken as two separate QTLRs. In the latter case, the exact boundary between the QTLRs was the window with lowest -LogP value.
Bioinformatics
The SNPchiMp database [70] was used to convert the Illumina SNP name to the rsID (the SNP accession number used to search a SNP in all public databases). The rsID was used in NCBI database (http://www.ncbi.nlm.nih.gov/) to verify the precise position (intronic-exonic position, close to gene) of each SNP in respect to a gene. With one exception, none of the analyzed SNPs was in a coding region. SNP location and gene annotation were according to the UMD3.1.1 assembly. The full Ensembl v79 gene set for the autosomal chromosomes was downloaded (http://www.ensembl.org/biomart/martview/76d1cab099658c68bde77f7daf55117e). To identify the genes located within the QTLRs and inside 0.5 Mb intervals flanking the position of each SNP marker that define the same regions, we created a consensus list (among QTLRs and downloaded genes) using the BedTools software [71].
GO and pathway analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (http://david.abcc.ncifcrf.gov/). GO terms were used to categorize candidate genes in terms of their functions.
Supporting Information
P values on all autosomes.
(PDF)
Sheet 1. SNPs report. Sheet 2. Genes clustered. Sheet 3: Genes not clustered. Sheet 4: Kegg pathways.
(XLSX)
Acknowledgments
We thank Dr. Wassim Geraisy, the Chief Veterinarian in "Adom Adom" abattoir for his collaboration, which enabled the collection of kosher phenotypes. We acknowledge with thanks the helpful comments of the reviewers.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL, Koohmaraie M, Dikeman ME. Bovine respiratory disease in feedlot cattle: phenotypic, environmental, and genetic correlations with growth, carcass, and longissimus muscle palatability traits. J Anim Sci. 2007; 85(8): 1885–92. [DOI] [PubMed] [Google Scholar]
- 2.Thompson PN, Stone A, Schultheiss WA. Use of treatment records and lung lesion scoring to estimate the effect of respiratory disease on growth during early and late finishing periods in South African feedlot cattle. J Anim Sci. 2006; 84(2): 488–498. [DOI] [PubMed] [Google Scholar]
- 3.Lekeux P, Hajer R, Breukink HJ. Effect of somatic growth on pulmonary function values in healthy Friesian cattle. Am J Vet Res. 1984; 45(10): 2003–2007. [PubMed] [Google Scholar]
- 4.Duff GC, Galyean ML. Board-invited review: recent advances in management of highly stressed, newly received feedlot cattle. J Anim Sci. 2007; 85(3):823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitam H, Vaya J, Brosh A, Orlov A, Khatib S, Izhaki I, et al. Differential stress responses among newly received calves: variations in reductant capacity and Hsp gene expression. Cell Stress Chap. 2010; 15(6): 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner BA, Dolezal HG, Bryant LK, Owens FN, Smith RA. Health of finishing steers: effects on performance, carcass traits, and meat tenderness. J Anim Sci. 1999; 77(12): 3168–3175. [DOI] [PubMed] [Google Scholar]
- 7.Roeber DL, Speer NC, Gentry JG, Tatum JD, Smith CD, Whittier JC, et al. Feeder cattle health management: effects on morbidity rates, feedlot performance, carcass characteristics, and beef palatability. Prof. Anim. Sci. 2001; 17:39–44. [Google Scholar]
- 8.Hodgson PD, Aich P, Manuja A, Hokamp K, Roche FM, Brinkman FS, et al. Effect of stress on viral-bacterial synergy in bovine respiratory disease: novel mechanisms to regulate inflammation. Comp Funct Genomics. 2005; 6(4): 244–250. 10.1002/cfg.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb JM, Steptoe A. Psychosocial stress and susceptibility to upper respiratory tract illness in an adult population sample. Psychosom. Med. 1996; 58: 404–412. [DOI] [PubMed] [Google Scholar]
