Abstract
Clubroot, caused by Plasmodiophora brassicae, is an important disease on Brassica species worldwide. A clubroot resistance gene, Rcr1, with efficacy against pathotype 3 of P. brassicae, was previously mapped to chromosome A03 of B. rapa in pak choy cultivar “Flower Nabana”. In the current study, resistance to pathotypes 2, 5 and 6 was shown to be associated with Rcr1 region on chromosome A03. Bulked segregant RNA sequencing was performed and short read sequences were assembled into 10 chromosomes of the B. rapa reference genome v1.5. For the resistant (R) bulks, a total of 351.8 million (M) sequences, 30,836.5 million bases (Mb) in length, produced 120-fold coverage of the reference genome. For the susceptible (S) bulks, 322.9 M sequences, 28,216.6 Mb in length, produced 109-fold coverage. In total, 776.2 K single nucleotide polymorphisms (SNPs) and 122.2 K insertion / deletion (InDels) in R bulks and 762.8 K SNPs and 118.7 K InDels in S bulks were identified; each chromosome had about 87% SNPs and 13% InDels, with 78% monomorphic and 22% polymorphic variants between the R and S bulks. Polymorphic variants on each chromosome were usually below 23%, but made up 34% of the variants on chromosome A03. There were 35 genes annotated in the Rcr1 target region and variants were identified in 21 genes. The numbers of poly variants differed significantly among the genes. Four out of them encode Toll-Interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat proteins; Bra019409 and Bra019410 harbored the higher numbers of polymorphic variants, which indicates that they are more likely candidates of Rcr1. Fourteen SNP markers in the target region were genotyped using the Kompetitive Allele Specific PCR method and were confirmed to associate with Rcr1. Selected SNP markers were analyzed with 26 recombinants obtained from a segregating population consisting of 1587 plants, indicating that they were completely linked to Rcr1. Nine SNP markers were used for marker-assisted introgression of Rcr1 into B. napus canola from B. rapa, with 100% accuracy in this study.
Introduction
Clubroot disease, caused by Plasmodiophora brassicae Woronin, can result in a 10–15% reduction in seed yield globally on Brassica species and related crops [1]. The pathogen belongs to the Infra Kingdom Rhizaria, which is a diverse group of amoeboid protists [2]. The characteristic symptom of clubroot is the development of large, disorganized growths (clubs) on the roots of susceptible plants, leading to wilting, stunted growth, and premature ripening [3]. The life cycle of P. brassicae consists of a primary phase in root hairs and a secondary phase in the cortex and stele [4].
On the Canadian prairies, clubroot was first identified on canola (B. napus L.) in 2003 and has spread rapidly. Genetic resistance is considered to be the most effective means for clubroot management [5–7]. Identification and genetic mapping of clubroot resistance genes have been carried out in B. rapa [8–23], B. oleracea [24–32] and B. napus [33–36]. It has also been assessed in Arabidopsis thaliana [37] and Raphanus sativus [38]. Two resistant genes, CRa and Crr1, have been isolated from Chinese cabbage lines of B. rapa. They encode Toll-Interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat (TIR-NBS-LRR; TNL) proteins [39, 40].
Lines of B. rapa with resistance to several pathotypes of P. brassicae [41] have been identified [42, 43] and could be used to broaden the genetic base of clubroot resistance in B. napus. Introgression of important agronomic traits from B. rapa into B. napus through conventional breeding methods is possible [44–48], so resistance to clubroot could be transferred from B. rapa into B. napus through interspecific crosses. Conventional methods are increasingly supplemented with marker-assisted selection (MAS) using single nucleotide polymorphisms (SNP) and insertion / deletion (InDel) markers because of their high efficiency [49, 50]. Also, SNP markers occur abundantly in genomes and are amenable for use in high-throughput detection platforms [51].
Next generation sequencing (NGS) has ignited a revolution in life sciences. NGS has had a major impact on crop improvement, allowing the rapid development and application of genomics tools in plant breeding [52]. It enables the inexpensive and rapid discovery of DNA variants in many crops, including Brassica species [53, 54]. Another important technique, bulked segregant analysis (BSA), which consists of genotyping two bulks of individual plants with extreme phenotypes, is used to identify markers linked to disease resistance genes [55]. BSA and NGS were recently applied to mapping and marker development for disease resistance loci in several crops [56–59]. To minimize costs for NGS sequencing of complex genomes, BSA has also been coupled with transcriptional profiles from RNA sequencing (RNA-seq) data for mapping of QTLs and genes of interest [60–62].
More than 10 clubroot resistance genes and more than 20 QTLs have been identified in Brassica species and close relatives. However, no robust SNP markers for MAS are available. Recently, Rcr1 was mapped to chromosome A03 of B. rapa based on resistance to pathotype 3 of P. brassicae, and differentially expressed genes associated with Rcr1 were identified via RNA-seq [11]. The objectives of this study were to assess the efficacy of Rcr1 against other pathotypes of interest to canola producers in Canada, and to use bulked segregant RNA-seq analysis to i) characterize genome-wide DNA variation; ii) examine DNA variation in the Rcr1 target region and identify the most probable candidates of Rcr1; iii) develop SNP markers associated with Rcr1; and iv) validate SNP markers to select for Rcr1 via MAS in canola (B. napus) breeding.
Materials and Methods
Development of F2 lines from a F1 segregating population and evaluation of the F2 lines for resistance to clubroot
Pak choy (B. rapa subsp. chinensis) cv. “Flower Nabana” (FN), a hybrid developed by Evergreen Y.H. Enterprises, Anaheim, CA, USA, is highly resistant to pathotype 3 of P. brassicae. Rcr1 was identified and genetically mapped in 1587 plants from a F1 population that segregated for resistance (R) and susceptibility (S) [11]. This population was derived from a cross between FN, which is heterozygous at Rcr1 locus, and canola line ACDC, which is a clubroot-susceptible B. rapa doubled haploid developed at the Saskatoon Research Centre of Agriculture and Agri-Food Canada. An additional 200 plants in the F1 population were self-pollinated, but most of the plants did not produce seed due to self-incompatibility in B. rapa. Seed was obtained from 38 plants, resulting in 38 F2 lines that could be evaluated for resistance to multiple pathotypes of P. brassicae. These lines were tested for resistance to pathotypes 2, 3, 5 and 6 under controlled conditions at the University of Guelph, ON, Canada. Plant seedlings were grown in soil-less mix in tall narrow plastic pots (164 mL conetainers, Stuewe & Sons Inc., Corvallis, OR, USA) and inoculated with 5 ml of resting spore suspension (1×106 spores ml−1) at 10 days after seeding. Roots of 7-14 plants per line were assessed for clubroot severity at 6 weeks after inoculation using a standard 0 to 3 scale where: 0 = no clubbing; 1 = small clubs only; 2 = moderate clubs; and 3 = severe clubbing. A disease severity index (DSI) [63, 64] was calculated using the following formula:
The highly susceptible Shanghai pak choy cv. “Mei Qing Choi” (Stokes Seeds, ON, Canada) was included as a susceptible control for each group to ensure that the inoculation was highly effective. Evaluation of the F2 lines and controls for resistance to the pathotypes of P. brassicae was replicated two times.
RNA-seq and sequence alignment
At 15-days post-inoculation, root tissue from 9 R plants was combined to form the R bulk and from 9 S plants to form the S bulk; together, the two bulks comprised one biological replicate as described by Chu et al. 2014 [11]. In total, three replicates were assessed. The total RNA from each replicate was isolated using an RNeasy Plant Mini Kit (Qiagen; Toronto, ON, Canada) with oncolumn deoxyribonuclease (DNase) digestion using a Qiagen RNase-Free DNase set following the manufacturer’s instruction. The cDNA library was prepared using TruSeq RNA Sample Preparation Kits v2 (Illumina; San Diego, CA, USA). RNA-seq was carried out using a Illumina Hiseq 2500 platform (Plant Biotechnology Institute, National Research Council, Saskatoon, SK, Canada) and global gene expression by the RNA-seq was conducted using the B. rapa reference genome v1.2 (B. rapa subsp. pekinensis cv. “Chiifu”) [11]. In the current study, the short reads from the RNA-seq were aligned to the B. rapa reference genome v1.5, which was downloaded at http://brassicadb.org/brad/downloadOverview.php. The reference genome consists of 10 chromosomes and 40,357 scaffolds. The total lengths of chromosomes and scaffolds are about 258 million bases (Mb) and 27 Mb, equivalent to about 90% and 10% of the reference genome, respectively. To simplify downstream data analysis, only the 258 Mb chromosome sequences were used in the current study, so the short reads from the RNA-seq project were assembled into 10 chromosomes A01 to A10 of B. rapa. The program SeqMan NGen 11 (DNASTAR, Madison, WI, USA) was used for short read assembly. Two methods were used: 1) short reads from three biological replicates each of the R and S bulks were assembled into the reference genome, resulting in six assembly files—hereafter, this method is called the single sample assembly (SSA); 2) short reads from a pool of the three R bulks and a pool of the three S bulks were assembled into the reference genome, resulting in two assembly files—hereafter called the pooled sample assembly (PSA). Standard assembling and filtering parameters were used.
Identification of variants
Discovery of variants (SNPs and InDels) in comparison with the DNA sequences in the B. rapa “Chiifu” [65] were performed using SeqMan Pro 11 (DNASTAR, Madison, WI, USA) with Q call ≥ 15 and depth ≥ 5. Analysis of variance and t-tests (LSD) were used to assess the variation in SNPs and InDels [66].
Genotyping SNP markers
Selected SNPs identified in the target region were confirmed using the Kompetitive Allele Specific PCR (KASP) method (http://www.lgcgroup.com/) following the manufacture’s instruction. PCR reactions were performed using a StepOne Plus Real Time PCR System (Applied Biosystem, Mississauga, ON, Canada). FN8, a line derived from the B. rapa cv. FN and homozygous at the Rcr1 locus, was included for this study.
