WDR92 is a highly conserved component of a prefoldin-like cochaperone complex that has a phylogenetic signature compatible with a role in motile cilia assembly or function. Lack of this component in planaria causes pleiomorphic defects in ciliary architecture, resulting in immotile or poorly motile structures.
Abstract
WDR92 is a highly conserved WD-repeat protein that has been proposed to be involved in apoptosis and also to be part of a prefoldin-like cochaperone complex. We found that WDR92 has a phylogenetic signature that is generally compatible with it playing a role in the assembly or function of specifically motile cilia. To test this hypothesis, we performed an RNAi-based knockdown of WDR92 gene expression in the planarian Schmidtea mediterranea and were able to achieve a robust reduction in mRNA expression to levels undetectable under our standard RT-PCR conditions. We found that this treatment resulted in a dramatic reduction in the rate of organismal movement that was caused by a switch in the mode of locomotion from smooth, cilia-driven gliding to muscle-based, peristaltic contractions. Although the knockdown animals still assembled cilia of normal length and in similar numbers to controls, these structures had reduced beat frequency and did not maintain hydrodynamic coupling. By transmission electron microscopy we observed that many cilia had pleiomorphic defects in their architecture, including partial loss of dynein arms, incomplete closure of the B-tubule, and occlusion or replacement of the central pair complex by accumulated electron-dense material. These observations suggest that WDR92 is part of a previously unrecognized cytoplasmic chaperone system that is specifically required to fold key components necessary to build motile ciliary axonemes.
INTRODUCTION
In metazoans, motile cilia play key roles in development, fertility, and organismal homeostasis. In mammals, for example, defects in ciliary motility compromise lung function, as mucus secreted as a protectant against pollutants and pathogens cannot be cleared; they also inhibit the flow of cerebrospinal fluid in the brain ventricles, resulting in hydrocephalus, and can even cause female infertility, as oocytes may fail to traverse the fallopian tubes to the uterus (Becker-Heck et al., 2012). In these cases, it is the flow of the external fluid bathing the ciliated epithelium that is required for normal physiology.
These motile structures have a highly conserved architecture built around nine outer doublet microtubules and usually a central pair of singlet microtubules. Dynein arms generate the motive force for ciliary beating, whereas the radial spokes, nexin–dynein regulatory complex, and Mia complex together provide a physical pathway that regulatory signals from the central pair can traverse to affect individual dynein motors, thereby controlling waveform and beat frequency (Porter, 2012; King, 2013; Yamamoto et al., 2013). Disruption of the intraflagellar transport system affects or negates the assembly of both motile and immotile sensory cilia and thus compromises both the motility and signaling properties of these organelles, with often devastating consequences (Rosenbaum and Witman, 2002; Hildebrandt et al., 2011). However, in mammals, other metazoans, and unicellular organisms, defects in the assembly of ciliary subcomplexes (e.g., dynein arms or radial spokes) lead only to poorly motile or immotile structures without altering cilia-based signaling (Kamiya, 1988; Neilsen et al., 2001; Austin-Tse et al., 2013). Although much has been learned about the identity of various proteins in cytoplasm required for the folding and assembly of dynein motors (Omran et al., 2008; Tarkar et al., 2013), even in that well-studied system the precise functional roles of many of these effectors is uncertain. What other factors might be necessary for cytoplasmic production of the myriad other components needed to build an intact functional motile cilium is almost completely unexplored.
Tryptophan/aspartate repeat 92 (WDR92; also known as Monad) is a WD-repeat protein that has been described as a proapoptotic factor based on the overexpression phenotype in a mammalian cell culture system (Saeki et al., 2006). It has been suggested that WDR92 interacts with RNA polymerase II–associated protein 3 (RPAP3) potentially through the RPAP3_C domain (Itsuki et al., 2008). Intriguingly, this domain is present in only three protein types: the eponymous RPAP3 and CCDC103 and SPAG1, both of which act as ciliary dynein assembly factors (Panizzi et al., 2012; Knowles et al., 2013; King and Patel-King, 2015). Consequently, we considered the possibility that WDR92 and RPAP3 might also act in the ciliary dynein assembly pathway. Furthermore, WDR92 interacts with several components of the prefoldin cochaperone complex to form a noncanonical chaperone that has been demonstrated to aid in the assembly of RNA polymerase II and have many other cellular activities (Boulon et al., 2010; Kakihara and Houry, 2012; Millán-Zambrano and Chávez, 2014). This occurs in conjunction with a second cochaperone, the R2TP complex, formed from the RuvBL1 (pontin) and RuvBL2 (reptin) AAA+ ATPases, a PIH domain–containing protein (PIHD1), and RPAP3 (also known as hSpagh; Kakihara and Houry, 2012). Prefoldin is believed to detect proteins that have folded incorrectly and target them to other chaperones that allow for repeat attempts at refolding. Previous studies also suggested a connection between prefoldin and the cytoskeleton, as this complex appears to be involved in generating correctly structured actin and tubulin (Vainberg et al., 1998; Hansen et al., 1999).
