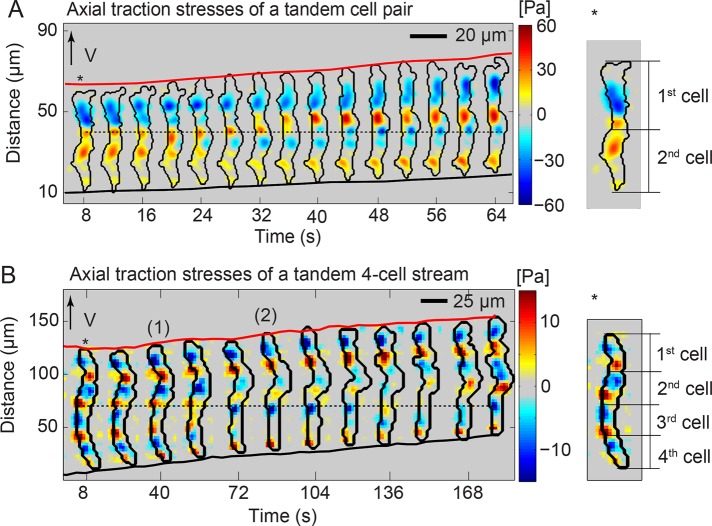
Tandem pairs of Dictyostelium cells migrate synchronously with an ~54-s time delay between the formation of their frontal protrusions. Each cell establishes two active adhesions, with the trailing cell reusing the location of the adhesions of the leading cell. This coordinated motility is mechanically driven and aided by cell–cell adhesions.
Abstract
Streams of migratory cells are initiated by the formation of tandem pairs of cells connected head to tail to which other cells subsequently adhere. The mechanisms regulating the transition from single to streaming cell migration remain elusive, although several molecules have been suggested to be involved. In this work, we investigate the mechanics of the locomotion of Dictyostelium tandem pairs by analyzing the spatiotemporal evolution of their traction adhesions (TAs). We find that in migrating wild-type tandem pairs, each cell exerts traction forces on stationary sites (∼80% of the time), and the trailing cell reuses the location of the TAs of the leading cell. Both leading and trailing cells form contractile dipoles and synchronize the formation of new frontal TAs with ∼54-s time delay. Cells not expressing the lectin discoidin I or moving on discoidin I–coated substrata form fewer tandems, but the trailing cell still reuses the locations of the TAs of the leading cell, suggesting that discoidin I is not responsible for a possible chemically driven synchronization process. The migration dynamics of the tandems indicate that their TAs’ reuse results from the mechanical synchronization of the leading and trailing cells’ protrusions and retractions (motility cycles) aided by the cell–cell adhesions.
INTRODUCTION
Directional cell migration is important in various physiological and pathological processes, ranging from wound healing to metastatic cancer invasion (Roussos et al., 2011). It is also essential for the survival of the social amoeba Dictyostelium discoideum (Bagorda et al., 2006). When starved, Dictyostelium cells become highly motile and enter a differentiation program that leads to the formation of long, tightly packed cell streams in which cells form head-to-tail attachments (Hirose et al., 2011). They do so by producing, secreting, and collectively responding to cAMP in a regulated manner (Driscoll et al., 2012). Further into their development, these streams converge into an aggregation center that gives rise to the fruiting body. The similarities Dictyostelium cells share with leukocytes and other highly motile cells make them an excellent model with which to study directional cell migration, as well as the transition from single-cell to collective-cell motility (Friedl et al., 2001).
Four key mechanisms drive the migration of Dictyostelium single cells and multiple-cell streams: 1) actin polymerization and/or 2) lateral contractions mediated by cortical tension promote protrusion of the cell’s leading edge; 3) actomyosin contractility powers the retraction of the back cell edge; and 4) cell–substratum adhesion enables the transmission of the necessary forces that drive cell movement (Friedl et al., 2001; Bastounis et al., 2014; Alvarez-Gonzalez et al., 2015). Unlike neutrophils, which adhere to their substratum via firm integrin-based focal adhesions, Dictyostelium cells form transient diffuse adhesions (Fey et al., 2002). Although various molecules have been shown to be involved in Dictyostelium adhesion, the precise adhesion mechanism is unknown, and there is controversy as to whether nonspecific van der Waals forces play a role in the process (Loomis et al., 2012). In addition, there is a lack of consensus on the nature of the interactions of the cell surface glycoproteins with the substratum and whether migration and streaming are guided by the externalization of proteins from cells that could form a “trail of breadcrumbs” behind them, allowing other cells to sense and follow them (Springer et al., 1984; Uchida and Yumura, 1999; Loomis et al., 2012). Besides the externalization of some matrix protein suggested by others, Hirose et al. (2011) showed that the pair of polymorphic genes, tiger gene B1 (tgrB1) and tgrC1, which mediate cell–cell adhesion through direct binding, are necessary for proper streaming to take place. However, it is not known whether these genes are responsible for the formation of tension-bearing elements linked to the cytoskeletons of neighbor migrating cells.