To validate the SNP markers for MAS selection with Rcr1, DNA samples from 38 F1 plants from the cross between FN and the susceptible B. rapa line ACDC and 26 recombinants from the B. rapa population that was originally used to identify Rcr1 [11] were assessed. The accuracy of the SNP markers for use in MAS was determined by testing the recombinants from a BC1 population derived from B. napus introgressed with Rcr1 described by Chu et al, 2014 [11].
Results
Resistance to selected pathotypes
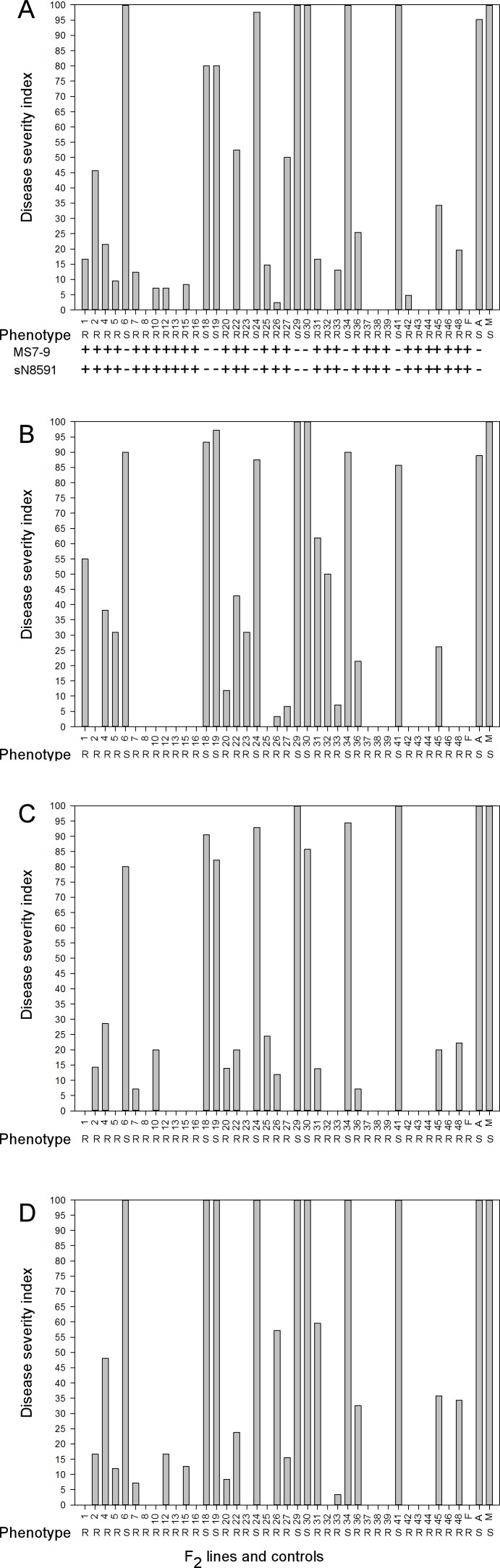
Rcr1 was initially identified based on resistance to pathotype 3 (Williams’ differentials) [41] of P. brassica, the most prevalent pathotype identified in western Canada [11]. The resistant donor FN was found to be resistant to multiple pathotyes of P. brassica [43]. To determine if resistance to other pathotypes in the R donor cv. FN was also associated with Rcr1, we tested 38 F2 lines derived from the F1 segregating population for resistance to pathotypes 2, 3, 5 and 6. As expected, FN conferred resistance to pathotype 3 (0 DSI), but ACDC (95 DSI) and “Mei Qing Choi” (100 DSI) were highly susceptible (Fig 1). Eight of the 38 lines (F2-6, 18, 19, 24, 29, 30, 34 and 41) were highly susceptible to pathotype 3 (severity > 80 DSI) so their parental F1 plants likely did not carry Rcr1. Severity was low to moderate in the other 30 lines (0-52 DSI), which indicates that the parental F1 plants for these lines likely carried the Rcr1 gene. To further confirm the F1 phenotypes determined by F2 lines, DNA samples from the original 38 F1 plants were assessed using two microsatellite markers, MS7-9 and sN8591, that flanked Rcr1 [11]. The 30 F1 parental plants of the F2 lines with low to moderate DSI carried the allele associated with resistance; the 8 F1 plants with high DSIs in their F2 lines carried the allele associated with susceptibility (Fig 1), confirming the phenotypes in the 38 F1 plants determined by DSIs in the F2 lines.
Fig 1. Evaluation of plants from 38 F2 lines for resistance to clubroot.
A, Plants were inoculated with pathotype 3 and confirmed with molecular markers MS7-9 and sN8591 flanking Rcr1 described by Chu et al [11]. “+”:heterozygous alleles, one allele from R parent and the other one from S parent, “-”:homozygous alleles from S parent; B, Inoculated with pathotype 2; C, Inoculated with pathotype 5; D, Inoculated with pathotype 6. Names of the lines indicate below X axis. Letters F, A, and M represent B. rapa cv. or breeding line FN, ACDC and “Mei Qing Choi”, respectively. Disease severity indexes (DSI) were calculated from clubroot severity ratings. R and S are the phenotypes assigned for each line based on DSIs. The experiments were replicated two times with similar results.
The clubroot reaction of the parental lines and the F2 lines to pathotypes 2, 5 and 6 was also assessed. The parental cv. FN was highly resistance to pathotypes 2, 5 and 6, but ACDC and “Mei Qing Choi” were highly susceptible (Fig 1). The eight F2 lines that were susceptible to pathotype 3 were also highly susceptible to pathotypes 2, 5 and 6 (> 80 DSI). In contrast, clubroot severity was low to moderate (0-55 DSI) in the other 30 lines (Fig 1). Therefore, we conclude that resistance to pathotypes 2, 5 and 6 of P. brassicae was associated with resistance to pathotype 3 that was used for identification of Rcr1 in the 38 lines.
RNA-seq and sequence alignment
The average sequence counts and average accumulated sequence length were 125.9 million (M) and 11,093.8 Mb per R bulk, and 114.0 M and 10,005.8 Mb per S bulk using the SSA method (Table 1). This provided an average depth of coverage of the reference genome of 43 fold in R bulks and 39 fold in S bulks. The average sequence length assembled into the reference genome was 88 bases. More sequences were assembled into the longer chromosomes A03 and A09, while fewer sequences were assembled into the shorter chromosomes A04 and A10 (Table 1). The sequence counts assembled into the genome for each chromosome were highly correlated to chromosome length (r = 0.90) for both R and S bulks.
Table 1. Short reads assembled into chromosomes of the reference genome B. rapa “Chiifu” in single sample assembly (SSA) and pooled sample assembly (PSA)a.
| Chromosome | Number of sequences (x 106) | Accumulated length of sequences (bases x 106) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Size (bases x 106) | SSA | PSA | SSA | PSA | ||||
| R | S | R | S | R | S | R | S | ||
| A01 | 26.9 | 12.9±1.4 | 11.8±3.1 | 35.9 | 33.2 | 1132.6±123.2 | 1029.6±271.6 | 3128.1 | 2881.3 |
| A02 | 27.0 | 11.6±1.2 | 10.4±2.7 | 32.1 | 29.1 | 1019.1±105.3 | 907.8±235.9 | 2807.9 | 2539.0 |
| A03 | 31.9 | 18.6±2.0 | 16.9±4.4 | 51.8 | 47.8 | 1627.9±173.8 | 1477.3±382.0 | 4516.7 | 4157.1 |
| A04 | 19.3 | 8.6±0.9 | 7.8±2.0 | 23.5 | 21.5 | 764.2±80.9 | 685.5±176.2 | 2070.0 | 1883.6 |
| A05 | 25.4 | 12.1±1.3 | 11.0±2.9 | 33.6 | 30.8 | 1067.5±116.3 | 963.7±251.3 | 2947.7 | 2695.6 |
| A06 | 25.3 | 13.9±1.4 | 12.5±3.3 | 38.8 | 35.7 | 1230.0±128.9 | 1098.6±291.1 | 3423.7 | 3133.5 |
| A07 | 25.9 | 12.0±1.3 | 10.9±2.8 | 33.9 | 31.1 | 1058.5±112.4 | 959.1±249.2 | 2977.3 | 2723.2 |
| A08 | 20.9 | 9.9±1.1 | 9.0±2.4 | 27.7 | 25.6 | 864.6±95.5 | 785.6±208.5 | 2420.6 | 2225.4 |
| A09 | 39.0 | 17.7±1.9 | 15.9±4.1 | 49.8 | 45.5 | 1561.7±169.9 | 1400.0±362.9 | 4369.2 | 3981.1 |
| A10 | 16.4 | 8.7±0.9 | 7.9±2.1 | 24.6 | 22.7 | 767.7±82.5 | 698.6±185.8 | 2175.4 | 1996.6 |
| Total | 257.9 | 125.9±13.4 | 114.0±29.8 | 351.8 | 322.9 | 11093.8±1188.2 | 10005.8±2614.0 | 30836.5 | 28216.6 |
a The data under SSA are mean ± SE.
Short reads from three R bulks and three S bulks were further assembled using the PSA method. As observed previously with the SSA method, more sequences were aligned into the longer chromosomes A03 and A09 and fewer sequences were aligned into the short chromosome A04 and A10 (Table 1). A total of 351.8 M sequences, 30,836.5 Mb in length, with coverage of 120 fold of the reference genome were assembled into B. rapa chromosomes from the pool of three R bulks, and 322.9 M sequences, 28,216.9 Mb in length, with 109 fold coverage were assembled from the pool of three S bulks (Table 1). About 50% of the aligned sequences were aligned to the top strand of the reference DNA and the other 50% to the bottom strand using the either the SSA or PSA methods (S1 Table).