Our initial phylogenetic analysis revealed that WDR92 has been highly conserved among organisms that build motile cilia but is completely and specifically absent in those that do not assemble cilia at all or build only immotile sensory structures. On the basis of this phylogenetic signature, we predicted that WDR92 might be specifically involved in motile cilia assembly or function; this suggestion is also consistent with the reported high-level expression in mammalian testis and ciliated tissues such as lung and the fallopian tubes. Consequently, we set out to test this hypothesis by performing an RNA interference (RNAi)–mediated knockdown of WDR92 gene expression in the planarian Schmidtea mediterranea. Here we demonstrate that reduction of WDR92 expression leads to aberrant ciliary assembly on the ventral epithelium, with pleiomorphic defects in ciliary architecture and poor motile activity, but has no apparent effect on the overall viability of the animal. On the basis of these results, we propose that WDR92 forms part of a novel cytoplasmic system potentially involving the R2TP complex that is specifically required for motile cilia assembly.
RESULTS AND DISCUSSION
Phylogenetic signature and structural conservation of WDR92
Homo sapiens WDR92 is a 357-residue protein with molecular weight of 39,740 and a calculated pI of 8.32. This protein is present throughout the Animalia, including chordates, echinoderms, arthropods, annelids, cnidarians, and placozoans (all WDR92 accession numbers are provided in Supplemental Table S1), with the notable exception of nematodes, which assemble only sensory cilia; for example, the best Caenorhabditis elegans hit (NM_065690) had an expect value of only 8 × 10−04 (21% sequence identity) when using the Schmidtea mediterranea orthologue as the query. WDR92 is also found in choanoflagellates, excavates such as Trypanosoma, alveolates (including Tetrahymena, Paramecium, and the oomycete Phytophthora), and the chytrid fungus Batrochochytrium, which has flagellated zoospores. In contrast, neither ascomycetes nor basidiomycetes encode this protein. The situation is more uncertain in the zygomycetes, as several reasonably strong hits were obtained, although they have probability scores greatly lower than those for other organisms (expect values were 5 × 10−180 for H. sapiens but only 4 × 10−77 for Lichtheimia ramosa, with sequence identity reduced to ∼37%); a reciprocal BLAST search using the zygomycete sequence returned WDR92 orthologues as the closest hits. A search for WDR92 within plants identified orthologues in the bryophyte Physcomitrella and the spike moss Selaginella, but only a single high-scoring sequence was found for any angiosperm, suggesting that the one identified (in the Chinese white pear interspecific hybrid Pyrus × bretschneideri genome) is most likely a result of contamination; although no protein identical to this is present in the database, the most closely related orthologues identified by a reciprocal BLAST search derived from insects and other arthropods. Within the green algae, WDR92 is present in Chlamydomonas, Volvox, and also Ostreococcus, which, although it apparently does not assemble cilia or flagella, does encode the two dynein heavy chains that comprise inner arm I1/f. WDR92 is also present in a brown alga (Ectocarpus) and diatoms (Thalassiosira) but appears to be completely missing from the aflagellate red algae. Thus, with the exception of a highly divergent protein present in zygomycetes and a single annotated angiosperm sequence that is likely contamination, this phylogenetic signature is generally compatible with a predicted role for WDR92 in the assembly or function of motile cilia and is also consistent with the reported high-level expression in mammalian testis (Saeki et al., 2006).
Sequence alignments revealed that WDR92 has been highly conserved during evolution and forms a discrete grouping within the WD-repeat protein family that is clearly distinct from other WD-repeat proteins such as dynein intermediate chains, Lis1, and the G protein β subunit (Figure 1a). For example, BLAST analysis revealed that the H. sapiens and Chlamydomonas reinhardtii proteins share 58% identity (72% similarity) with an expect value of 2 × 10−149. The 1.95-Å crystal structure of H. sapiens WDR92 was previously solved by the Structural Genomics Consortium at the University of Toronto (PDB accession 3I2N; Xu and Min, 2011). This protein consists of 28 β-strands and a single short α-helix (Figure 1b). There are seven WD repeats, each of which forms a four-stranded β-sheet and which are arranged as a classic toroidal β-propeller, with a single large protrusion derived from two extended loops located between β20/β21 and β24/β25. The ribbon structure and van der Waal surface, painted with full Poisson–Boltzmann electrostatics, are shown in Figure 2, a–d; WDR92 has several highly positively and negatively charged exposed patches. Furthermore, the molecular surface of WDR92 has been highly conserved between H. sapiens, S. mediterranea, and C. reinhardtii (Figure 2, e and f).