Using traction force microscopy, we previously showed that single Dictyostelium cells contract axially by exerting traction forces on their substratum at two regions (traction adhesions [TAs]) localized at their front and back halves, thereby forming a contractile dipole (del Álamo et al., 2007; Meili et al., 2010; Bastounis et al., 2011; Álvarez-González et al., 2014; Figure 1A). We also showed that cells prevalently move in a stepwise manner, with their TAs remaining stationary in space and stable in time (Bastounis et al., 2014). TA dynamics are in concert with the motility cycle of the cell, in which a cell periodically creates a new pseudopod (forming a new TA at its front) and then retracts its back edge forward (breaking an old TA at the rear of the cell), while the old TA at the front becomes the new rear TA. We also found that neutrophil-like dHL-60 cells move in a similar manner.
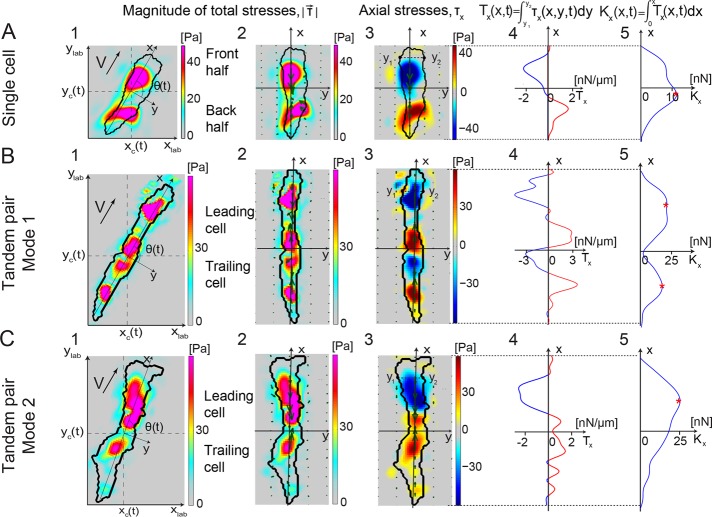
FIGURE 1:
Characterization of the locomotion dynamics of amoeboid tandem pairs with high spatiotemporal resolution. Traction stresses (force/area; 1–3), tension (force/length; 4), and cumulative integral of the tension (force; 5) along the major axis for a representative single cell (A) and pairs of tandem cells (B, C) at a given instant of time. Color bars at the right side of each traction stress map indicate stress values in pascals. 1) Color contours mapping the instantaneous magnitude of the traction stress,  , in the lab reference frame (xlab, ylab). The black contour shows the cell (or cell pair) outline. The axes (x, y) are aligned with the principal axes of the cell (or cell pair), and their origin is located at the center of mass of the cell (or cell pair) (xc, yc). The thick black arrow shows the direction of motion of the cell (or cell pair). 2) Instantaneous magnitude of the traction stresses in a reference frame rotated to coincide with (x, y). Superimposed green arrows show the intensity and direction of the traction stresses. 3) Axial traction stress, τx. Stresses pointing toward the cell (or cell pair) front are considered positive and are indicated with red, and negative stresses are shown in blue. 4) Integral of the axial traction stress across the cell (or cell pair) width (axial tension, Tx) as a function of the position along the length. The horizontal axis displays the tension value in nanonewtons/millimeter. Positive and negative values are indicated by red and blue, respectively. 5) Integral of the axial tension across the cell (or cell pair) length (cumulative integral of tension or internal axial tension, Kx) as a function of the position along the length. The horizontal axis displays the Kx values in nanonewtons. The number of maxima of Kx (red asterisks) indicates whether the cell (or cell pair) moves as one contractile dipole or two. (A) A single cell. (B, C) A cell pair using two different modes of motility: mode 1 (B), in which the pair acts as two contractile dipoles (with four TAs), and mode 2 (C), in which the pair acts as one dipole (with three TAs).
, in the lab reference frame (xlab, ylab). The black contour shows the cell (or cell pair) outline. The axes (x, y) are aligned with the principal axes of the cell (or cell pair), and their origin is located at the center of mass of the cell (or cell pair) (xc, yc). The thick black arrow shows the direction of motion of the cell (or cell pair). 2) Instantaneous magnitude of the traction stresses in a reference frame rotated to coincide with (x, y). Superimposed green arrows show the intensity and direction of the traction stresses. 3) Axial traction stress, τx. Stresses pointing toward the cell (or cell pair) front are considered positive and are indicated with red, and negative stresses are shown in blue. 4) Integral of the axial traction stress across the cell (or cell pair) width (axial tension, Tx) as a function of the position along the length. The horizontal axis displays the tension value in nanonewtons/millimeter. Positive and negative values are indicated by red and blue, respectively. 5) Integral of the axial tension across the cell (or cell pair) length (cumulative integral of tension or internal axial tension, Kx) as a function of the position along the length. The horizontal axis displays the Kx values in nanonewtons. The number of maxima of Kx (red asterisks) indicates whether the cell (or cell pair) moves as one contractile dipole or two. (A) A single cell. (B, C) A cell pair using two different modes of motility: mode 1 (B), in which the pair acts as two contractile dipoles (with four TAs), and mode 2 (C), in which the pair acts as one dipole (with three TAs).