Identification of variants
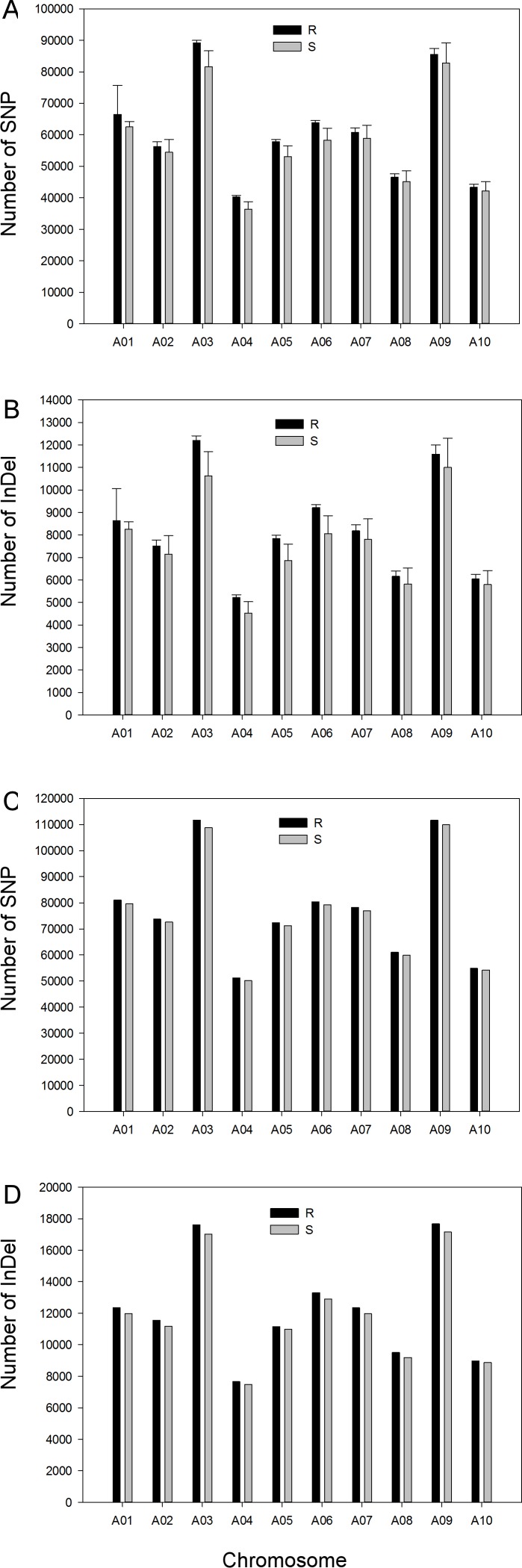
Variants in both R and S samples were very frequent, with means of about 605.9 K SNPs and 82.2 K InDels per R bulk and 579.4 K SNPs and 76.3 K InDels per S bulk by using the SSA method. There was a strong positive correlation between the numbers of SNPs compared to InDels (r = 0.99 in both R and S bulks). The numbers of SNPs and InDels varied among the chromosomes (Fig 2), indicating that the longer chromosomes (A03, A09) carried more variants than the shorter chromosomes (A04, A10). However, the proportion of SNPs and InDels was similar among chromosomes, with about 88% and 12%, respectively, in both R and S bulks (S1 Fig).
Fig 2. Numbers of variants (SNPs and InDels) identified in R and S bulks in comparing with the DNA sequence of the B. rapa cv. “Chiifu”.
A, Number of SNPs by single sample assembly (SSA); B, Number of InDels by SSA; C, Number of SNPs by pooled sample assembly (PSA); D, Number of InDel by PSA.
The numbers of SNPs and InDels identified using the PSA method were higher than with the SSA method, with 776.2 K SNPs and 122.2 K InDels in the R bulk, and 762.8 K SNPs and 118.7 K InDels in the S bulk. As with the SSA method, there was a strong positive correlation between the numbers of SNPs and InDels with the correlation coefficient (r = 0.99) in either R or S bulks. The proportion of SNPs and InDels on each chromosome was 87% and 13% in both the R and S bulks (S1 Fig), which is a slightly lower proportion of SNP and higher proportion of InDel identified using the PSA method than the SSA method. As observed with the SSA method, the numbers of SNPs and InDels identified using PSA method (Fig 2) were higher on the longer chromosomes.
Comparison of variants in R and S bulks
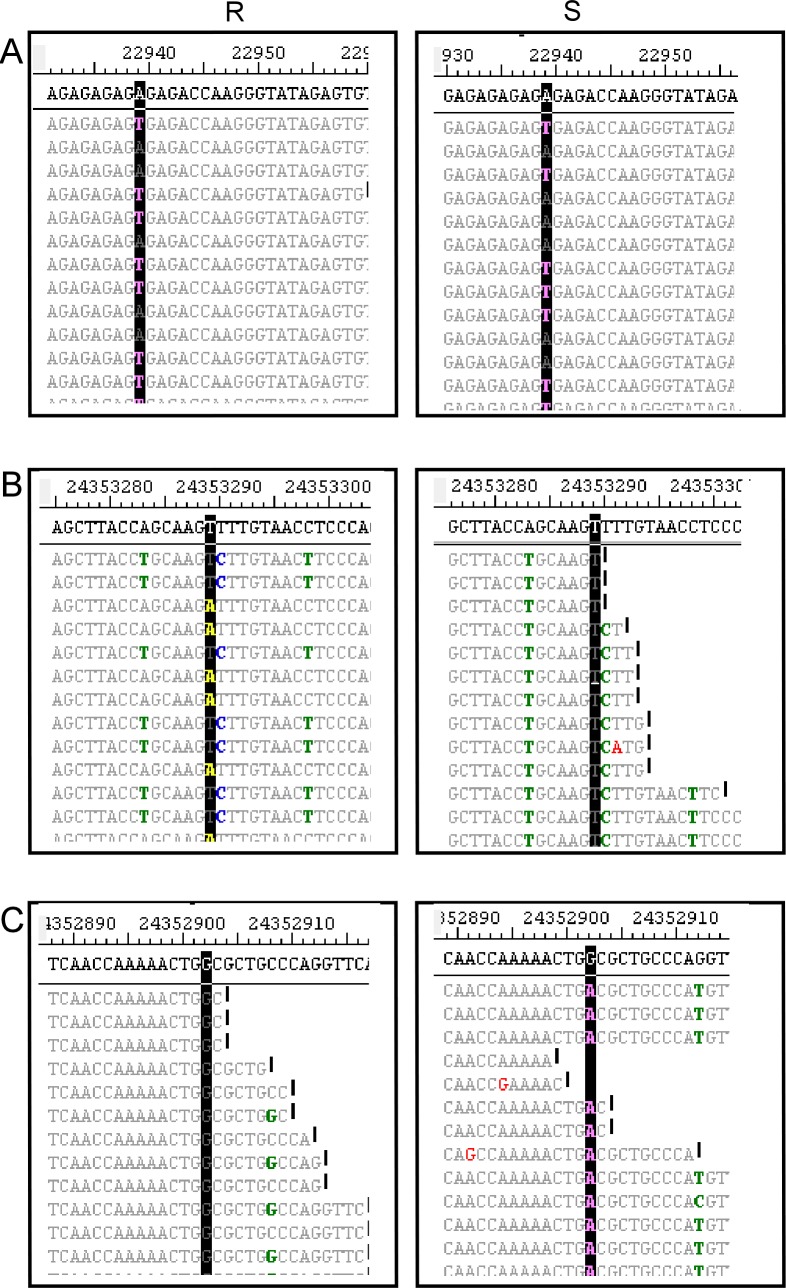
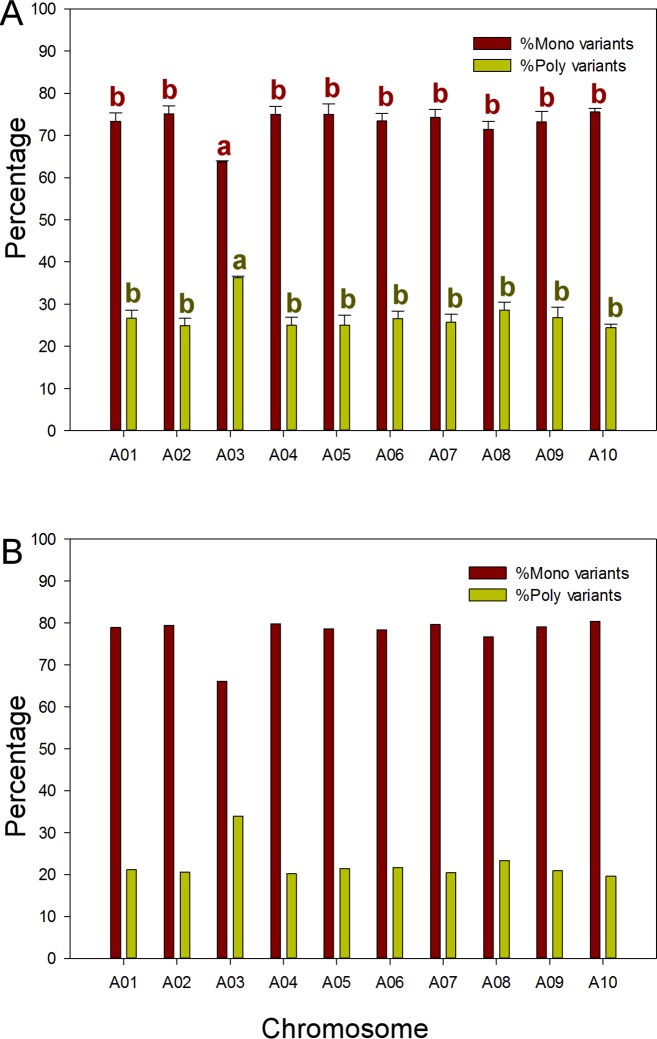
Variants identified in the R and S samples could be the same (monomorphic, mono) or different (polymorphic, poly) (Fig 3). Mono variants comprised 73% (range 63.7-75.6%) of the variants identified across the B. rapa genome using the SSA method, and poly variants 27% (range 24.4%-36.3%). There were no differences among chromosomes in the proportion of mono and poly variants except for chromosome A03, where the clubroot resistance gene Rcr1 is located, which carried more ploy variants and fewer mono variants (64%, P ≤ 0.05) than the other chromosomes (Fig 4).
Fig 3. Examples of polymorphic and monomorphic variants on B. rapa chromosome A03 displayed by SeqMan Pro (DNASTAR, Madison, WI, USA).