FIGURE 1:
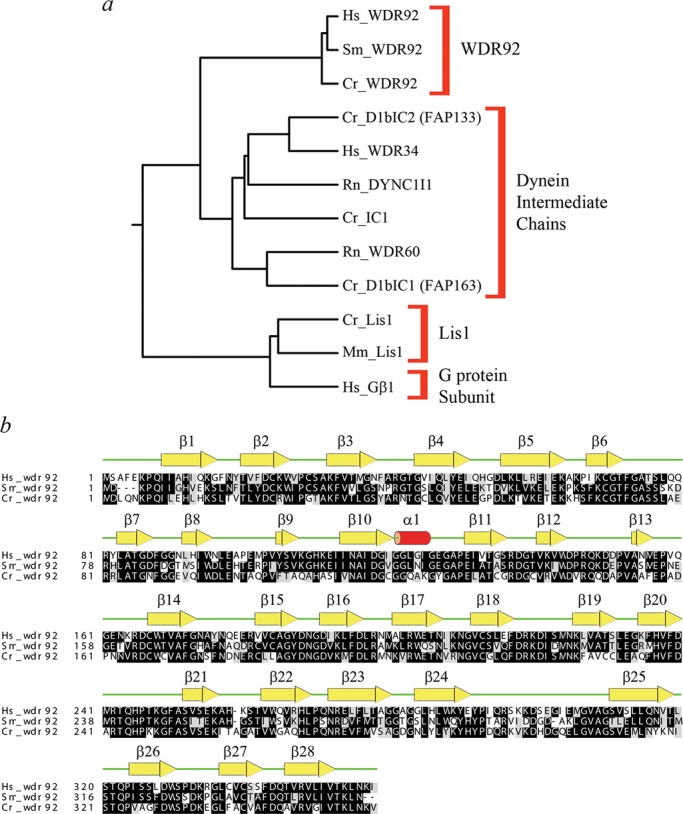
Phylogeny and structural alignment of WDR92 orthologues. (a) Neighbor-joining phylogenetic tree constructed following a CLUSTALW alignment of members from several WD-repeat protein families, including WDR92, dynein intermediate chains, Lis1, and a G protein β-subunit. Organism abbreviations: Cr, Chlamydomonas reinhardtii; Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Sm, Schmidtea mediterranea. (b) CLUSTALW alignment of WDR92 proteins from H. sapiens, C. reinhardtii, and S. mediterranea: residues conserved in two or more sequences are shaded. The secondary structural elements determined from the crystal structure (PDB accession 3I2N) are shown above the alignment. The WDR92 family proteins exhibit a high degree of sequence conservation over their entire length; the most divergent regions are near the extreme N-terminus and in the extended loop between strands β24 and β25.
FIGURE 2:
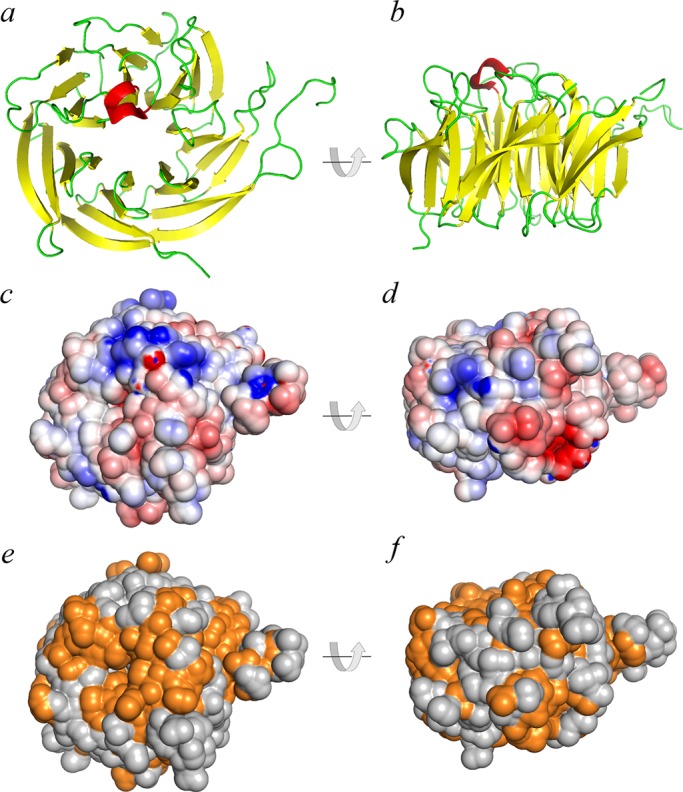
Structural features of WDR92. (a, b) Ribbon diagrams of H. sapiens WDR92 from the 1.95-Å crystal structure (3I2N; Xu and Min, 2011); the β−strands are indicated in yellow and the single short α-helix is in red. The two views are related by a 90° rotation. (c, d) The same orientations of WDR92, shown with the van der Waal molecular surface displayed and painted based on a full Poisson–Boltzmann electrostatics calculation; negatively charged regions are in red and positively charged segments in blue. (e, f) Residues on the WDR92 molecular surface are colored orange to illustrate those regions that are completely conserved between the H. sapiens, S. mediterranea, and C. reinhardtii orthologues; orientation is the same as before.