To shed light onto the first steps of the transition between single and collective cell migration, we examined Dictyostelium cell tandem pairs moving during early streaming while linked in a head-to-tail manner. We determined the coordination between the motion of the cells in each pair by analyzing the dynamics of the cells’ TAs. We first classified movement into two modes, depending on whether or not both cells of the pair maintained their single-cell traction force signature (i.e., the contractile dipole). We report that 80% of the time, both cells maintained their single-cell signature, and leading cells formed stable TAs that were reused by trailing cells. The remaining 20% of the time, the TAs generated by the two cells fused into a single contractile dipole. This behavior is associated with an increase in the cell–cell tensional force and was found to lower their migration speed. Remarkably, when the two cells moved in tandem, there was a time delay between the formation of their protrusions. We examined mutants lacking the cell–cell adhesion molecules TgrB1 and TgrC1, which are necessary for stable tandem streaming, to assess their role in the coordinated movement of tandem pairs (Hirose et al., 2011). Cell pairs lacking these proteins adhered poorly to the substratum, had considerably reduced cell–cell forces, and disintegrated into single cells much more often than wild-type pairs. Thus these proteins could act as tension-bearing elements between adjacent cells. We investigated the role of discoidin I in TA reuse, since externalization of this lectin by cells was previously implicated in streaming (Barondes et al., 1985; Crowley et al., 1985; Alexander et al., 1992). However, we found no evidence corroborating that discoidin I secretion was responsible for the reuse of TAs. Taken together, our findings indicate that the mechanics govern the coordination of the motion of cells during the first steps of their transition from single to collective cell migration.
RESULTS
Cells migrating in tandem exhibit two distinct TA patterns
We performed traction force microscopy experiments to examine the dynamics of the TAs of connected pairs of wild-type cells migrating in tandem on collagen I–coated substrata (Supplemental Video S1). We previously found that a single moving cell contracts inward in a dipole manner by exerting traction forces on the substratum on two diffuse areas located along the cell’s major axis (Figure 1A1). Plotting these traction forces in a reference frame in which the coordinate axes (x, y) coincide with the major and minor axes of the cell (Figure 1A2) allows us to study the axial and lateral components of the traction stresses (Figure 1A3). In this coordinate system, in which the cell moves along the x-direction, the axial stresses are negative at the front TA and positive at the rear one, consistent with a dipole of inward contraction (Figure 1A3). Similarly, the axial traction tension Tx (obtained by integrating the axial traction stresses over the cell’s width) is negative at the front half of the cell and positive at the back half (Figure 1A4). The cumulative integral of Tx along the length of the cell provides the internal axial tension, Kx (Lee et al., 2013), which is zero at the edges of the cell and reaches its maximum near its center (Figure 1A5). These mechanical quantities are defined in detail in the Supplemental Materials and Methods.
Using the foregoing methodology, we examined the axial traction stresses and tension and the intercellular tension generated by chemotaxing tandem pairs during early streaming. We found that the pairs can alternate between two different modes of locomotion (Figure 1, B and C). In mode 1, each cell in the pair generates a contractile dipole, thereby maintaining the TA signature of the single-cell case. Remarkably, the internal tension of the pair at the location of the cell–cell junction is almost zero (Figure 1B), suggesting that there is little transmission of mechanical forces between the front and back cells. In contrast, the two cells fuse into a single contractile dipole in mode 2, which causes the internal tension to increase at the cell–cell junction, implying tight mechanical coupling between the two cells (Figure 1C). In this second mode, there is usually one strong TA pointing backward (blue) at the leading cell, which is balanced by two weaker TAs that point forward (red; Figure 1C3).
Wild-type cell pairs moving in tandem maintain their single-cell traction force signature
To investigate the spatiotemporal dynamics of TAs implemented by tandem pairs during locomotion, we plotted kymographs of axial traction stresses, traction tension, and internal tension by stacking consecutive temporal measurements of the spatial distribution of these variables (Figure 2, A and B, and Supplemental Video S2; Bastounis et al., 2014). Using the kymograph of internal tension Kx as input, we performed an automatic identification of the different modes implemented by the cell pairs in time (Figure 2, C and D, and Supplemental Figure S2B). The criterion for mode identification was chosen consistent with Figure 1; if Kx peaks at two locations for each instant of time, the pair is in mode 1, whereas if Kx peaks at only one location, the cell is in mode 2. We applied this classification to 14 wild-type tandem pairs and determined the traction stress maps and motility parameters statistically associated with each mode, normalized by the mean value for the whole cell population (Figure 3). We found that cell pairs adopted mode 1 much more frequently than mode 2 (∼80 vs. ∼20% of the time; Figure 3B). Compared to single cells, tandems considerably increased their strain energy (work done by the cells in deforming the elastic substratum; see Eq. 6 of the Supplemental Materials and Methods), but their migration speed and the ratio of axial to lateral contractility did not change significantly (Supplemental Figure S2, A–D).