- 10.Aich P, Wilson HL, Kaushik RS, Potter AA, Babiuk LA, Griebel P. Comparative analysis of innate immune responses following infection of newborn calves with bovine rotavirus and bovine coronavirus. J Gen Virol. 2007; 88(Pt 10):2749–2761. [DOI] [PubMed] [Google Scholar]
- 11.Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. Influence of breed, heterozygosity, and disease incidence on estimates of variance components of respiratory disease in preweaned beef calves. J Anim Sci. 2005; 83(6): 1247–61. [DOI] [PubMed] [Google Scholar]
- 12.Schneider MJ, Tait RG Jr, Ruble MV, Busby WD, Reecy JM. Evaluation of fixed sources of variation and estimation of genetic parameters for incidence of bovine respiratory disease in preweaned calves and feedlot cattle. J Anim Sci. 2010; 88(4):1220–1228. 10.2527/jas.2008-1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muggli-Cockett NE, Cundiff LV, Gregory KE. Genetic analysis of bovine respiratory disease in beef calves during the first year of life. J Anim Sci. 1992; 70(7): 2013–2019. [DOI] [PubMed] [Google Scholar]
- 14.Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser J, et al. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned holstein calves. BMC Genomics. 2014; 15(1): 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittum TE, Woollen NE, Perino LJ, Littledike ET. Relationships among treatment for respiratory tract disease, pulmonary lesions evident at slaughter, and rate of weight gain in feedlot cattle. J Am Vet Med Assoc. 1996; 209(4): 814–818. [PubMed] [Google Scholar]
- 16.Bishop SC. Disease resistance: Genetics In: Pond W. G & Bell A. W. (eds) Encyclopedia of Animal Science. Marcel Dekker, Inc; New York, 288–290; 2005. [Google Scholar]
- 17.Bishop SC, Axford RFE, Nicholas FW, Owen JB. Breeding for Disease Resistance in Farm Animals. 3rd Ed CABI, UK; 2011. [Google Scholar]
- 18.Hanotte O, Ronin Y, Agaba M, Nilsson P, Gelhaus A, Horstmann R, et al. Mapping of quantitative trait loci controlling trypanotolerance in a cross of tolerant West African N'Dama and susceptible East African Boran cattle. Proc Natl Acad Sci U S A 2003; 100(13):7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heifetz EM, Fulton JE, O'Sullivan NP, Arthur JA, Wang J, Dekkers JC, et al. Mapping quantitative trait loci affecting susceptibility to Marek's disease virus in a backcross population of layer chickens. Genetics. 2007; 177(4): 2417–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H, Niikura M, Kim T, Mao W, MacLea KS, Hunt H, et al. Using integrative genomics to elucidate genetic resistance to Marek's disease in chickens. Dev Biol (Basel) 2008; 132:365–72. [DOI] [PubMed] [Google Scholar]
- 21.Tal-Stein R, Fontanesi L, Dolezal M, Scotti E, Bagnato A, Russo V, et al. A genome scan for quantitative trait loci affecting milk somatic cell score in Israeli and Italian Holstein cows by means of selective DNA pooling with single- and multiple-marker mapping. J Dairy Sci. 2010; 93(10): 4913–4927. 10.3168/jds.2010-3254 [DOI] [PubMed] [Google Scholar]
- 22.Goddard ME, Hayes BJ. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009; 10 (6): 381–391. 10.1038/nrg2575 [DOI] [PubMed] [Google Scholar]
- 23.Druet T, Georges M. A hidden Markov model combining linkage and linkage disequilibrium information for haplotype reconstruction and quantitative trait locus fine mapping. Genetics. 2010; 184(3): 789–798. 10.1534/genetics.109.108431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010; 42(12): 1118–1125. 10.1038/ng.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neibergs H, Zanella R, Casas E, Snowder GD, Wenz J, Neibergs JS, et al. Loci on Bos taurus chromosome 2 and Bos taurus chromosome 26 are linked with bovine respiratory disease and associated with persistent infection of bovine viral diarrhea virus. J Anim Sci. 2011; 89(4): 907–915. 10.2527/jas.2010-3330 [DOI] [PubMed] [Google Scholar]