WDR92 is required for cilia-mediated gliding locomotion in planaria
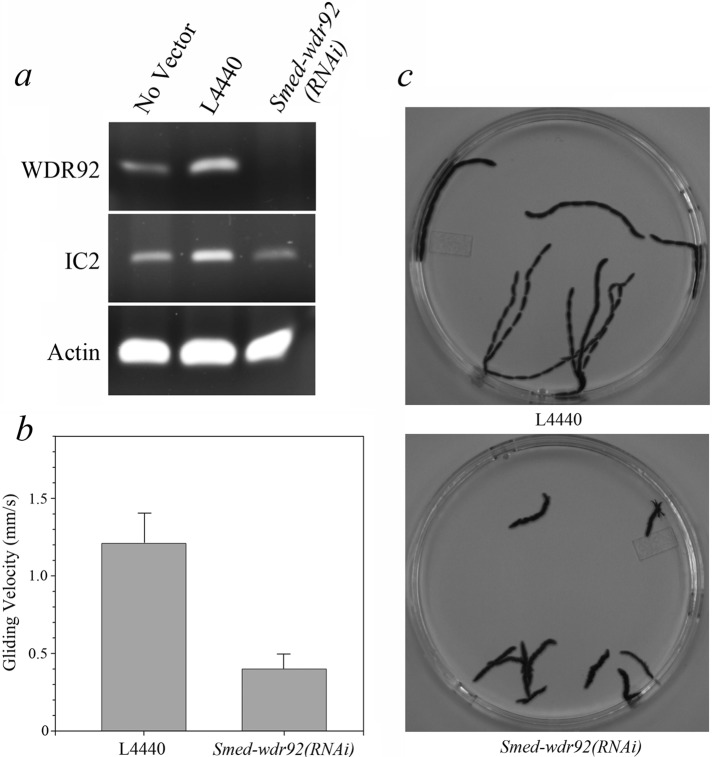
To test whether WDR92 might be involved in ciliary assembly or motility, we fed planaria every 2–3 d for 3 wk with bacteria expressing a 317–base pair double-stranded RNA (dsRNA) encoding a portion of WDR92 or with bacteria containing the empty L4440 vector or a no-vector control. Phenotypes began to appear after 2 wk, and animals were analyzed in week 3. Ingestion of the 317–base pair dsRNA resulted in a reduction in WDR92 mRNA to levels that could not be detected by reverse transcriptase PCR (RT-PCR) after 25 cycles; in contrast, mRNA encoding outer arm dynein IC2 and actin were readily detected in both control and experimental samples (Figure 3a). The control planaria exhibited normal, smooth, cilia-driven gliding motility and moved at 1.21 ± 0.18 mm/s. However, the motility mode of Smed-wdr92(RNAi) animals was completely different, involving peristaltic, muscle-driven body contractions that essentially squeezed the animal forward at a rate of only 0.40 ± 0.09 mm/s (Figure 3, b and c, and Supplemental Movie S1). We found previously that this switch in locomotive behavior is a defining characteristic of planaria exhibiting defects in the assembly or motility of the cilia lining the ventral surface (Rompolas et al., 2010).