FIGURE 2:
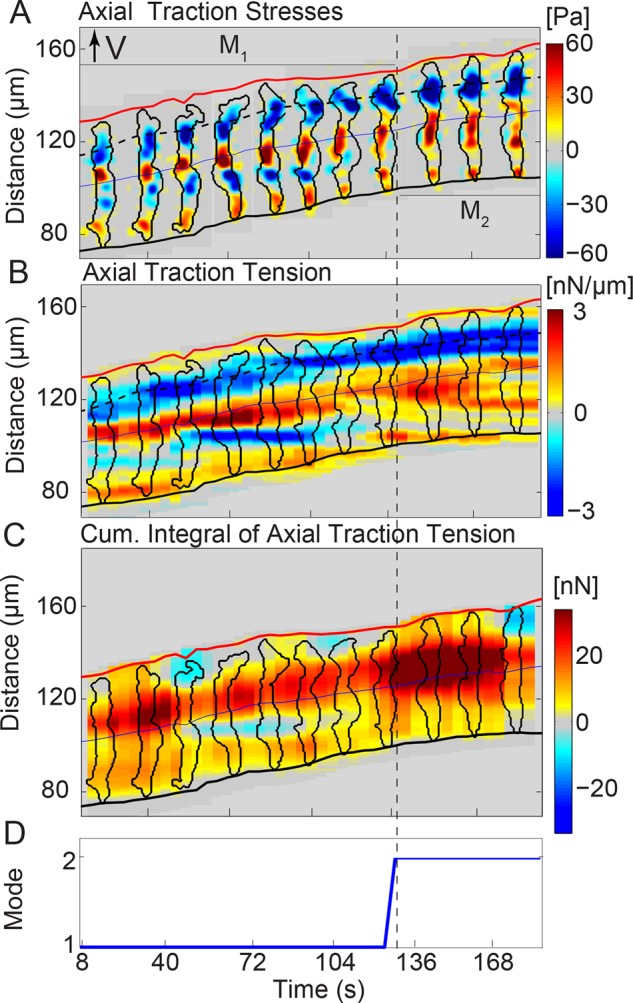
Tandem wild-type pairs migrate by switching between two motility modes with distinct TA dynamics. Spatiotemporal kymographic representation of the axial stresses (A), axial traction tension (Tx; B), and cumulative integral of the axial tension along the cell pair length (Kx; C) as a function of the position along the pair’s trajectory and time. At any given instant of time, the centroid of the pair is displaced vertically according to its motion, so that the pair is moving from bottom to top. The blue line indicates the centroid of the pair, which coincides roughly with the border between the two cells. The inclined red and black lines indicate the instantaneous positions of the front and back pair edges. The black contours are the pair outlines, shown every 16 s. In B, the color map represents Tx in nanonewtons/micrometer plotted every 4 s. The dashed black line shows the location of the minimum traction tension at the leading cell’s front. Red or blue patches represent positive or negative values of Tx, respectively, and correspond to tensions aligned with the direction of the pair’s motion or pointing in the opposite direction. In C, Kx tracks whether the pair forms one contractile dipole (three TAs, one red patch in the Kx kymograph) or two (four TAs, two red patches in the Kx kymograph). (D) On the basis of the number of red patches (local maxima), one can categorize the pair’s TA dynamics into two modes: the pair migrates forming two contraction units (mode 1) or one (mode 2). The dashed vertical lines indicate the respective modes 1 and 2 (M1, M2; see also Supplemental Figure S1).
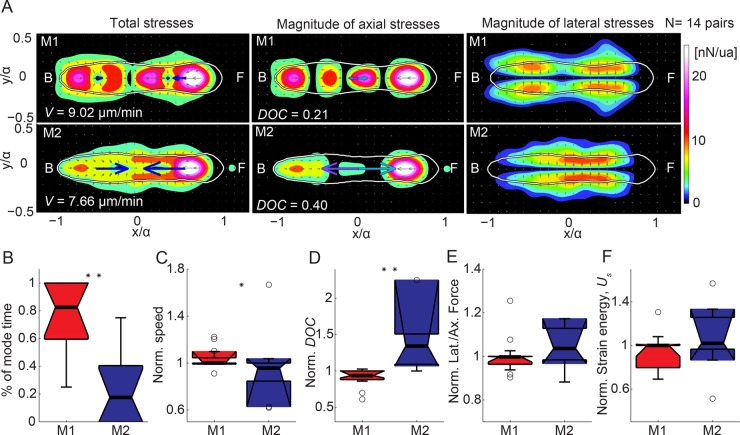
FIGURE 3:
Average motility characteristics of locomoting tandem pairs in modes 1 and 2. (A) Mode-average traction stress maps (nanonewtons/unit area) in the pair-based reference frame for 14 pairs migrating by alternating between modes 1 and 2. Each row corresponds to a different mode as indicated. Left, middle, and right: magnitude of the total, axial, and lateral traction stresses, respectively, for each mode. The color patches indicate the magnitude of the stresses, and the gray arrows denote their direction. Blue arrows indicate the mean net axial force generated by each cell of the pair assuming that the contact line between cells is in the middle of the pair. Purple and cyan arrows indicate the mean force exerted by the leading cell on the trailing cell and vice versa. The white contours show the average shape of the pair. The front (F) and back (B) of the pair are indicated. Mode average speed (V) and degree of coupling (DOC). (B–F) Boxplots of motility parameters corresponding to modes 1 (M1, red) and 2 (M2, blue) for 14 pairs. Each parameter is normalized with its mean value for each pair. (B) Percentage of time spent in mode 1 vs. 2. (C) Speed of migration (V). (D) DOC between the cells of the pair. (E) Ratio of lateral to axial force (Fy/Fx). (F) Total strain energy imparted by the pair (Us). Circles represent outliers, and the notched section of the boxplots shows the 95% confidence interval around the median (Wilcoxon–Mann–Whitney test). One or two asterisks denote statistically significant differences between the medians of two distributions (<0.05 or <0.01, respectively; Wilcoxon rank sum test; see also Supplemental Figure S2).
