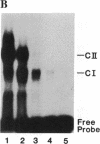
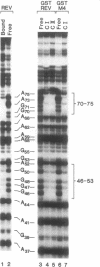
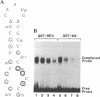
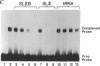
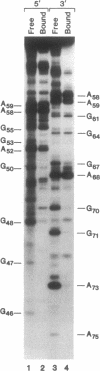
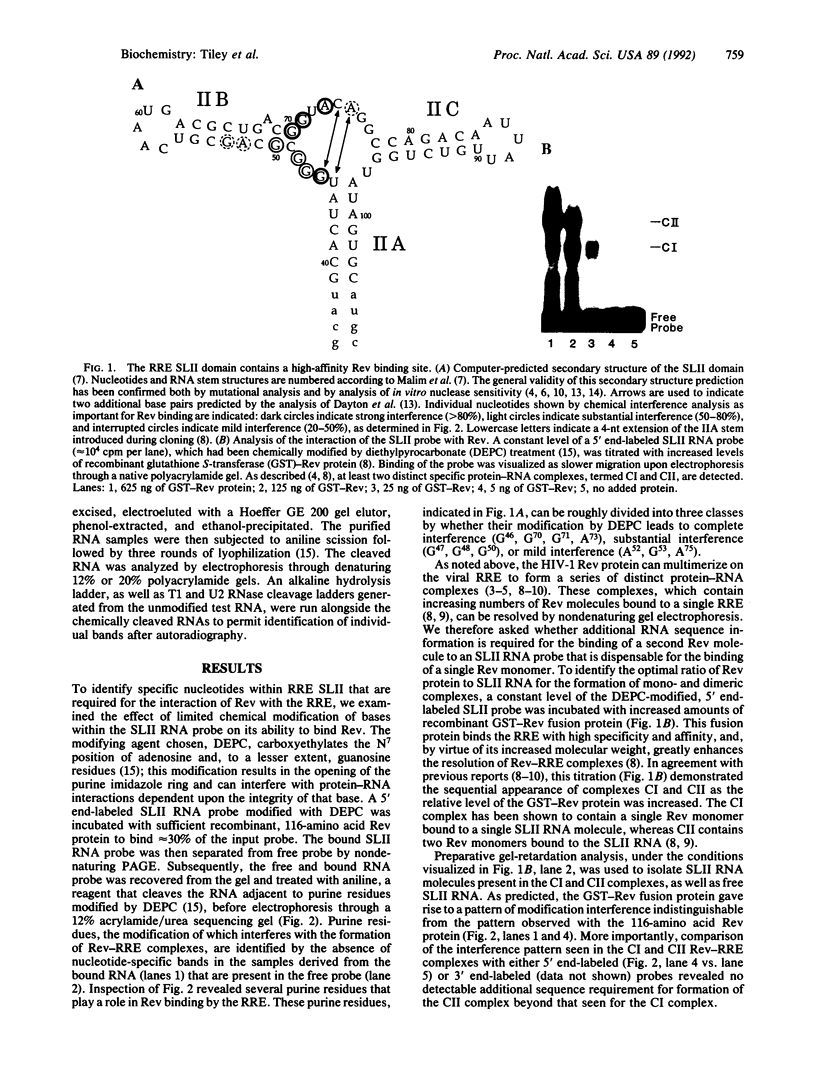
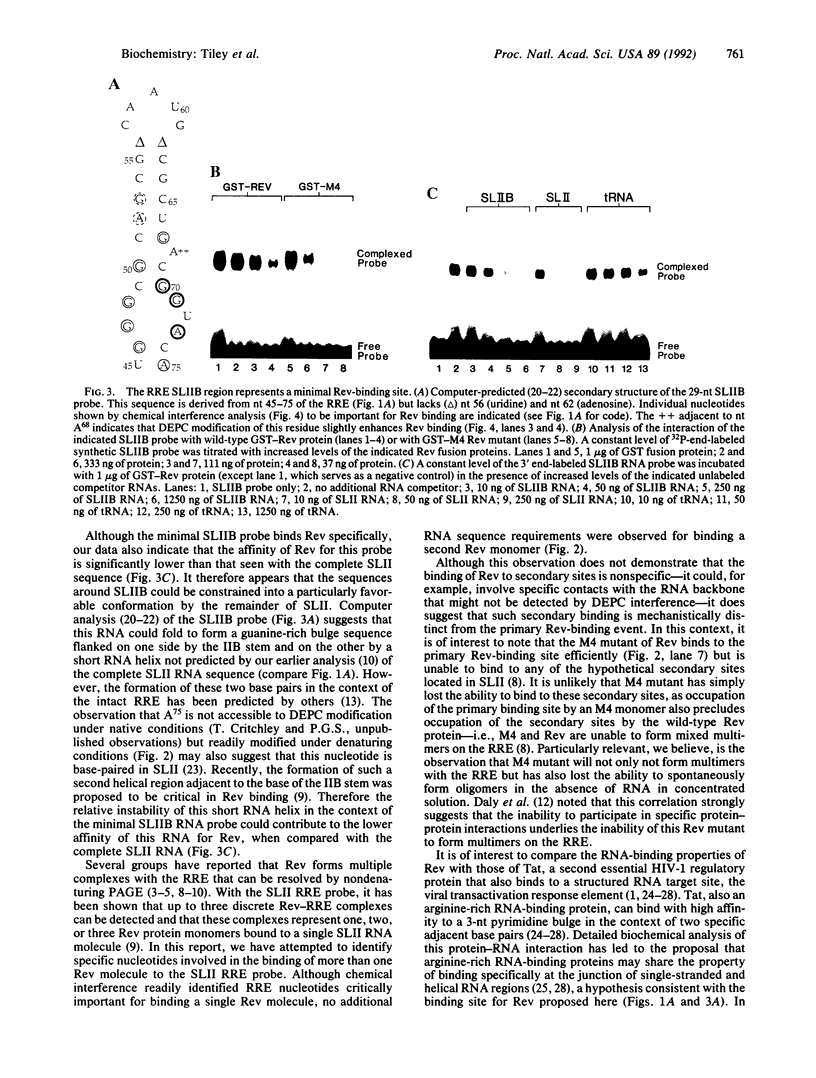
Abstract
Expression of the structural proteins of human immunodeficiency virus type 1 requires the direct interaction of multiple copies of the viral Rev protein with its highly structured RNA target sequence, the Rev response element (RRE). Nucleotides critical for Rev monomer binding have been mapped by chemical interference to a single site flanking the base of an RNA helix (stem IIB) located within the 234-nucleotide RRE. Binding of additional Rev molecules to an RRE probe did not require any RNA primary sequence information detectable by modification interference beyond that required for binding of a single Rev protein molecule. A synthetic 29-nucleotide RNA molecule designed to incorporate nucleotides identified as critical for Rev binding retained the ability to bind Rev specifically and, therefore, represents a minimal Rev-binding site. We propose that Rev binding to the RRE initiates with the direct interaction of a Rev monomer with a high-affinity binding site located at the base of the IIB stem of the RRE. The subsequent formation of Rev multimers on the RRE appears, in contrast, primarily driven by specific protein-protein interactions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Cook K. S., Fisk G. J., Hauber J., Usman N., Daly T. J., Rusche J. R. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991 Apr 11;19(7):1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Greene W. C. Regulatory pathways governing HIV-1 replication. Cell. 1989 Aug 11;58(3):423–426. doi: 10.1016/0092-8674(89)90420-0. [DOI] [PubMed] [Google Scholar]
- Daly T. J., Cook K. S., Gray G. S., Maione T. E., Rusche J. R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989 Dec 14;342(6251):816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- Dayton E. T., Powell D. M., Dayton A. I. Functional analysis of CAR, the target sequence for the Rev protein of HIV-1. Science. 1989 Dec 22;246(4937):1625–1629. doi: 10.1126/science.2688093. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy S., Dingwall C., Ernberg I., Gait M. J., Green S. M., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990 Feb 23;60(4):685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- Heaphy S., Finch J. T., Gait M. J., Karn J., Singh M. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich "bubble" located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S. M., Ahmad N., Maitra R. K., Wingfield P., Venkatesan S. Human immunodeficiency virus rev protein recognizes a target sequence in rev-responsive element RNA within the context of RNA secondary structure. J Virol. 1990 Dec;64(12):5966–5975. doi: 10.1128/jvi.64.12.5966-5975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. J., Hope T. J., Bond B. L., McDonald D., Grahl K., Parslow T. G. Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol. 1991 Apr;65(4):2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J., Brown M., Chang D. D., Sharp P. A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Malim M. H., Cullen B. R., Maizel J. V. A highly conserved RNA folding region coincident with the Rev response element of primate immunodeficiency viruses. Nucleic Acids Res. 1990 Mar 25;18(6):1613–1623. doi: 10.1093/nar/18.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M. H., Cullen B. R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991 Apr 19;65(2):241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Tiley L. S., McCarn D. F., Rusche J. R., Hauber J., Cullen B. R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990 Feb 23;60(4):675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- Olsen H. S., Cochrane A. W., Dillon P. J., Nalin C. M., Rosen C. A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990 Aug;4(8):1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- Olsen H. S., Nelbock P., Cochrane A. W., Rosen C. A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990 Feb 16;247(4944):845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Talbot S. J., Goodman S., Bates S. R., Fishwick C. W., Stockley P. G. Use of synthetic oligoribonucleotides to probe RNA-protein interactions in the MS2 translational operator complex. Nucleic Acids Res. 1990 Jun 25;18(12):3521–3528. doi: 10.1093/nar/18.12.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Wu T., Ogilvie K. K., Pon R. T. Prevention of chain cleavage in the chemical synthesis of 2'-silylated oligoribonucleotides. Nucleic Acids Res. 1989 May 11;17(9):3501–3517. doi: 10.1093/nar/17.9.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp M. L., Green M. R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989 Dec 7;342(6250):714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]