FIGURE 3:
RNAi-mediated knockdown of WDR92 expression inhibits planarian gliding locomotion. (a) RT-PCR analysis of mRNA levels of WDR92, IC2, and actin in control (no vector or empty L4440 vector) and Smed-wdr92(RNAi) animals. The mRNA for WDR92 is reduced to levels undetectable after 25 cycles of RT-PCR, whereas IC2 and actin mRNAs are not. (b) Bar chart indicating the velocity of L4440 control and Smed-wdr92(RNAi) planarian movement. (c) Overlays (of every 100th frame) prepared from deconvolved movies (taken at 30 frames/s) illustrating the tracks taken by L4440 control and Smed-wdr92(RNAi) planaria during a 30-s period (Supplemental Movie S1).
Smed-wdr92(RNAi) planaria have malformed, poorly motile cilia
Examination of the cilia on the ventral epithelium of Smed-wdr92(RNAi) planaria using high-speed videomicroscopy revealed that while many cilia were either immotile or merely twitched, others beat in an uncoordinated manner with frequencies ranging from ∼5.5 to 12 Hz (Supplemental Movie S2). In contrast, the L4440-fed control animals exhibited coordinated ciliary motion with beat frequencies >20 Hz (Supplemental Movie S3), as found previously (Rompolas et al., 2010; Patel-King et al., 2013).
In an attempt to ascertain why Smed-wdr92(RNAi) cilia exhibited a range of motility phenotypes and determine the structural consequences of RNAi-mediated reduction of WDR92, we examined the ventral epithelial surface by scanning electron microscopy (EM) and found that the Smed-wdr92(RNAi) planaria assembled cilia of normal length and at a density similar to that observed in control animals (Figure 4). However, transmission electron microscopy of cilia in cross section revealed a complex pleiomorphic array of aberrant structures (Figure 5, a–c). Although the axonemes in many ciliary cross sections (44% of the total; n = 118) appeared to be of normal morphology, others (11%) showed a clear loss of outer dynein arms on some doublet microtubules. In addition, we routinely observed defects in the doublet microtubules themselves, including A-tubules completely occluded with an electron-dense material (11%) and B-tubules that were incompletely formed, resulting in a C-shaped structure (22%); in all cases observed, these aberrant B-tubules failed to close at the outermost junction with the A-tubule near the ciliary membrane. In other axonemal cross sections (12%), amorphous electron-dense material obscured or replaced the central pair complex and/or packed into the luminal region between the central pair and doublet microtubules. The pleiomorphic nature of the phenotypes observed in Smed-wdr92(RNAi) planaria was quite different from that found previously for animals defective for intraflagellar transport (IFT) or outer arm dynein components, where phenotypes appeared to be almost completely penetrant (Rompolas et al., 2010; Patel-King et al., 2013). For example, reduction of IFT88 mRNA resulted in animals totally devoid of ventral cilia; similarly, IC2- knockdown planaria had no detectable outer dynein arms incorporated. These previous results strongly suggest that all of the cilia on the ventral surface are being completely remodeled over the time frame of these experiments.
FIGURE 4:
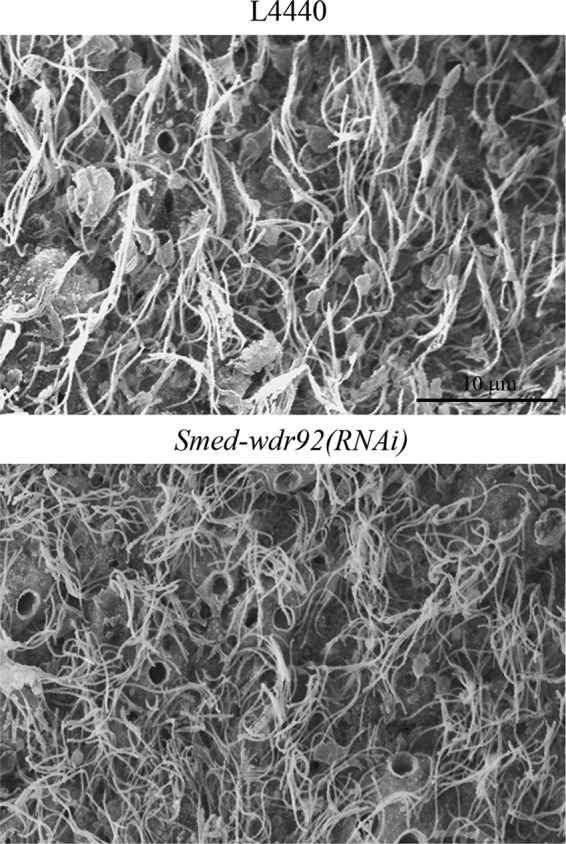
Smed-wdr92(RNAi) planaria assemble full-length cilia. Scanning electron microscopic images of the ventral surface of vector control (L4440) and Smed-wdr92(RNAi) planaria. Cilia on the knockdown animals are of similar length and density as the controls. Bar, 10 μm.