- 26.Casas E, Garcia MD, Wells JE, Smith TP. Association of single nucleotide polymorphisms in the ANKRA2 and CD180 genes with bovine respiratory disease and presence of Mycobacterium avium subsp. paratuberculosis. Anim Genet. 2011; 42(6): 571–577. 10.1111/j.1365-2052.2011.02189.x [DOI] [PubMed] [Google Scholar]
- 27.Tizioto PC, Kim J, Seabury CM, Schnabel RD, Gershwin LJ, Van Eenennaam AL, et al. Immunological Response to Single Pathogen Challenge with Agents of the Bovine Respiratory Disease Complex: An RNA-Sequence Analysis of the Bronchial Lymph Node Transcriptome. PLoS One. 2015; 10(6): e0131459 10.1371/journal.pone.0131459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galyean ML, Perino LJ, Duff GC. Interaction of cattle health/immunity and nutrition. J Anim Sci. 1999; 77(5): 1120–1134. [DOI] [PubMed] [Google Scholar]
- 29.Bryant LK, Perinol LJ, Griffin D, Doster AR, Wittum TEA. Method for recording pulmonary lesions of beef calves at slaughter, and the association of lesions with average daily gain. Bovine Pract. 1999; 33: 163–173. [Google Scholar]
- 30.Schneider MJ, Tait RG Jr, Busby WD, Reecy JM. An evaluation of bovine respiratory disease complex in feedlot cattle: Impact on performance and carcass traits using treatment records and lung lesion scores. J Anim Sci. 2009; 87(5): 1821–1827. 10.2527/jas.2008-1283 [DOI] [PubMed] [Google Scholar]
- 31.Sulaiman H, Dawson L, Laurent GJ, Bellingan GJ, Herrick SE. Role of plasminogen activators in peritoneal adhesion formation. Biochem Soc Trans. 2002; 30(2): 126–131. [DOI] [PubMed] [Google Scholar]
- 32.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008; 214(2): 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busby D. Factors that impact profit in feeder cattle–TCSCF data summary. Cattle Feeder Clinics. 2014; 24–27. [Google Scholar]
- 34.Agbeniga B, Webb EC, O’Neill HA. Influence of Kosher (Shechita) and conventional slaughter techniques on shear force, drip and cooking loss of beef. S. Afr. J. Anim. Sci. 2013; 43:S98–S102. [Google Scholar]
- 35.Regenstein JM, Chaudry MM, Regenstein CE. The Kosher and Halal food laws. CRFSFS 2003; 2(3): 111–127. 10.1111/j.1541-4337.2003.tb00018.x [DOI] [PubMed] [Google Scholar]
- 36.Mosig MO, Lipkin E, Khutoreskaya G, Tchourzyna E, Soller M, Friedmann A. A whole genome scan for quantitative trait loci affecting milk protein percentage in Israeli-Holstein cattle, by means of selective milk DNA pooling in a daughter design, using an adjusted false discovery rate criterion. Genetics. 2001; 157(4):1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernando RL, Nettleton D, Southey BR, Dekkers JCM, Rothschild MF, Soller M. Controlling the proportion of false positives in multiple dependent tests. Genetics. 2004; 166(1): 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moiseeva EP, Roach KM, Leyland ML, Bradding P. CADM1 is a key receptor mediating human mast cell adhesion to human lung fibroblasts and airway smooth muscle cells. PLoS One. 2013; 8(4): e61579 10.1371/journal.pone.0061579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setta-Kaffetzi N, Simpson MA Navarini AA Patel VM Lu HC, Allen MH, Duckworth M, et al. AP1S3 mutations are associated with pustular psoriasis and impaired Toll-like receptor 3 trafficking. Am J Hum Genet. 2014; 94(5): 790–797. 10.1016/j.ajhg.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Hsu LA, Teng MS, Lin JF, Chang HH, Sun YC, et al. Association of SELE genotypes/haplotypes with E-selectin levels in Taiwanese individuals: interactive effect of MMP9 level. BMC Med Genet. 