A tandem pair in mode 1 has higher velocity and lower intercellular forces than a pair in mode 2
While migrating in tandem, the leading and trailing cells of each pair adhere to the substratum and to each other. In contrast to single cells, cells migrating in tandem can exert a nonzero net force on the substratum, which is balanced by the force transmitted to adjacent cells. In our experiments, we found that the leading cell of the tandem generated a forward force on the rear cell and vice versa (Figure 3A). To characterize the mechanical connection between the cells in each pair, we calculated the ratio between the intercellular force and the magnitude of traction forces exerted on the substratum (Figure 3D). We named this ratio the degree of coupling (DOC) of the two cells in the pair. We found that the DOC in mode 2 was twofold higher than in mode 1 (Figure 3, A and D). This difference follows from the fact that in mode 1, the two cells formed two separate contraction dipoles, whereas in mode 2, the pair formed a merged contraction pattern (Supplemental Videos S3 and S4). Of interest, we found that the average velocity of the pairs in mode 1 was higher than in mode 2, implying that both the independent cell–substratum adhesions of each cell of the pair and the reduced cell–cell force transmission result in more effective locomotion (Figure 3C). There were no significant differences in the overall strain energy or in the ratio of lateral to axial contractility when comparing the two modes (Figure 3, E and F).
Cells of tandem pairs undergo motility cycles of similar period but exhibit a 3/4-cycle lag in the formation of leading-edge protrusions
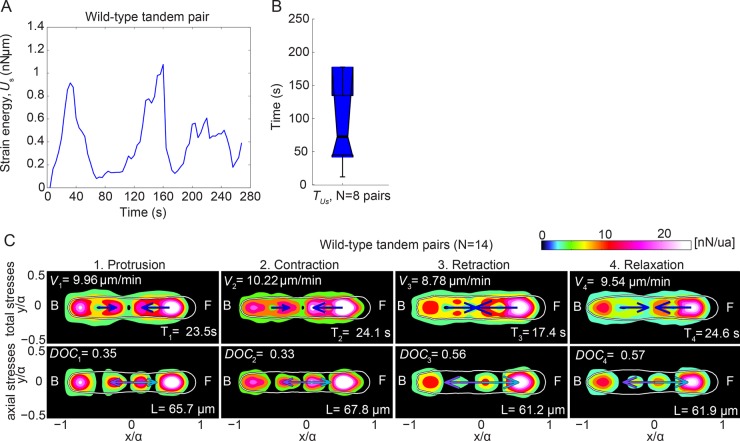
We calculated the autocorrelation function of the strain energy for eight pairs and found that it oscillates quasiperiodically (motility cycle) with a mean period of ∼70 s, similar to single chemotaxing cells (Figure 4, A and B; Weber et al., 1995; Lauffenburger and Horwitz, 1996; del Álamo et al., 2007; Meili et al., 2010). In our previous work for single cells (Meili et al., 2010), we showed that motility can be split into different cycles, using the strain energy time record, and each cycle can be further split into four phases: 1) protrusion of the cell’s front; 2) contraction of the cell’s body; 3) retraction of the rear of the cell; and 4) relaxation (Supplemental Figure S3, A and B). Because the total strain energy of tandem pairs also oscillates quasiperiodically, we split the time records of the movement of cell pairs into cycles and the cycles into phases as we previously did for single cells. We then compiled averaged stress maps for each phase of the cycle (Figure 4C).
FIGURE 4:
Strain energy applied by a tandem pair is quasiperiodic. (A) Quasiperiodic oscillations over 270 s of the total strain energy (Us) imparted by a representative wild-type tandem pair. (B) Boxplots for eight pairs of the period of the oscillations of Us, TUs. Circles represent outliers, and the notched section of the boxplots shows the 95% confidence interval around the median (Wilcoxon–Mann–Whitney test). (C) Phase-average traction stress maps (nanonewtons/unit area) in the pair-based reference frame for Ax2 wild-type pairs (N = 14). The splitting into phases is performed using Us(t). Each column corresponds to a different phase. First and second rows show the magnitude of the total and axial stresses, respectively, for each phase. Front (F) and back (B) of the pairs are indicated. Blue arrows indicate the mean net axial force generated by each cell of the pair, assuming the contact line between cells is in the middle of the pair. Purple and cyan arrows indicate the mean force exerted by the leading cell on the trailing cell and vice versa. Phase-average duration (T), speed (V), DOC between the pair’s cells, and pair length (L; see also Supplemental Figure S3).
We found that the traction stresses of the leading cell increase as it establishes its front TA in the protrusion phase (Figure 4C1). During the ensuing contraction phase, the stresses increase further at both the front and back TAs of the leading cell (Figure 4C2). During the retraction phase, the back TA of the leading cell is unloaded. Finally, during relaxation, the stresses decrease further in both the front and back TAs, and the leading cell’s front elongates, indicating the reinitiation of a new motility cycle (Figure 4C, 3 and 4). In contrast, the traction stresses exerted by the trailing cell are highest during protrusion and then progressively decrease in all the subsequent phases. Interestingly, we found that the DOC is significantly higher during retraction and relaxation than during the contraction and protrusion phases (Figure 4C).