A, Monomorphic variant at the location 22939; SNP “T” in both R and S bulks; B, Polymorphic variant at the location 24353288; SNP “A” in R, but not in S; C, Polymorphic variant at the location 24352902; SNP “A” in S, but not in R. SNPs are highlighted in black. The numbers in the first row of each snapshot are the physical location on chromosome A03 in the B. rapa reference genome. The DNA sequences shown in the second rows are the reference genome sequences. The sequences below the reference genome are the short reads from either R or S bulks assembled into the reference genome.
Fig 4. The proportion (%) of monomorphic and polymorphic variants on each chromosome.
A, Using the single sample assembly method; and B, Using the pooled sample assembly method. Bars capped with the same letter do not differ at P ≤ 0.05.
With the PSA method, the proportion of mono variants was higher than for the SSA method for each chromosome (Fig 4). Overall, approximately 5% of the poly variants identified by SSA method were identified as mono using the PSA method. On chromosome A03, there were 66% mono variants and 34% poly variants, very similar to the results from SSA (Fig 4).
Analysis of variants in the target region
A previous study reported that Rcr1 was mapped in an interval of 0.28 centi-Morgan on B. rapa chromosome A03 and located in the 24.26-24.54 Mb of A03 [11]. In the current study, the target region was first analyzed using the SSA method. There were 35 genes annotated in the region, based on the reference genome v1.5 (Table 2). Three genes (Bra038750, Bra019396 and Bra019394) did not show any expression and no short reads were assembled into the reference genome, so no variants could be identified. Short reads with sequence counts < 20 and number of variants < 1 was associated with 11 genes (Bra038751, Bra038747, Bra019411, Bra019408, Bra019404, Bra019402, Bra019401, Bra019399, Bra019397, Bra019393 and Bra019392). More than 20 sequence counts with > 1 variants were associated with 21 genes in both R and S bulks (Table 2). Information on BLASTX (best hit) to A. thaliana and gene ontology annotation for the genes was obtained from http://brassicadb.org/brad/index.php (S2 Table). Four genes (Bra019409, Bra019410, Bra019412 and Bra019413) encode TNL-class disease resistance proteins.
Table 2. Characterization of genes in the Rcr1 target region based on location, number of sequences, and number of variants in the resistant (R) and susceptible (S) bulked samples.
| Genea | Location on A03 | Number of sequencesb | No. of variantsb | ||
|---|---|---|---|---|---|
| R | S | R | S | ||
| Bra038758 | 24263024…24264157 | 193.3±4.5 | 118.3±3.9 | 1.7±0.1 | 0.3±0.1 |
| Bra038757 | 24275881…24279414 | 289.0±5.7 | 248.3±9.8 | 28.3±0.6 | 19.0±0.9 |
| Bra038756 | 24280484…24283179 | 1610±12.6 | 2170.0±125.4 | 18.3±0.1 | 10.0±0.2 |
| Bra038755 | 24284960…24288356 | 287.7±6.2 | 481.0±23.4 | 14.0±0.5 | 7.0±0.4 |
| Bra038754 | 24290155…24292149 | 5085.7 ±81.4 | 3473.0±156.9 | 33.7±0.2 | 26.0±0.5 |
| Bra038753 | 24294019…24295276 | 2640±63.3 | 2456±100.3 | 19.7±0.2 | 17.7±0.1 |
| Bra038751 (L) | 24296257…24296659 | 4.3±0.2 | 3.7±0.1 | 0.0±0.0 | 0.3±0.1 |
| Bra038750(N) | 24303342…24304157 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra038749 | 24306906…24310339 | 629.3±18.6 | 700.3±28.8 | 9.3±0.1 | 8.7±0.2 |
| Bra038748 | 24323537…24327067 | 3393.3±84.2 | 2970.0±145.3 | 6.3±0.1 | 11.7±0.1 |
| Bra038747 (L) | 24339450…24340813 | 12.0±0.7 | 32.7±1.8 | 0.0±0.0 | 0.3±0.1 |
| Bra019413 | 24350950…24353977 | 615.3±14.4 | 101.7±5.6 | 37.3±0.2 | 14.0±0.9 |
| Bra019412 | 24370531…24371199 | 351.3±2.0 | 115.7±6.4 | 5.0±0.1 | 4.3±0.2 |
| Bra019411 (L) | 24372280…24372516 | 0.7±0.1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019410 | 24373815…24379176 | 1156±18.4 | 493.7±25.6 | 94.7±0.5 | 45.3±1.8 |
| Bra019409 | 24381590…24386315 | 548.0±10.5 | 710.7±36.3 | 71.3±0.5 | 57.7±1.4 |
| Bra019408 (L) | 24388976…24389449 | 1.3±0.1 | 1.3±0.2 | 0.0±0.0 | 0.0±0.0 |
| Bra019407 | 24391878…24392811 | 745.3±20.3 | 951.0±47.4 | 3.3±0.1 | 2.7±0.1 |
| Bra019406 | 24393210…24394793 | 4919.3±199.0 | 4304.3±212.0 | 46.7±0.1 | 30.0±0.0 |
| Bra019405 | 24403694…24404722 | 35.7±1.9 | 73.3±4.6 | 0.7±0.1 | 2.0±0.1 |
| Bra019404 (L) | 24409283…24411164 | 10.0±0.4 | 6.7±0.4 | 0.0±0.0 | 0.3±0.1 |
| Bra019403 | 24413728…24415552 | 70.3±1.2 | 2.0±0.2 | 16.3±0.3 | 0.3±0.1 |
| Bra019402 (L) | 24429989…24430549 | 1.3±0.2 | 2.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019401 (L) | 24436080…24436250 | 0.7±0.2 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019400 | 24437662…24439848 | 32.3±4.8 | 325.0±25.0 | 0.7±0.2 | 6.0±0.5 |
| Bra019399 (L) | 24444666…24444989 | 0.0±0.0 | 1.0±0.3 | 0.0±0.0 | 0.0±0.0 |
| Bra019398 | 24454780…24457677 | 45.3±2.3 | 22.3±2.0 | 3.7±0.4 | 0.3±0.1 |
| Bra019397 (L) | 24480924…24483948 | 4.0±1.0 | 2.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019396(N) | 24501547…24502262 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019395 | 24510450…24512273 | 1327.0±43.9 | 1127±99.0 | 11.3±0.1 | 2.3±0.1 |
| Bra019394(N) | 24517143…24519546 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 |
| Bra019393 (L) | 24520116…24521078 | 20.0±0.6 | 1.0±0.4 | 0.7±0.3 | 0.0±0.0 |
| Bra019392 (L) | 24525560…24526147 | 12.7±0.9 | 10.7±2.3 | 0.0±0.0 | 0.0±0.0 |
| Bra019391 | 24529596…24530189 | 57.3±8.0 | 83.7±10.7 | 1.0±0.3 | 4.7±0.7 |
| Bra019390 | 24534962…24537043 | 7449.0±1215.0 | 6775.0±1473.0 | 32.0±1.2.0 | 24.7±2.0.0 |
a(L) = Little expression; (N) = No expression
bMean ± SE.
The numbers of poly variants between the R and S bulks among the genes in the coding regions was assessed because the poly variants represent differences in the DNA sequences between the R and S bulks. The numbers of poly variants differed significantly among the 21 genes (Table 3).Three TNL genes (Bra019409, Bra019410, and Bra019413) had the most poly variants, with means of 81.7, 75.0 and 34.7 poly variants per gene, respectively, followed by four genes (Bra019406, Bra038754, Bra019390 and Bra038757) with means of 33.3, 31.3, 29.0 and 24.0. These seven genes exhibited more poly variants than the other 14 genes (P ≤ 0.05). The fourth TNL-class gene (Bra019412), with mean of 4.0 poly variants, was one of the 14 genes that showed few polymorphic variants (Table 3).
Table 3. Number of polymorphic variants identified in each gene located in the Rcr1 interval in the simple sequence assembly (SSA) and pooled sequence assembly (PSA) analysis.
| Gene | No. of variants (SSA) a | No of variants (PSA) |
|---|---|---|
| Bra019409 | 81.7±1.2 a | 115 |
| Bra019410 | 75.0±0.9 b | 85 |
| Bra019413 | 34.7±0.4 c | 52 |
| Bra019406 | 33.3±0.1 cd | 37 |
| Bra038754 | 31.3±0.5 cd | 39 |
| Bra019390 | 29.0±0.8 de | 38 |
| Bra038757 | 24.0±0.3 e | 31 |
| Bra019403 | 16.7±0.5 f | 18 |
| Bra038753 | 14.3±0.3 fg | 20 |
| Bra038755 | 13.0±0.6 fgh | 17 |
| Bra038756 | 11.3±0.2 gh | 9 |
| Bra019395 | 9.7±0.3 ghi | 12 |
| Bra038748 | 8.0±0.1 hij | 11 |
| Bra019391 | 5.7±0.8 ijk | 10 |
| Bra038749 | 5.3±0.2 ijk | 5 |
| Bra019400 | 5.3±0.9 ijk | 4 |
| Bra019412 | 4.0±0.4 jk | 4 |
| Bra019398 | 4.0±0.5 jk | 9 |
| Bra038758 | 2.0±0.1 k | 3 |
| Bra019407 | 1.7±0.1 k | 3 |
| Bra019405 | 1.3±0.1 k | 2 |
a Mean (± SE) followed by the same letter do not differ at P ≤ 0.05.
The PSA method identified more poly variants among the 21 genes in the target region than the SSA method (Table 3). For both methods, three TNL genes (Bra019409, Bra019410, and Bra019413) consistently carried the highest number of poly variants among the 21 genes.