FIGURE 5:
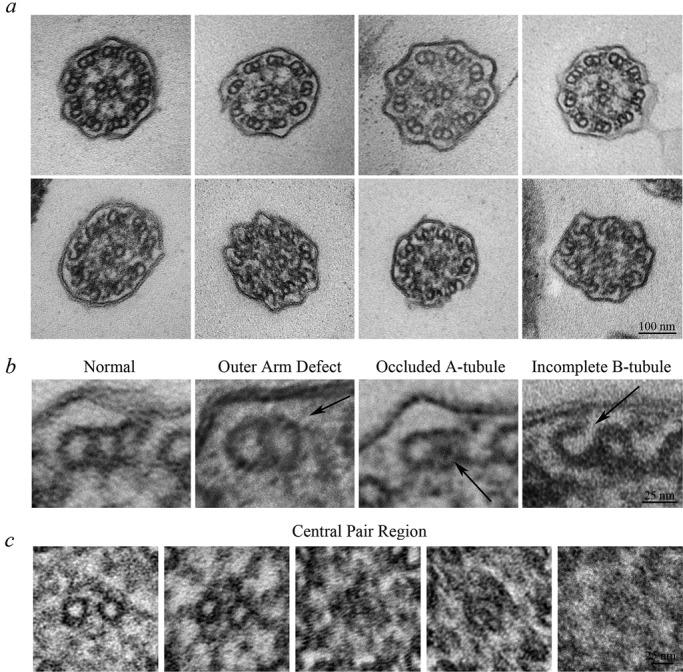
Smed-wdr92(RNAi) planaria have defective ciliary architecture. (a) Transmission electron micrographs of transverse sections through cilia of Smed-wdr92(RNAi) animals. Although many cilia appear morphologically normal (top left), others exhibit a variety of structural defects, including missing outer dynein arms, occlusion of the A-tubule lumen, accumulation of electron-dense material within the central region of the axoneme, where it obscures (or replaces) the central pair microtubules, and the incomplete closure of B-tubules at the outermost A/B-tubule junction. Bar, 100 nm. (b) Magnified views of individual microtubule doublets illustrating the variety of structural defects observed. Bar, 25 nm. (c) Magnified views of the central pair region of the axonemal lumen. Although most axonemes contained a morphologically normal central complex (left), others lacked the central pair and/or contained amorphous material that either obscured or replaced it. In these cases, the assembly status of the radial spokes was also unclear. Bar, 25 nm.
The observed pattern of ciliary defects strongly supports the idea that multiple steps in ciliary assembly are at least partially dependent upon the activity of WDR92. Although the knockdown animals were clearly capable of generating assembly-competent outer dynein arms that can generate force, axonemal incorporation was at least partially disrupted, suggesting defects in the docking site or possibly in the rate at which the motors are trafficked into the cilium. Similarly, the failure of B-tubule closure supports the hypothesis that some components required for assembly of the outer doublet wall are at least partially defective in the absence of WDR92. Although we obtained a highly efficient knockdown of gene expression, it is possible that some WDR92 protein remains functional over the time course of these experiments. Intriguingly, previous studies showed that nodal cilia of mice bearing an Arl13b-null allele also show variable defects in ciliary architecture, including failure of the B-tubule to close at the inner junction (Caspary et al., 2007).
WDR92, prefoldin, and the R2TP complex
The R2TP complex is a multicomponent system built around the RuvBL1 (pontin) and RuvBL2 (reptin) AAA+ ATPases (Kakihara and Houry, 2012). Reptin and its binding partner Lrrc6/Seahorse were shown to be involved in cilia and dynein arm formation but also to have cilia-independent functions (Zhao et al., 2013). In association with Hsp90, R2TP has been found to be involved in multiple other cellular activities, including the cytoplasmic preassembly of RNA polymerase II (Boulon et al., 2010), apoptosis, phosphatidylinositol kinase signaling pathways (Izumi et al., 2010), and small nucleolar ribonucleoprotein complex formation (Watkins et al., 2004). WDR92 in the prefoldin-like complex is known to associate in cytoplasm with several components of R2TP, including RPAP3, RuvBL2 (reptin), and PIH1D1 (Itsuki et al., 2008; Inoue et al., 2010). Intriguingly, two other PIH domain proteins, including Ktu/PF13 and Mot48, are specifically required for the cytoplasmic assembly of axonemal dynein heavy chains (Omran et al., 2008; Yamamoto et al., 2010). Thus potentially all of the phenotypes we observed after WDR92 depletion might be explained by a reduction in cytoplasmic noncanonical prefoldin activity, thereby leading to reduced levels of properly folded or assembled key axonemal components necessary for the correct architectural organization of the motile cilium. This might be achieved directly through the failure of prefoldin-like chaperone activity to help fold specific components needed for motile cilia formation or more indirectly by altering the activity of the R2TP complex. To fully understand the biochemical mechanisms involved, it will be essential to identify the key axonemal molecules involved and determine how they interact with WDR92 and potentially other cytoplasmic factors. Another possible explanation is that lack of WDR92 and defective prefoldin activity might have a global effect on some posttranslational modification(s) necessary for these various aspects of motile ciliary assembly.