The average stress maps suggest that, although the leading and trailing cells of each pair follow a motility cycle with a common period, the phases of the two cycles are not synchronized. Consistently, when we superimposed information about the phases of the leading cell on the kymographs, we observed that while the leading cell establishes a new frontal TA during protrusion, the trailing cell does not establish its frontal TA concurrently (Supplemental Figure S3C). To quantify the time delay between the initiation of the cycle of the two cells, we analyzed the dynamics of the axial traction tension generated at the frontal TAs. For cell pairs in mode 1, we identified the events when the leading cell establishes a frontal TA (Figure 5, A and B; Supplemental Materials and Methods). We then compiled average maps of the axial traction stress at the instants of time when the leading cell is establishing a new TA, as well as at 4, 8, 12, and 16 s before and after. These data showed that there is an ∼16-s lag between the establishment of a new TA at the front of the leading and trailing cells (Figure 5C). The trailing cell has two TAs at −16 s: the old TA near the middle of the cell that is about to become a back TA and the newly formed TA. Then, at 4 s, the old front TA of the trailing cells is unloaded, and only the new TA is observed. The same process is observed for the leading cell between 0 and 16 s. Given that the mean period of the motility cycle T was 70 s, this finding implies that there was a ¾-cycle delay in the protrusion of the front of the trailing cell compared with the leading cell. Thus the formation of the frontal TA for the trailing cell occurred during the retraction (or at the beginning of the relaxation) phase of the leading cell, which was the phase of maximum transmission of force between the two cells. Overall these results indicate that the rear of the leading cell pulls the front of the trailing cell forward, suggesting that the coordination of migration of the two cells in the pair is mechanical and that the trailing cell is dragged by the leading one.
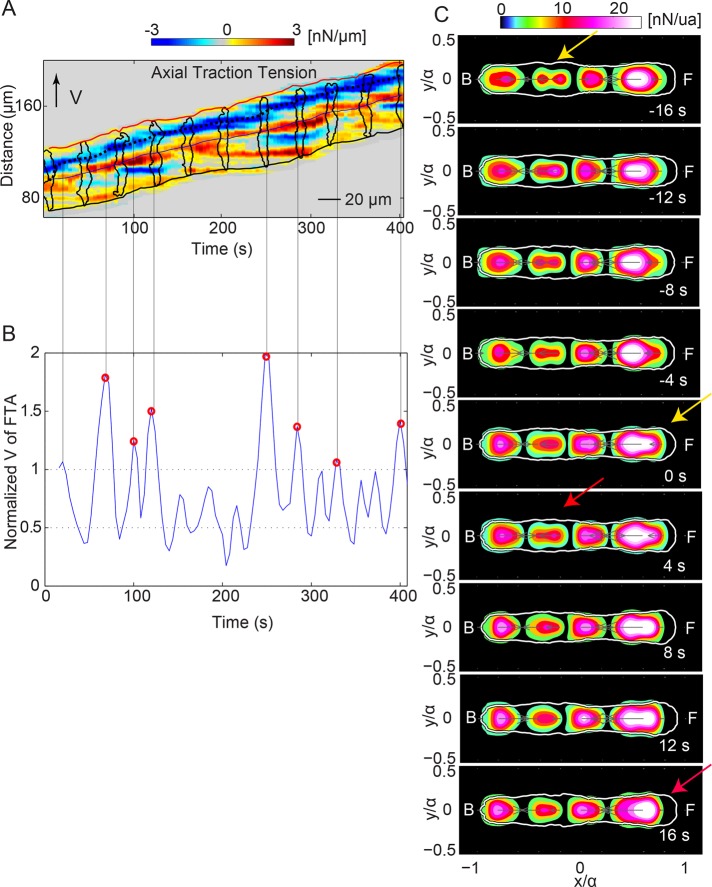
FIGURE 5:
When the leading cell is retracting, the trailing cell is protruding. (A) Axial tension kymograph of a representative wild-type tandem pair. Black dotted line tracks the spatiotemporal evolution of the minimum value of the traction tension, Tx, at the front of the pair’s leading cell. The line is roughly parallel to the time axis, indicating that the front TA of the leading cell is stationary until a new TA is formed. The inclined red and black lines indicate the instantaneous position of the front and back pair edges. The black outlines show the pair contours displayed every 40 s. (B) Identification of the instants of time when a new TA is established at the front of the leading cell, using the peaks of the speed of the position of minimum tension at its front (blue) normalized with the mean velocity of the pair’s centroid (VF/<VCM>). Vertical black lines originate from the peaks of VF/<VCM> and show the time points when a new TA is formed at the leading cell’s front (i.e., (VF/<VCM>) > 1). Red circles show the peaks that coincide with the tandem pair being in mode 1. (C) Average magnitude of the axial traction stresses before, during, and after the establishment of a new TA at the leading cell’s front (six pairs). The color contours display the magnitude of the axial stresses (nanonewtons/unit area), and the white contour shows the average pairs’ outline. Front (F) and back (B) of the pairs are indicated. Average stress maps in a time interval of 16 s before and after the leading cell’s front TA establishment. Yellow and red arrows indicate the instants of time when a new TA is formed at the front of either the trailing or leading cell.
The trailing cell uses the same adhesion sites as the leading cell
We previously observed that the TAs of chemotaxing single cells remain stationary in space and stable in time as the cell body translocates forward (Supplemental Figure S3A; Bastounis et al., 2014). Similarly, during migration of tandem pairs, all of the TAs remained stationary, and the TAs of the leading cell became the TAs of the trailing cell (Figure 6A). We observed that this pattern persists even in streams longer than two cells (three, four, and five cells), suggesting that the movements of all the cells are coordinated when streaming. We also found that, even when the tandem pairs do not follow straight trajectories, they still reuse their stationary TAs (Supplemental Video S8). Further, we have evidence that the migration modes followed by wild-type pairs moving on stiff, 4.5-kPa substrata are the same as for wild-type pairs moving on soft, 1.2-kPa (Supplemental Figure S4), suggesting that substratum stiffness does not influence the migration pattern and the coordinated force generation in the tandem pairs.