2012; 13:115 10.1186/1471-2350-13-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M, Fang L, Tan C, Long T, Chen H, Xiao S. Understanding Streptococcus suis serotype 2 infection in pigs through a transcriptional approach. BMC Genomics. 2011; 12:253 10.1186/1471-2164-12-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckham Sporer KR, Burton JL, Earley B, Crowe MA. Transportation stress in young bulls alters expression of neutrophil genes important for the regulation of apoptosis, tissue remodeling, margination, and anti-bacterial function. Vet Immunol Immunopathol. 2007; 118(1–2): 19–29. [DOI] [PubMed] [Google Scholar]
- 43.Sakata D, Yao C, Narumiya S. Emerging roles of prostanoids in T cell-mediated immunity. IUBMB Life. 2010; 62(8): 591–596. 10.1002/iub.356 [DOI] [PubMed] [Google Scholar]
- 44.Daniłowicz E, Martinez-Arias R, Dolf G, Singh M, Probst I, Tümmler B, et al. Characterization of the porcine transferrin gene (TF) and its association with disease severity following an experimental Actinobacillus pleuropneumoniae infection. Anim Genet. 2010; 41(4): 424–427. 10.1111/j.1365-2052.2009.02012.x [DOI] [PubMed] [Google Scholar]
- 45.Fu Y, Mukhamedova N, Ip S, D'Souza W, Henley KJ, DiTommaso T, et al. ABCA12 regulates ABCA1-dependent cholesterol efflux from macrophages and the development of atherosclerosis. Cell Metab. 2013; 18(2): 225–238 10.1016/j.cmet.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 46.Brickner AG, Warren EH, Caldwell JA, Akatsuka Y, Golovina TN, Zarling AL, et al. The immunogenicity of a new human minor histocompatibility antigen results from differential antigen processing. Journal of Experimental Medicine 193: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinfelder S, Floess S, Engelbert D, Haeringer B, Baron U, Rivino L, et al. Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood. 2011; 117(10): 2839–2846. 10.1182/blood-2010-06-293027 [DOI] [PubMed] [Google Scholar]
- 48.Catz SD. The role of Rab27a in the regulation of neutrophil function. Cell Microbiol. 2014; 16(9): 1301–1310. 10.1111/cmi.12328 [DOI] [PubMed] [Google Scholar]
- 49.Ruteshouser EC, Ashworth LK, Huff V. Absence of PPP2R1A mutations in Wilms tumor. Oncogene. 2001; 20(16): 2050–2054. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013; 39(2): 211–227. 10.1016/j.immuni.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 51.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010; 189(6): 925–935. 10.1083/jcb.201002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.John LB, Yoong S, Ward AC. Evolution of the Ikaros gene family: implications for the origins of adaptive immunity. J Immunol. 2009; 182(8): 4792–4799. 10.4049/jimmunol.0802372 [DOI] [PubMed] [Google Scholar]
- 53.Burgner D, Davila S, Breunis WB, Ng SB, Li Y, Bonnard C, et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 2009; 5(1): e1000319 10.1371/journal.pgen.1000319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitaker HC, Shiong LL, Kay JD, Grönberg H, Warren AY, Seipel A, et al. N-acetyl-L-aspartyl-L-glutamate peptidase-like 2 is overexpressed in cancer and promotes a pro-migratory and pro-metastatic phenotype. Oncogene. 2014; 33(45): 5274–5287. 10.1038/onc.2013.464 [DOI] [PubMed] [Google Scholar]