In conclusion, we demonstrate here that reduction in WDR92 gene expression leads directly and specifically to defects in axonemal architecture and consequently to compromised ciliary motility. Together with the described phylogenetic signature and mammalian expression pattern, these observations support the hypothesis that a noncanonical prefoldin complex plays a key and previously unrecognized role in the assembly of motile cilia and that this activity specifically requires WDR92.
MATERIALS AND METHODS
Computational methods
WDR92 sequences were identified using BLAST and aligned with CLUSTALW. The ribbon diagram, van der Waals surface, and full Poisson–Boltzmann electrostatics calculations for the WDR92 crystal structure (PDB accession 3I2N; Xu and Min, 2011) were performed and displayed using PyMOL (Schrödinger, New York, NY). The planarian sequence of WDR92 was obtained from the SmedGD Genome Database (http://smedgd.neuro.utah.edu/; Robb et al., 2008).
Planarian culture
The hermaphroditic sexual strain of S. mediterranea was kept in dark conditions in plastic boxes containing a 1× solution of Montjuïc salts as described previously (Rompolas and King, 2009). The colony was expanded as needed by slicing larger animals into several pieces and then allowing them to regenerate their original body plan over a period of several weeks. Planaria were allowed to feed approximately twice a week on calf liver.
RNA interference
A 317–base pair region of S. mediterranea WDR92 encompassing a region encoding residues 23–133 was obtained by RT-PCR and subcloned into plasmid L4440. Expression of dsRNA in Escherichia coli strain HT115(DE3), which lacks RNase III and cannot degrade dsRNA, was as described previously (Newmark et al., 2003; Rompolas and King, 2009). Bacterial pellets were mixed with liver homogenate, and animals were fed for 1 h every 3 d. For RNAi-based knockdowns, three groups of animals were used. Controls were fed either liver alone or mixed with bacteria containing the empty vector control. The experimental group was fed with liver mixed with bacteria expressing WDR92 dsRNA. The degree of gene knockdown was assessed by our standard RT-PCR methods using DNase I–treated RNA as template after 2–3 wk, using outer arm dynein IC2 and actin as reference controls (Rompolas et al., 2010).
Analysis of organismal and ciliary motility
Planaria were placed in a 9-cm Petri dish, and their movement over a 30- to 60-s period was recorded at 30 frames/s using a USB 2.0 1024 × 768 pixel color camera (DFK31BU03.H; Imaging Source, Charlotte, NC) with a progressive scan charge-coupled device image sensor and IC Capture 2.2 software. Individual frames from decompiled videos were overlaid using Photoshop CS4 (Adobe, San Jose, CA) to provide a complete record of the path taken by each animal. High-speed videomicroscopy of ciliary motility on the ventral surface of control and experimental planaria was performed essentially as described previously (Rompolas et al., 2010) using an X-PRI high-speed complementary metal–oxide–semiconductor camera (AOS Technologies, Switzerland) operating at 250 frames/s and differential interference contrast optics.
Electron microscopy
For scanning EM, planaria were placed in relaxant solution (1% HNO3, 0.85% formaldehyde, 50 mM MgSO4) overnight and then fixed with 2.5% glutaraldehyde in 0.1 M Na cacodylate, pH 7.4. After osmication and dehydration in an ethanol series, animals were dried at the critical point and imaged using a JEOL JSM5900 SEM. For transmission EM, animals were fixed with multiple changes of 1% glutaraldehyde in phosphate-buffered to avoid fixative depletion by secreted mucus and then washed into cacodylate buffer. Postfixation was with 1% OsO4, and samples were then stained en bloc with methanolic uranyl acetate, dehydrated in ethanol, and embedded in Epon resin. Ultrathin sections were poststained with uranyl acetate and lead citrate and imaged using a Hitachi H-7650 operating at 80 kV.