FIGURE 6:
TAs of the leading cell are reused by the trailing cell. (A) Kymograph of the instantaneous axial traction stresses for a representative tandem pair. The instantaneous axial stresses and pair contours (black) are displayed every 4 s. Black dotted lines indicate the conversion of the back TAs of the leading cell to front TAs of the trailing cell. Right, approximate location of each cell of the pair based on the TAs and the differential interference contrast image. (B) Kymograph of the instantaneous axial traction stresses for a representative four-cell tandem stream. The instantaneous axial stresses and stream’s contours (black) are displayed every 8 s. Black dotted lines indicate the conversion of the back TA of the stream’s second cell to 1) the front TA of the stream’s third cell and 2) the back TA of the stream’s third cell. Right, approximate location of each cell of the stream (see also Supplemental Figure S4).
Discoidin I plays a role in cell–substratum adhesion and stream formation but not in the reuse of the TAs
The lectin discoidin I (DiscI) is believed to be externalized by developing cells and necessary for cell streaming (Barondes et al., 1985; Crowley et al., 1985; Mathieu et al., 2010). Thus the discoidin I gene (dscA) could play a role in coordinating the motility of tandem pairs by modifying the extracellular matrix. To investigate this hypothesis, we examined the migration of pairs not expressing dscA (dscA− pairs) and the migration of wild-type and dscA− pairs on substrata coated with DiscI.
We first characterized the frequency of tandem pair formation of wild-type and dscA− cells migrating on either collagen-coated or DiscI-coated substrata. We found that an average of 19.57% wild-type cells formed aligned pairs per hour on collagen-coated substrata (Supplemental Figure S5A and Supplemental Videos S5 and S6) versus 5.79% on DiscI-coated substrata (Supplemental Figure S5A and Supplemental Video S7). On the other hand, we found that migrating dscA− cells formed aligned pairs with similar frequency on collagen- and DiscI–coated substrata (Supplemental Figure S5A). We then quantified the time that pairs remained together once they were formed and found that, on collagen-coated substrata, this time was significantly longer for wild-type pairs than for dscA− pairs (Supplemental Figure S5B). Furthermore, dscA− cell pairs moving on DiscI-coated substrata remained together significantly longer than when moving on collagen-coated substrata (Supplemental Figure S5B). Although dscA− cell pairs attached more weakly on their substratum and migrated more slowly than wild-type pairs, we found that single dscA− cells as well as cells that do not express both discoidin I and II (dscA−/dscE− cells) migrated as rapidly as wild-type cells despite the weak forces they establish with their substratum (Supplemental Figure S6).
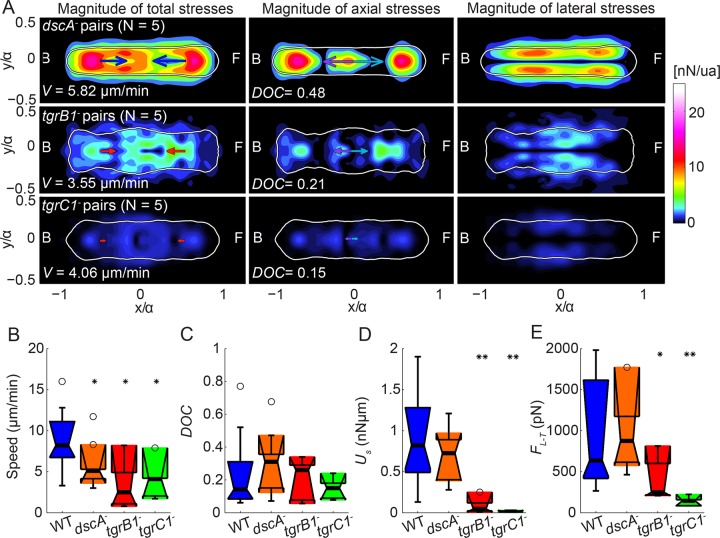
Overall these data suggest that DiscI plays a role in stabilizing the cell–cell connections in migrating tandem pairs in a substratum-dependent manner. Consistently, dscA− cell pairs preferentially established only one contractile dipole and were slower than wild-type pairs (Figure 7). However, inspection of traction-stress kymographs suggests that tandem dscA− pairs still form stationary TAs and reuse their TA locations, particularly the stronger ones (Supplemental Figure S5, D–F, and Supplemental Video S8).