Analysis of variant types in the TNL genes
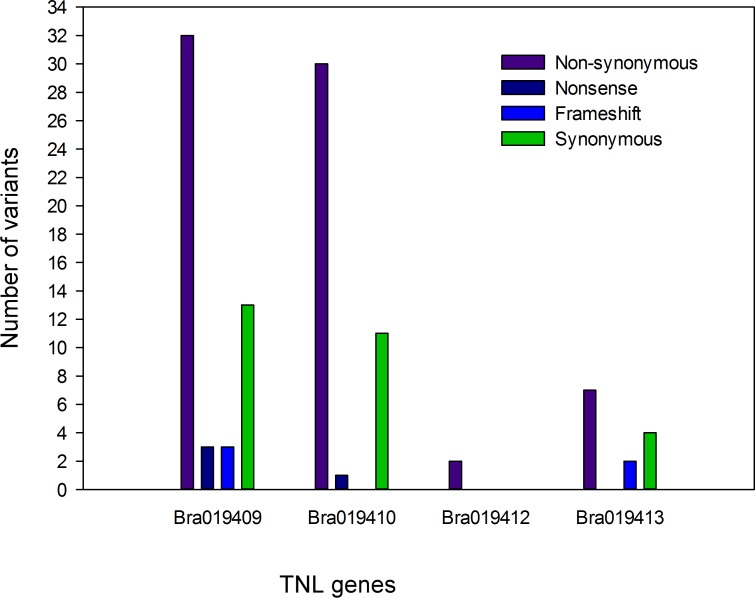
The poly variants that uniquely occurred in the R bulks but not in the S bulks (Fig 3) with depth > 5 in both samples were further assessed using the PSA method. A total of 108 poly variants were identified from the coding sequences in the four TNL genes (S3 Table). SeqMan Pro software was used to sort the 108 variants that affect amino acid sequences into four groups: non-synonymous, nonsense, frameshift and synonymous variants. Non-synonymous variants occurred in each of the TNL genes, but Bra019409 and Bra019410 carried many more non-synonymous variants than the other two TNL genes (Fig 5). Three nonsense variants were identified in Bra019409 and one in Bra019410, but none were identified in the other two genes. Frameshift variants caused by InDels were found in Bra019409 and Bra019413, and synonymous variants occurred in each of the genes except Bra019412 (Fig 5). Overall, Bra019409 and Bra019410 carried higher number of poly variants that uniquely occurred in the R samples and could affect the amino acid sequence in their proteins.
Fig 5. Number of unique poly variants in the four TIR-NBS-LRR genes from the resistant bulked samples.
Genotyping SNP markers identified in the Rcr1 region
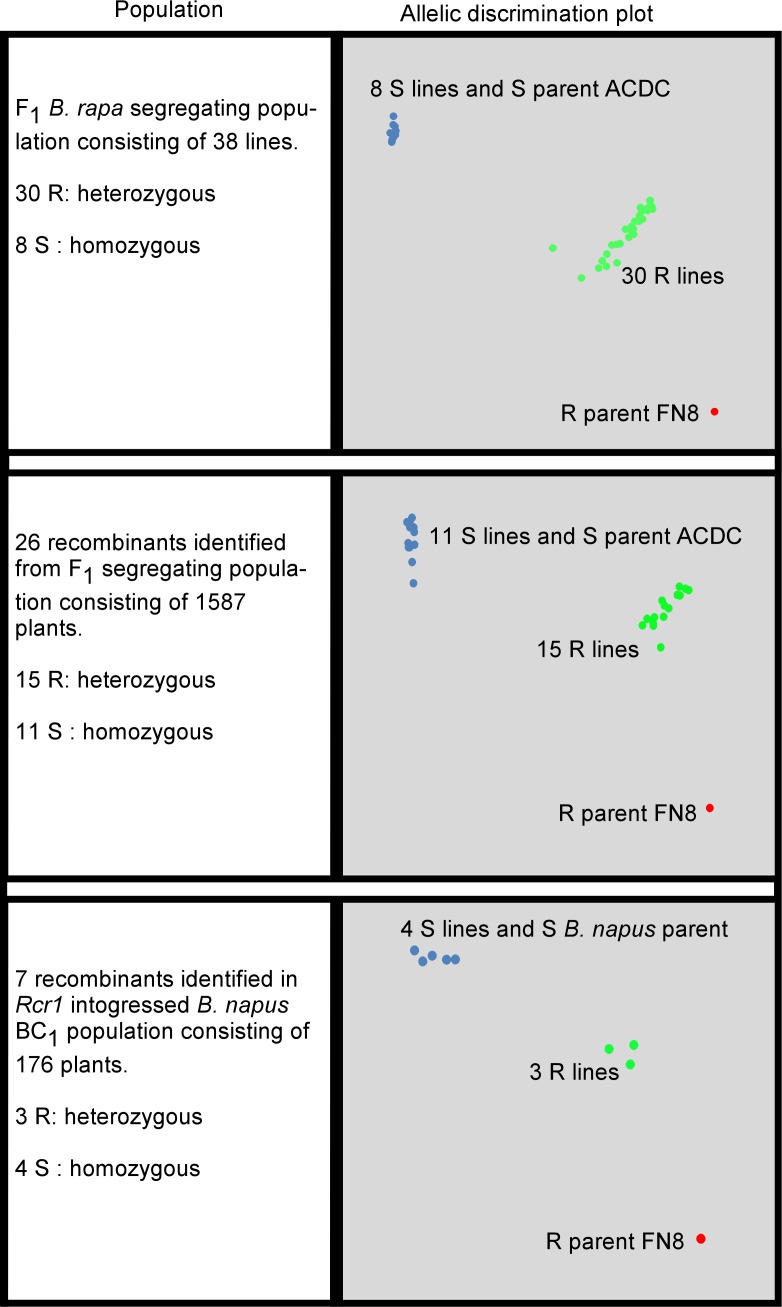
The DNA sequences flanking 11 SNPs in the four TNL genes (S3 Table) and three SNPs (SNP_A03_18, 19, and 32) in the gene Bra019406 that was one of genes harboring more poly variants (Table 3) were obtained from the B. rapa reference genome (S4 Table). These SNP markers, which spanned about 42 Kb in the target region (Table 4), were genotyped using the KASP method. As expected, each SNP marker was polymorphic between the R parental line FN8 and the S parental line ACDC (Fig 6), which confirmed that a SNP was present at each locus. In an allelic discrimination plot, PCR products from the FN8 (R) were usually close to the right bottom and ACDC (S) to the left top (Fig 6). This indicates that they carried homozygous alleles that might be associated with R and S, respectively. In contrast, when DNA samples from the 38 F1 plants (Fig 1) were analyzed using the 14 SNP markers, the PCR products from 30 R plants were in the middle (Fig 6). This indicates that these plants were heterozygous at this locus (Table 4), with one allele from the R parent and the other one from the S parent. PCR products from the eight S plants were close to the right top (Fig 6), where the PCR product from the susceptible line ACDC was located, indicating that they carried homozygous alleles from the S parent (Table 4). These results confirm that the 14 SNP markers identified by RNA-seq were completely associated with Rcr1 on chromosome A03 in this population.
Table 4. Association between disease reaction phenotype and SNP markers in the F1 segregating populations.
| SNP marker | Gene | A03 locationa | Phenotypeb | Number of plants with markerc | |
|---|---|---|---|---|---|
| + | - | ||||
| SNP_A03_11 | Bra019413 | 24352616 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_104 | Bra019413 | 24352908 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_08 | Bra019413 | 24353289 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_09 | Bra019412 | 24371044 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_102 | Bra019410 | 24375018 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_12 | Bra019410 | 24375572 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_13 | Bra019410 | 24375979 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_14 | Bra019410 | 24376507 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_103 | Bra019410 | 24378671 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_16 | Bra019409 | 24382012 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_101 | Bra019409 | 24382587 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_18 | Bra019406 | 24393429 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_19 | Bra019406 | 24393825 | R | 30 | 0 |
| S | 0 | 8 | |||
| SNP_A03_32 | Bra019406 | 24394435 | R | 30 | 0 |
| S | 0 | 8 | |||
aPhysical location on chromosome A03 based on Brassica rapa reference genome v1.5.
bPhenotypes in 38 F1 plants were determined in F2, as shown in Fig 1.
c Genotyping SNP markers in the Rcr1 target region using KASP method: “+” allele from the resistant parent FN; “-” allele from the susceptible parent ACDC.
Fig 6. Allelic discrimination plots show the genotypes of three populations based on SNP marker, SNP_A03_12, using the KASP method.
ACDC and FN8 are homozygous susceptible and resistant lines, respectively.
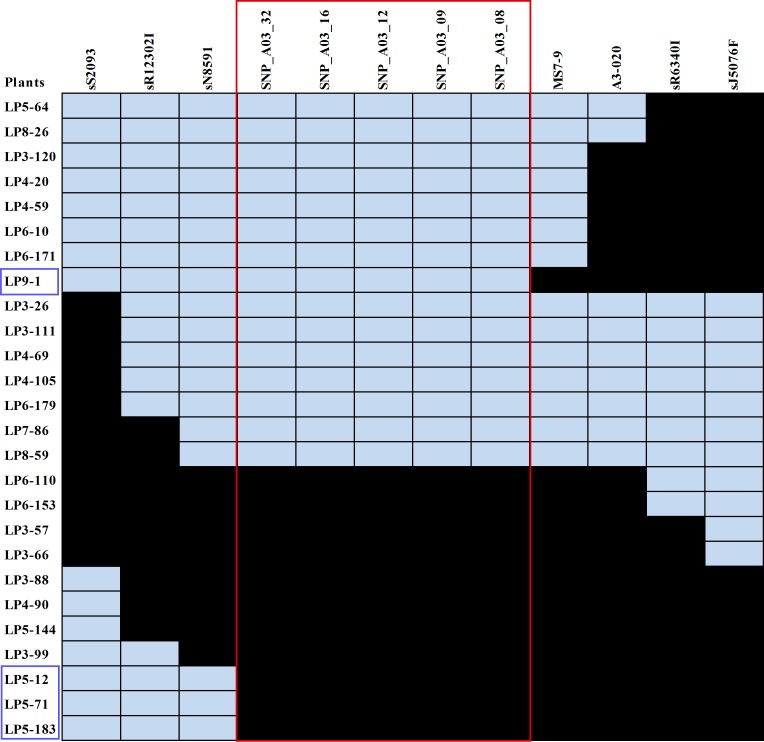
To determine if the SNP markers were located in the target region, we analyzed 26 recombinants, including LP9-1, LP5-12, LP5-71 and LP5-183 that were identified by the closest flanking markers, MS7-9 and sN8591 from 1587 F1 plants with five SNP markers (SNP_A03_08, 09, 12, 16, and 32) that reside in the genes Bra019413, Bra019412, Bra019410, Bra019409 and Bra019406 respectively (Fig 6). Each SNP marker co-segregated with phenotypes and no recombination event was found in these plants (Fig 7). This indicates that the SNP markers were completely associated with the clubroot resistance gene Rcr1.