Supplementary Material
Acknowledgments
We thank Adam Schuyler for assistance with the Poisson–Boltzmann electrostatics calculation and Maya Yankova for help with electron microscopy. This study was supported by Grant GM051293 from the National Institutes of Health (to S.M.K.).
Abbreviations used:
- EM
electron microscopy
- IFT
intraflagellar transport
- RT-PCR
reverse transcriptase PCR
- WDR
tryptophan/aspartate repeat.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0040) on February 24, 2016.
REFERENCES
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and ccdc65 defects as causing primary ciliary dyskinesia. Am J Hum Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Heck A, Loges NT, Omran H. Dynein dysfunction as a cause of primary ciliary dyskinesia and other ciliopathies. In: King SM, editor. Dyneins: Structure, Biology and Disease. Waltham, MA: Elsevier; 2012. pp. 603–627. [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to sonic hedgehog depends on ciliary architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Saeki M, Egusa H, Niwa H, Kamisaki Y. PIH1D1, a subunit of R2TP complex, inhibits doxorubicin-induced apoptosis. Biochem Biophys Res Commun. 2010;403:340–344. doi: 10.1016/j.bbrc.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Itsuki Y, Saeki M, Nakahara H, Egusa H, Irie Y, Terao Y, Kawabata S, Yatani H, Kamisaki Y. Molecular cloning of novel Monad binding protein containing tetratricopeptide repeat domains. FEBS Lett. 2008;582:2365–2370. doi: 10.1016/j.febslet.2008.05.041. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kutrata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Houry WA. The R2TP complex: discovery and functions. Biochim Biophys Acta. 2012;1823:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM. A solid-state control system for dynein-based ciliary/flagellar motility. J Cell Biol. 2013;201:173–175. doi: 10.1083/jcb.201302077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Patel-King RS. The oliogmeric outer arm dynein assembly factor CCDC103 is tightly integrated within the ciliary axoneme and exhibits periodic binding to microtubules. J Biol Chem. 2015;290:7388–7401. doi: 10.1074/jbc.M114.616425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Ostrowski LE, Loges NT, Hurd T, Leigh MW, Huang L, Wolf WE, Carson JL, Hazucha MJ, Yin W, et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am J Hum Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán-Zambrano G, Chávez S. Nuclear functions of prefoldin. Open Biol. 2014;4:140085. doi: 10.1098/rsob.140085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilsen M, Turner F, Huchens J, Raff E. Axoneme-specific beta tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr Biol. 2001;11:529–533. doi: 10.1016/s0960-9822(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Newmark P, Reddien P, Cebria F, Sanchez-Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planaria. Proc Natl Acad Sci USA. 2003;100:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O’Toole E, Hara C, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzi J, Becker-Heck A, Castleman V, Al-Mutairi D, Liu Y, Loges NT, Pathak N, Austin-Tse C, Sheridan E, Schmidts M, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, Gilberti RM, Hom EF, King SM. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell. 2013;24:2668–2677. doi: 10.1091/mbc.E13-05-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME. Flagellar motility and the dynein regulatory complex. In: King SM, editor. Dyneins: Structure, Biology and Disease. Waltham, MA: Elsevier; 2012. pp. 337–365. [Google Scholar]
- Robb S, Ross E, Sanchez-Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36(Database issue):D599–D606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, King SM. Schmidtea mediterranea: a model system for analysis of motile cilia. Methods Cell Biol. 2009;93:81–98. doi: 10.1016/S0091-679X(08)93004-1. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Patel-King RS, King SM. An outer arm dynein conformational switch is required for metachronal synchrony of motile cilia in planaria. Mol Biol Cell. 2010;21:3669–3679. doi: 10.1091/mbc.E10-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Saeki M, Irie Y, Ni L, Yoshida M, Itsuki Y, Kamisaki Y. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-α. Biochem Biophys Res Commun. 2006;342:568–572. doi: 10.1016/j.bbrc.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Tarkar A, Loges NT, Slagle CE, Francis R, Dougherty GW, Tamayo JV, Shook B, Cantino M, Schwartz D, Jahnke C, et al. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Lührmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Hirono M, Kamiya R. Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J Cell Biol. 2010;190:65–71. doi: 10.1083/jcb.201002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa H, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol. 2013;201:263–278. doi: 10.1083/jcb.201211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Yuan S, Cao Y, Kallakuri S, Li Y, Kishimoto N, DiBella L, Sun Z. Reptin/Ruvbl2 is a Lrrc6/Seahorse interactor essential for cilia motility. Proc Natl Acad Sci USA. 2013;110:12697–12702. doi: 10.1073/pnas.1300968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.