- 55.Vannahme C, Gösling S, Paulsson M, Maurer P, Hartmann U. Characterization of SMOC-2, a modular extracellular calcium-binding protein. Biochem J. 2003; 373(Pt 3): 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi W, Chen F, Cardoso WV. Mechanisms of lung development: contribution to adult lung disease and relevance to chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009; 6(7): 558–563 10.1513/pats.200905-031RM [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010; 11(7): 502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- 58.Paschoud S, Jond L, Guerrera D, Citi S. PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers. 2014; 2(1): e28755 10.4161/tisb.28755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maier S, Paulssona M, Hartmanna U. The widely expressed extracellular matrix protein SMOC-2 promotes keratinocyte attachment and migration. Exp. Cell Res. 2008; 314: 2477–2487. 10.1016/j.yexcr.2008.05.020 [DOI] [PubMed] [Google Scholar]
- 60.Fulton JE, Soller M, Lund AR, Arango J, Lipkin E. Variation in the ovocalyxin-32 gene in commercial egg-laying chickens and its relationship with egg production and egg quality traits. Anim Genet. 2012; 43 Suppl 1:102–113. 10.1111/j.1365-2052.2012.02384.x [DOI] [PubMed] [Google Scholar]
- 61.Höglund JK, Guldbrandtsen B, Su G, Thomsen B, Lund MS. Genome scan detects quantitative trait loci affecting female fertility traits in Danish and Swedish Holstein cattle. J Dairy Sci. 2009; 92(5): 2136–2143. 10.3168/jds.2008-1104 [DOI] [PubMed] [Google Scholar]
- 62.Howard JT, Kachman SD, Snelling WM, Pollak EJ, Ciobanu DC, Kuehn LA, et al. Beef cattle body temperature during climatic stress: a genome-wide association study. Int J Biometeorol. 2014; 58(7): 1665–1672. 10.1007/s00484-013-0773-5 [DOI] [PubMed] [Google Scholar]
- 63.Zanella R, Casas E, Snowder G, Neibergs HL. Fine Mapping of Loci on BTA2 and BTA26 Associated with Bovine Viral Diarrhea Persistent Infection and Linked with Bovine Respiratory Disease in Cattle. Front Genet. 2011; 2:82 10.3389/fgene.2011.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saatchi M, Schnabel RD, Taylor JF, Garrick DJ. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics. 2014; 15:442 10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayes NS, Schwartz CA, Phelps KJ, Borowicz P, Maddock-Carlin KR, Maddock RJ. The relationship between pre-harvest stress and the carcass characteristics of beef heifers that qualified for kosher designation. Meat Sci. 2015; 100: 134–138. [DOI] [PubMed] [Google Scholar]
- 66.Strillacci MG, Frigo E, Schiavini F, Samoré AB, Canavesi F, Vevey M, et al. Genome-wide association study for somatic cell score in Valdostana Red Pied cattle breed using pooled DNA. BMC Genet. 2014; 15: 106 10.1186/s12863-014-0106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bagnato A, Schiavini F, Rossoni A, Maltecca C, Dolezal M, Medugorac I,et al. Quantitative trait loci affecting milk yield and protein percent in a three-country Brown Swiss population. J Dairy Sci 2008; 91:767–783. 10.3168/jds.2007-0507 [DOI] [PubMed] [Google Scholar]
- 68.Lipkin, E, Dolezal MA, Bagnato A, Fulton JE, Settar P, Arango J, et al. Linkage Disequilibrium and Haplotype Blocks in Cattle and Chicken Populations. Proceedings of the Plant & Animal Genomes XXI Conference, San Diego USA, P0680: Genome Mapping, Tagging & Characterization: General Comparative; 2013.
- 69.Weller JI. Quantitative Trait Loci Analysis in Animals, second edition CABI Publishing; London; 2009. [Google Scholar]
- 70.Nicolazzi EL, Picciolini M, Strozzi F, Schnabel RD, Lawley C, Pirani A, et al. SNPchiMp: a database to disentangle the SNPchip jungle in bovine livestock. BMC Genomics. 2014; 15:123 10.1186/1471-2164-15-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan AR and Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26(6): 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P values on all autosomes.
(PDF)
Sheet 1. SNPs report. Sheet 2. Genes clustered. Sheet 3: Genes not clustered. Sheet 4: Kegg pathways.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.