Fig 7. Analysis of the recombinants selected from the F1 population derived from the cross ACDC x FN.
Phenotypes (R for resistant, S for susceptible) on the right, molecular markers (sS2093, sR12302I, sN8591, MS7-9, A3-020, sR6340I and sJ5076F) on the top were described previously [11]. The recombinants were analyzed with five SNP markers (SNP_A03_08, 09, 12, 16 and 32) residing in the genes Bra019413, Bra019412, Bra019410, Bra019409 and Bra019406 respectively using the KASP method. They are highlighted in the red rectangle. PCR products amplified from R alleles are denoted in light grey and those from S alleles in black. The four recombinants in blue rectangles were identified with the closest flanking markers sN8591 and MS7-9 [11].
Validation of SNP markers for MAS of Rcr1 in canola breeding
To assess the potential use of the SNP markers in MAS for introgression of Rcr1 into canola, DNA samples were assessed from seven recombinants in the BC1 populations consisting of 176 plants, which were previously used for validating Rcr1 linked to microsatellite markers for MAS [11], with the 14 SNP markers (Table 4). Five SNP markers (SNP_A03_16, 18, 19, 32, 103) were polymorphic in the B. rapa population; however, they were monomorphic in the B. napus population. Polymorphism was identified in the rest of 9 SNP markers (Table 5). The SNP alleles from the R parent occurred in 3 R plants (BN-4, 27, and 58) and the alleles from the S parent occurred in 4 S plants (BN-1, 86, 103, and 113) (Fig 6; Table 5). The accuracy for MAS in the BC1 population using the 9 KASP SNP markers was 100%, and the SNP markers completely co-segregated with the phenotypes.
Table 5. Validation of SNP markers for detecting Rcr1 in BC1 populations derived from a cross of FN x Brassica napus described by Chu et al [11] a.
| Plant | FN | B. napus | BN-01 | BN-04 | BN-27 | BN-58 | BN-86 | BN-103 | BN-113 |
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | R | S | S | R | R | R | S | S | S |
| SNP_A03_11 | + | - | - | + | + | + | - | - | - |
| SNP_A03_104 | + | - | - | + | + | + | - | - | - |
| SNP_A03_08 | + | - | - | + | + | + | - | - | - |
| SNP_A03_09 | + | - | - | + | + | + | - | - | - |
| SNP_A03_102 | + | - | - | + | + | + | - | - | - |
| SNP_A03_12 | + | - | - | + | + | + | - | - | - |
| SNP_A03_13 | + | - | - | + | + | + | - | - | - |
| SNP_A03_14 | + | - | - | + | + | + | - | - | - |
| SNP_A03_101 | + | - | - | + | + | + | - | - | - |
a “+” allele from the R parent FN; “-” from S parent B. napus.
Discussion
More than 10 clubroot resistance genes including CRa, CRb, CRc, CRk, Crr1, Crr2, Crr3 and Crr4 were identified previously in vegetable cultivars of B. rapa [9, 10, 12–23]. These genes were identified based on the reaction to the P. brassicae pathotypes prevalent in Japan, Korea and China, which would likely be classed into pathotypes 2 or 4 based on Williams’ differential hosts [41]. At least five pathotypes of P. brassicae (2, 3, 5, 6 and 8, again based on Williams’ differentials) occur in Canada. Pathotype 3 is the most prevalent on canola [67, 68].
Rcr1 was previously mapped on chromosome A03 based on resistance to pathotype 3 [11], but the donor cv. FN was resistant to each of the pathotypes previously identified in Canada [43]. To determine if Rcr1 confers resistance against the range of Canadian pathotypes, a F2 population consisting of 38 lines was developed. The F2 lines were equivalent to self-pollinated BC1 populations (BC1S1). There were two genotypes “Rr”, heterozygous at Rcr1 locus, which should be resistant and “rr”, homozygous susceptible alleles, which should be susceptible phenotype in F1. Theoretically, segregation for resistance and susceptibility from “Rr” plants in the F2 lines is 3:1 and that from “rr” should be 0:1. Ideally, X2 analysis would have been performed. Unfortunately, the small number of plants available for evaluation of plant resistance in this study made X2 analysis impractical. We speculated that the eight F2 lines with high severity were from “rr” F1 plants and the 30 F2 lines with low to moderate severity were from “Rr” F1 plants. Similar results were obtained by repeating the experiments. In addition, the results were confirmed by analysis of the molecular markers (Fig 1, Table 4). Thus, the genotypes of the F1 plants determined by the phenotypes in F2 lines in this study were reliable, even though 7 to 14 plants each line for each pathotype were tested per replicate. In some of the F2 lines that were derived from “Rr” F1 plants, their DSIs were as low as zero. This could be caused by segregating distortion so no “rr” plants appeared in the small populations. In addition, disease escapes from “rr” plants could contribute to the zero DSIs. Resistance to pathotypes 2, 3, 5 and 6 was completely linked in each of the 38 lines. However, variation was usually found among DSIs of the same F2 lines responding to different pathotypes. The reason for the variation could be that the proportions of the genotypes (RR, Rr and rr) for those “Rr” derived F2 lines against each pathotype were not exactly same. It also could be caused by some escapes from “rr” plants especially for those “rr” derived F2 lines. The original intent was to assess 200 F1 plants in this study, but only these 38 plants produced sufficient seed in the F2 generation for assessment. Resistance to pathotype 8 was also associated with resistance to pathotype 3 in one iteration of the assessment (data not show), but there was not sufficient seed to repeat that portion of the study. Resistance to this range of pathotypes indicates that Rcr1 could confer broad spectrum resistance to many of the pathotypes of P. brassicae that are present in Canada. Another possibility is that the resistance to this range of pathotypes could be controlled by linked genes. Clearly, this still needs to be determined.
RNA-seq generates a wealth of short DNA sequence reads from random places in the transcriptome. Transcriptome analysis and identification of SNPs based on RNA-seq have been carried out recent years in many organisms, including Brassicas such as B. napus [53, 69, 70] and B. rapa [11, 54]. The current study focused on characterization of variants in the B. rapa population that carried Rcr1, and identified the most probable candidates for Rcr1 based on DNA variants and development of SNP markers tightly linked to the gene for MAS. Also, SNP variants were found to be the most common DNA sequence variation, accounting for over 80% of the B. rapa transcriptome in the current study. Similar results were reported in rice, with an average of 99,955 putative SNPs and 14,617 putative InDels [71] and the human genome, where SNPs made up about 3 million of the 4 million DNA sequence [72]. Recently, RNA-seq of human airway epithelial cells revealed that over 90% of variants were SNPs [73].
A wealth of SNPs and InDels were identified in the B. rapa genome using both the SSA and PSA approaches, which generally produced similar results. The SSA method, in which three biological replicates of R and S bulks were assessed, provided estimates of variance that could be used to assess the statistical significance of variation of SNPs and InDels. This approach is very useful for identifying differentially expressed genes through RNA-seq. In contrast, the PSA approach, which merged the data across biological replicates, is much less amenable to statistical analysis. However, it produced a greater depth of sequencing data, providing more reliable results in discovery of variants. Therefore, this approach would be preferred in future SNP discovery through bulked segregant RNA sequencing.
In both R and S samples, variants from the reference genome DNA sequences in B. rapa Chinese cabbage “Chiifu” v1.5 [65] were very frequent. One factor that likely contributed to this high frequency of variants was that the parental lines were pak choy and canola, rather than Chinese cabbage. Deep sequencing, with coverage of 120 fold of the reference genome in R and 109 fold in S samples, could also have contributed to the wealth of variants in the current study. More than 70% of the variants between R and S bulks were monomorphic in all chromosomes except A03, where a significantly higher percentage of polymorphic variants were present. The high frequency of poly variants could indicate that there was a higher level of difference between the R and S bulks in Rcr1 and its surrounding region than in other parts of genome.
Rcr1 was fine mapped in an interval of 0.28 centi-Morgan, flanking by microsatellite markers MS7-9 and sN8591 on B. rapa chromosome A03 [11]. In this study, we identified more than 400 variants (Table 3) in the target region. Fourteen SNP markers that reside in five genes (Bra019406, Bra019409, Bra019410, Bra019412 and Bra019413) in the target region were genotyped using KASP method. Each of the markers co-segregated with Rcr1, indicating that they were completely linked to Rcr1 and closer to Rcr1 than the markers previously described by Chu et al 2014 [11]. However, it was not possible to define the Rcr1 location further with the recombinants in the mapping population, because no recombination events could be identified in the set of recombinants. Therefore, it was essential to obtain accurate SNP and Indel profile on the genes of interest through analysis of RNA-seq data. Four of the five genes identified encode proteins related to disease resistance that belong to TNL families. Non-synonymous, nonsense, frameshift and synonymous variants were identified in the TNL genes. It is unlikely that the genes Bra019412 and Bra019413 are the candidates for Rcr1; although they showed differential expression between R and S bulks [11], few poly variants, especially variants that cause changes in amino acid sequence, were identified in these genes. Bra019409 and Bra019410 carried higher number of variants that could cause changes in amino acid sequence and higher number of synonymous variants, previously considered as 'silent', but which can cause changes in protein expression, conformation and function [74]. Therefore, it is more likely that either of these genes is a candidate for Rcr1. It is also possible that a combination of the TNL genes is required for the expression of Rcr1 resistance. Rcr1 was mapped in the same interval as one of the cloned clubroot resistance gene CRa [11]. However, it was not illustrated how CRa was related to the TNL genes in the target region [38]. The complete CRa coding sequence is 4223 bp (GenBank: AB751517.1). We searched the B. rapa genome using the 4223 bp sequence at http://brassicadb.org/brad/blastPage.php and found that CRa was homologous to the four TNL genes Bra019410, Bra019412, Bra019409 and Bra019413 with score and E value at 1905, 0.0; 823, 0.0; 739, 0.0 and 504, e-141 respectively. This indicates that CRa is a gene homologous to the TNL genes especially Bra019410, but obviously different from any of the TNLs. Identifying the TNL gene that corresponds with Rcr1 and relationship of Rcr1 with CRa will be addressed after Rcr1 has been cloned.
Breeding for clubroot resistance in canola at many locations in Canada is severely constrained because the pathogen, or the pathotype of interest, does not occur at or near the site of established breeding institutions. Use of highly specific markers in MAS could be to accurately assess the reaction of lines under controlled (i.e. indoor) conditions. A number of techniques are available for genotyping SNPs. NGS is an emerging method of SNP genotyping that is being increasingly adopted for discovery applications. However, this method can be expensive and time-consuming in terms of informatics needs, and currently generates datasets with a large proportion of missing data [75]. KASP offers cost-effective and scalable flexibility in applications [76]. In total, 14 robust SNP markers were confirmed that were completely associated with Rcr1, based on testing the recombinants identified from 1587 plants. Most of the SNPs proved to be very reliable markers in use for introgression of Rcr1 into elite canola breeding lines. Furthermore, MAS with the polymorphic SNP markers was confirmed to have detected Rcr1 in the BC1 population described by Chu et al [11] with 100% accuracy in this study. Therefore, we conclude that these SNP markers could provide an effective and robust basis for introgression of Rcr1 into canola using MAS.
Supporting Information
A, R bulks by SSA; B, S bulks by SSA; C. R bulks by PSA; D. S bulks by PSA.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Mr. Adrian Chang for assistance on developing the F2 population. An anonymous mathematician and software engineer provided great help with analyzing variants and designing KASP primers.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by http://www.fundingconsortium.ca/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dixon GR. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul 2009; 28; 194–202. 10.1007/s00344-009-9090-y [DOI] [Google Scholar]
- 2.Nikolaev SI, Berney C, Fahrni JF, Bolivar I, Polet S, Mylnikov AP, et al. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc Natl Acad Sci USA 2004; 101:8066–8071. 10.1073/pnas.0308602101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard RJ, Strelkov SE, Hardinget MW.Clubroot of cruciferous crops–new perspectives on an old disease. Can. J. Plant Pathol 2010; 32: 43–57. 10.1080/07060661003621761 [DOI] [Google Scholar]
- 4.Kageyama K, Asano T. Life cycle of Plasmodiophora brassicae. J Plant Growth Regul 2009; 28: 203–11. 10.1007/s00344-009-9101-z [DOI] [Google Scholar]
- 5.Diederichsen E, Frauen M, Linders EGA, Hatakeyama EK, Hirai EM. Status and perspectives of clubroot resistance breeding in crucifer crops. J Plant Growth Regul 2009; 28:265–281. 10.1007/s00344-009-9100-0 [DOI] [Google Scholar]
- 6.Strelkov SE, Hwang SF. Clubroot in the Canadian canola crop: 10 years into the outbreak. Can J Plant Pathol 2014; 36:27–36. 10.1080/07060661.2013.863807 [DOI] [Google Scholar]
- 7.Rahman H, Shakir A, Hasan MJ. Breeding for clubroot resistant spring canola (Brassica napus L.) for the Canadian prairies: can the European winter canola cv. Mendel be used as a source of resistance? Can J Plant Sci 2011; 91, 447–458. [Google Scholar]
- 8.Gao F, Hirani AH, Liu J, Liu Z, Fu G, Wu C, et al. Fine mapping a clubroot resistance locus in Chinese cabbage. J Amer Soc Hort Sci 2014; 139:247–252. [Google Scholar]
- 9.Chen J, Jing J, Zhan Z, Zhang T, Zhang C, et al. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. 2013; PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho KH, Park SH, Kim KT, Kim S, Kim JS, Park BS, Woo JG, Lee HJ. Mapping quantitative trait loci (QTL) for clubroot resistance in Brassica rapa L. J Hortic Sci Biotechnol 2012; 87: 325–333 [Google Scholar]
- 11.Chu M, Song T, Falk K, Zhang X, Liu X, Chang A, et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. 2014; BMC Genomics, 15(1: Article number 1166). 10.1186/1471-2164-15-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashida N, Takabatake Y, Nakazawa N, Aruga D, Nakanishi H, Taguchi G, Sakamoto K, Matsumoto E: Construction of a practical SCAR marker linked to clubroot resistance in Chinese cabbage, with intensive analysis of HC352b genes. J. Japan. Soc. Hort. Sci. 2008, 77:150–154 [Google Scholar]
- 13.Hirai M, Harada T, Kubo N, Tsukada M, Suwabe K, Matsumoto S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor Appl Genet 2004; 108:639–643. 10.1007/s00122-003-1475-x [DOI] [PubMed] [Google Scholar]
- 14.Kato T, Hatakeyama K, Fukino N, Matsumoto S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa Breed Sci. 2013. March; 63(1): 116–124. Published online 2013 Mar 1. 10.1270/jsbbs.63.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuginuki Y, Ajisaka H, Yui M, Yoshikawa H, Hida K, Hirai M. RAPD markers linked to a clubroot resistance locus in Brassica rapa L. Euphytica 1997; 98:149–154. 10.1023/A:1003147815692 [DOI] [Google Scholar]
- 16.Matsumoto E, Yasui C, Ohi M, Tsukada M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage. Euphytica 1998; 104:79–86. 10.1023/A:1018370418201 [DOI] [Google Scholar]
- 17.Pang W, Liang S, Li X, Li P, Yu S, Lim YP et al. Genetic detection of clubroot resistance loci in a new population of Brassica rapa. Hortic Environ Biotechnol 2014; 55:540–547. 10.1007/s13580-014-0079-5 [DOI] [Google Scholar]
- 18.Piao ZY, Deng YQ, Choi SR, Park YJ, Lim YP. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor Appl Genet 2004; 108:1458–1465. 10.1007/s00122-003-1577-5 [DOI] [PubMed] [Google Scholar]
- 19.Saito M, Kubo N, Matsumoto S, Suwabe K, Tsukada M, Hirai M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor Appl Genet 2006; 114:81–91. 10.1007/s00122-006-0412-1 [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K, Saito A, Hayashida N, Taguchi G, Matsumoto E. Mapping of isolate-specific QTL for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor Appl Genet 2008; 117:759–767. 10.1007/s00122-008-0817-0 [DOI] [PubMed] [Google Scholar]
- 21.Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, et al. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet 2003; 107:997–1002. 10.1007/s00122-003-1309-x [DOI] [PubMed] [Google Scholar]
- 22.Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, et al. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006; 173: 309–319. 10.1534/genetics.104.038968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Zhao Z, Zhang C, Pang W, Choi SR, Lim YP, et al. Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa. Mol Breeding 2014; 34:1173–1183. 10.1007/s11032-014-0108-1 [DOI] [Google Scholar]
- 24.Grandclément C, Thomas G. Detection and analysis of QTL based on RAPD markers for polygenic resistance to Plasmodiophora brassicae Wor. in Brassica oleracea L. Theor Appl Genet 1996; 93:86–90. 10.1007/BF00225731 [DOI] [PubMed] [Google Scholar]
- 25.Figdore SS, Ferreira ME, Slocum MK, Williams PH. Association of RFLP markers with trait loci affecting clubroot resistance and morphological characters in Brassica oleracea L. Euphytica 1993; 69:33–44. 10.1007/BF00021723 [DOI] [Google Scholar]
- 26.Landry BS, Hubert N, Crete R, Chiang MS, Lincoln SE, Etoh T. A genetic map for Brassica oleracea based on RFLP markers detected with expressed DNA sequences and mapping of resistance gene to race 2 of Plasmodiophora brassicae. Genome 1992; 35:409–420. 10.1139/g92-061 [DOI] [Google Scholar]
- 27.Moriguchi K, Kimizuka-Takagi C, Ishii K, Nomura K. A genetic map based on RAPD, RFLP, isozyme, morphological markers and QTL analysis for clubroot resistance in Brassica oleracea. Breeding Sci 1999; 49:257–265. 10.1270/jsbbs.49.257 [DOI] [Google Scholar]
- 28.Nagaoka T, Doullah MAU, Matsumoto S, Kawasaki S, Ishikawa T, Hori H, et al. Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theor Appl Genet 2010; 120:1335–1346. 10.1007/s00122-010-1259-z [DOI] [PubMed] [Google Scholar]
- 29.Nomura K, Minegishi Y, Kimizuka-Takagi C, Fujioka T, Moriguchi K, Shishido R, et al. Evaluation of F2 and F3 plants introgressed with QTL for clubroot resistance in cabbage developed by using SCAR markers. Plant Breeding 2005; 124:371–375. 10.1111/j.1439-0523.2005.01105.x [DOI] [Google Scholar]
- 30.Rocherieus J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G. Isolate-specific and broad spectrum QTL are involved in the control of clubroot in Brassica oleracea. Theor Appl Genet 2004; 108:1555–1563. 10.1007/s00122-003-1580-x [DOI] [PubMed] [Google Scholar]
- 31.Tomita H, Shimizu M, Doullah MA, Fujimoto R, Okazaki K. Accumulation of quantitative trait loci conferring broadspectrum clubroot resistance in Brassica oleracea. Mol Breeding 2013; 32:889–900. 10.1007/s11032-013-9918-9 [DOI] [Google Scholar]
- 32.Voorrips RE, Jongerius MC, Kanne HJ. Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of doubled haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor Appl Genet 1997; 94:75–82. 10.1007/s001220050384 [DOI] [PubMed] [Google Scholar]
- 33.Diederichsen E, Beckmann J, Schondelmeier J, Dreyer F. Genetics of clubroot resistance in Brassica napus ‘Mendel’. Acta Hort 2006; 706:307–312. 10.17660/ActaHortic.2006.706.35 [DOI] [Google Scholar]
- 34.Manzanares-Dauleux MJ, Deloureme R, Baron F, Thomas G. Mapping of one major gene and of QTL involved in resistance to clubroot in Brassica napus. Theor Appl Genet 2000; 101:885–891. 10.1007/s001220051557 [DOI] [Google Scholar]
- 35.Werner S, Diederichsen E, Frauen M, Schondelmaier J, Jung C. Genetic mapping of clubroot resistance genes in oilseed rape. Theor Appl Genet 2008; 116:363–372. 10.1007/s00122-007-0674-2 [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Feng J, Hwang SF, Strelkov SE, Falak I, Huang X, et al. Mapping of clubroot (Plasmodiophora brassicae) resistance in canola (Brassica napus). Plant Pathol 2015 [Google Scholar]
- 37.Fuchs H, Sacristán MD. Identification of a gene in Arabidopsis thaliana controlling resistance to clubroot (Plasmodiophora brassicae) and characterization of the resistance response. Mol Plant Microbe Interact 1996; 9:91–97. 10.1094/MPMI-9-0091 [DOI] [Google Scholar]
- 38.Kamei A, Tsuro M, Kubo N, Hayashi T Wang N, Fujimura T, et al. QTL mapping of clubroot resistance in radish (Raphanus sativus L.). Theor Appl Genet 2010; 120:1021–1027. 10.1007/s00122-009-1230-z [DOI] [PubMed] [Google Scholar]
- 39.Ueno H, Matsumoto E, Aruga D, Kitagawa S, Matsumura H, Hayashida N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol Biol 2012; 80:621–629. 10.1007/s11103-012-9971-5 [DOI] [PubMed] [Google Scholar]
- 40.Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, et al. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 2013; 8:e54745 10.1371/journal.pone.0054745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams PH. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 1966; 56:624–626. [Google Scholar]
- 42.Hasan MJ, Strelkov SE, Howard RJ, Rahman H. Screening of Brassica germplasm for resistance to Plasmodiophora brassicae pathotypes prevalent in Canada for broadening diversity in clubroot resistance. Can J Plant Sci 2012; 92:501–515. 10.4141/cjps2010-006 [DOI] [Google Scholar]
- 43.Peng G, Falk KC, Gugel RK, Franke C, Yu F, James B, et al. Sources of resistance to Plasmodiophora brassicae (clubroot) pathotypes virulent on canola. Can J Plant Pathol 2014; 36: 89–99. 10.1080/07060661.2013.863805 [DOI] [Google Scholar]
- 44.Johnston TD. Transfer of disease resistance from Brassica campestris L. to rape (B. napus L.). Euphytica 1974; 23:681–683. 10.1007/BF00022490 [DOI] [Google Scholar]
- 45.Gowers S. The transfer of characters from Brassica campestris l. to Brassica napus L.: Production of clubroot-resistant oil-seed rape (B. napus ssp oleifera). Euphytica 1982; 31: 971–976. 10.1007/BF00039237 [DOI] [Google Scholar]
- 46.Yu F, Lydiate DJ, Gugel R, Sharpe A, Rimmer SR. Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus through marker assisted selection. Mol Breeding 2012; 30:1495–1506. 10.1007/s11032-012-9735-6 [DOI] [Google Scholar]
- 47.Gautam M, Ge X, Li Z. 2013. Brassica (Book Chapter). Alien Gene Transfer in Crop Plants. 2: 207–229 [Google Scholar]
- 48.Li Q, Mei J, Zhang Y, Li J, Ge X, Li Z, et al. A large-scale introgression of genomic components of Brassica rapa into B. napus by the bridge of hexaploid derived from hybridization between B. napus and B. oleracea. Theorl Appl Genet 2013; 126: 2073–2080. 10.1007/s00122-013-2119-4 [DOI] [PubMed] [Google Scholar]
- 49.Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 2002; 5:94–100. 10.1016/S1369-5266(02)00240-6 [DOI] [PubMed] [Google Scholar]
- 50.Riahi L, Zoghlami N, Fournier-Level A, Dereeper A, Le Cunff L, Laucou V, et al. Characterization of single nucleotide polymorphism in Tunisian grapevine genome and their potential for population genetics and evolutionary studies. Genet Resour Crop Evol 2013; 60:1139–1151. 10.1007/s10722-012-9910-y [DOI] [Google Scholar]
- 51.Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S. SNP markers and their impact on plant breeding. International Journal of Plant Genomics 2012; Article ID:728398. 10.1155/2012/728398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger ME, Weisshaar B, Scholz U, Stein N, Usadel B. Mayer FX: Plant genome sequencing–applications for crop improvement. Curr Opin Biotechnol 2014; 26:31–37. 10.1016/j.copbio.2013.08.019 [DOI] [PubMed] [Google Scholar]
- 53.Bancroft I, Morgan C, Fraser F, Higgins J, Wells R, Clissold L, et al. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol 2011; 29:762–766. 10.1038/nbt.1926 [DOI] [PubMed] [Google Scholar]
- 54.Paritosh K, Yadava SK, Gupta V, Panjabi-Massand P, Sodhi YS, Pradhan AK. RNA-seq based SNPs in some agronomically important oleiferous lines of Brassica rapa and their use for genome-wide linkage mapping and specific-region fine mapping. BMC Genomics 2013; 14:463 10.1186/1471-2164-14-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations. Proc Natl Acad Sci USA 1991; 88:9828–9832. 10.1073/pnas.88.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfender WF, Saha MC, Johnson EA, Slabaugh MB. Mapping with RAD (restrictionsite associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor Appl Genet 2011; 122:1467–1480. 10.1007/s00122-011-1546-3 [DOI] [PubMed] [Google Scholar]
- 57.Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, et al. QTL-seq: Rapid mapping of quantitative trait loci by whole genome resequencing of DNA from two bulked populations. Plant J 2013; 74:174–183. 10.1111/tpj.12105 [DOI] [PubMed] [Google Scholar]
- 58.Yang H, Tao Y, Zheng Z, Li C, Sweetingham MW, Howieson JG. Application of next generationsequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genomics 2012; 13: 318 10.1186/1471-2164-13-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H, Tao Y, Zheng Z, Shao D, Li A, Sweetingham MW, et al. Rapid development of molecular markers by next-generation sequencing linked to a gene conferring phomopsis stem blight disease resistance for marker-assisted selection in lupin (Lupinus angustifolius L.) breeding. Theor Appl Genet 2013; 126:511–522. 10.1007/s00122-012-1997-1 [DOI] [PubMed] [Google Scholar]
- 60.Hu Z, Hua W, Huang S, Yang H, Zhan G, Wang X, et al. Discovery of pod shatter-resistant associated SNPs by deep sequencing of a representative library followed by bulk segregant analysis in rapeseed. PLoS One 2012; 7: e34253 10.1371/journal.pone.0034253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 2012; 7:e36406 10.1371/journal.pone.0036406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trick M, Admski NM, Mugford SG, Jiang CC, Febrer M, Uauy C. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploidy wheat. BMC Plant Biol 2012; 12:14 10.1186/1471-2229-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crête R, Laliberté J, Jasmin JJ. Lutte chimique contre la hernie, Plasmodiophora brassicae Wor., des cruciferes en sols mineral et organique. Can J Plant Sci 1963; 43:349–354. [Google Scholar]
- 64.Strelkov SE, Tewari JP, Smith-Degenhardt E. Characterization of Plasmodiophora brassicae populationsfrom Alberta, Canada. Can J Plant Pathol 2006; 28:467–474. 10.1080/07060660609507321 [DOI] [Google Scholar]
- 65.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 2011; 43:1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 66.Iversen GR, Gergen M. Statistics: The conceptual approach (Springer undergraduate textbooks in Statistics) 1997.
- 67.Strelkov SE, Manolii VP, Cao T, Xue S, Hwang SF. Pathotype classification of Plasmodiophora brassicae and its occurrence in Brassica napus in Alberta, Canada. J Phytopathology 2007; 155: 706–712. 10.1111/j.1439-0434.2007.01303.x [DOI] [Google Scholar]
- 68.Xue C, Cao T, Howard RJ, Hwang SF and Strelkov SE. Isolation and variation in virulence of single-spore isolates of Plasmodiophora brassicae from Canada. Plant Dis 2008; 92:456–462. 10.1094/PDIS-92-3-0456 [DOI] [PubMed] [Google Scholar]
- 69.Trick M., Long Y, Meng J; Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J 2009; 7:334–346. 10.1111/j.1467-7652.2008.00396.x [DOI] [PubMed] [Google Scholar]
- 70.Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, et al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 2012; 30:798–802. 10.1038/nbt.2302 [DOI] [PubMed] [Google Scholar]
- 71.Takano S, Matsuda S, Kinoshita N, Shimoda N, Sato T, Kato K. Genome-wide single nucleotide polymorphisms and insertion–deletions of Oryza sativa L. subsp. japonica cultivars grown near the northern limit of rice cultivation. Mol Breeding 2014; 34:1007–1021. 10.1007/s11032-014-0093-4 [DOI] [Google Scholar]
- 72.Marian AJ. Editorial Review: DNA sequence variants and the practice of medicine. Curr Opin Cardiol 2010; 25: 182–185. 10.1097/HCO.0b013e3283389683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sathya B, Dharshini AP, Kumar GR. NGS meta data analysis for identification of SNP and INDEL patterns in human airway transcriptome: A preliminary indicator for lung cancer. Applied & Translational Genomics 2015; 4:4–9 10.1016/j.atg.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. 10.1038/nrg3051 [DOI] [PubMed] [Google Scholar]
- 75.Beissinger TM, Hirsch CN, Sekhon RS, Foerster JM, Johnson JM, Muttoni G, et al. Marker density and read-depth for genotyping populations using genotyping-by-sequencing. Genetics 2013;193:1073–1081. 10.1534/genetics.112.147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol Breeding 2014; 33:1–14. 10.1007/s11032-013-9917-x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, R bulks by SSA; B, S bulks by SSA; C. R bulks by PSA; D. S bulks by PSA.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.