FIGURE 7:
dscA−, tgrC1−, and tgrB1− cell pairs migrate more slowly than wild type. (A) Average traction stress maps (nanonewtons/unit area) in the pair-based reference frame for dscA− (top; N = 5), tgrB1− (middle; N = 5), and tgrC1− (bottom; N = 5) cell pairs. Right, middle, left: magnitude of total, axial, and lateral traction stresses, respectively. The color patches indicate the magnitude of the stresses. Blue or red arrows indicate the mean net axial force generated by each cell of the pair, assuming the contact line between cells is in the middle of the pair. Purple and cyan arrows indicate the mean force exerted by the leading cell on the trailing cell and vice versa. The white contours show the average shape of the pairs. Front (F) and back (B) of the pairs are indicated. Average speed (V) and DOC. (B–E) Boxplots of motility parameters corresponding to wild-type (blue; N = 14), dscA− (orange; N = 5), tgrB1− (red; N = 5), and tgrC1− (green, N = 5) pairs. (B) Speed of migration (V). (C) DOC between the cells’ pairs. (D) Total strain energy imparted by the pairs (Us). (E) Intercellular force (FLT). Circles represent outliers, and the notched section of the boxplots shows the 95% confidence interval around the median (Wilcoxon–Mann–Whitney test). One or two asterisks denote statistically significant differences between the medians of two distributions (<0.05 or <0.01, respectively; Wilcoxon rank sum test; see also Supplemental Figures S5–S7).
Cell pairs that lack the cell–cell adhesion and signaling proteins TgrB1 and TgrC1 still reuse their TAs but do not form stable tandem pairs
To further investigate the possible mechanisms used by the cells to reuse their TAs and the origin of the time delay between the frontal protrusion in the two cells of the tandem pairs, we examined the traction force dynamics of mutant pairs lacking the cell–cell adhesion and signaling molecules TgrB1 and TgrC1 (Dynes et al., 1994; Hirose et al., 2011; Chen et al., 2013). We confirmed that these transmembrane proteins are essential for the cells to form stable tandem pairs (Supplemental Videos S9 and S10; Hirose et al., 2011). We also found that tgrB1− and especially tgrC1− cells, as well as cell pairs, adhered poorly to the substratum and that cell pairs had considerably reduced cell–cell forces. Thus these proteins could act as tension-bearing elements between adjacent cells (Figure 8, A, D, and E, and Supplemental Figure S7, C and D). Although the pairs formed between tgr mutant cells are transient, inspection of short time–history traction-stress kymographs while they move in tandem suggests that the trailing cells still reused the TAs of the leading ones (Supplemental Figure S7, C and D).
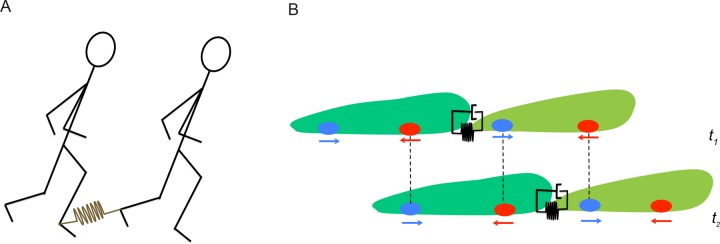
FIGURE 8:
Synchronization of a mechanically linked pair. (A) Schematic representation of two individuals moving one behind the other with their right and left ankles respectively linked through an elastic rope. The rope is shown in brown and represented by a linear spring. (B) Schematic representation of two cells following each other at two different instants of time, t1 and t2. The leading and trailing cells are shown in light and dark green, respectively. The cells are linked together via adhesion molecules modeled as a linear spring parallel to a dashpot (viscoelastic material). Ovals and arrows at the ventral surface of the cells represent TAs and the direction of the net force they exert, respectively. Dashed vertical lines show the reuse of these TAs as the pair proceeds forward.
DISCUSSION
To investigate the mechanics of collective amoeboid cell locomotion, we analyzed the dynamics of the TAs and intercellular forces exerted by Dictyostelium pairs migrating in tandem. We found that both cells of each pair maintain their single-cell traction force signature ∼80% of the time, leading to two contractile dipoles per tandem (mode 1). Then the two cells in the pair move synchronously, with the trailing cell reusing the location of the TAs of the leading cell, which remain spatially stationary. The remaining 20% of the time, the cells in each pair merge their traction forces into a unique contractile dipole (mode 2), increasing their intercellular tensional forces and decreasing their speed. The increased cell–cell tension of this second mode could allow streaming cells to slow and eventually form stalled tight aggregates later during development.
We previously observed that when a single cell closely followed another one without being attached to it, the TAs of the leading cell were not reused by the follower cell (Bastounis et al., 2014). Thus the coordination and reuse of TAs that we observed in migration of tandems likely requires cell–cell adhesions. For the sake of completion, we examined all plausible hypotheses regarding how this synchronization is achieved: 1) mechanically, 2) chemically, 3) by geometrical constraint, or 4) by a combination of these.
We first examined whether this process is mechanically driven by intercellular tensional forces via adhesion molecules connecting the leading and trailing cells (i.e., the springs and dashpots in Figure 8B). Dictyostelium cells use three different cell–cell adhesion systems: csA; DdCAD-1, and the Tgr protein pair TgrB1 and TgrC1 (Coates and Harwood, 2001). Because it is linked to the actin cytoskeleton, csA could be a tension-bearing element. However, cells lacking csA still stream, suggesting that this protein is not essential for tandem coordination (Harloff et al., 1989; Harris et al., 2001). Similarly, cells lacking DdCAD-1 can form streams, which brings into question the need for DdCAD-1 in coordinating early streaming (Wong et al., 2002). However, cells lacking the polymorphic cell–cell adhesion proteins TgrB1 and TgrC1 are unable to form stable tandem streams (Hirose et al., 2011; Chen et al., 2013). Eventually, these mutants can establish short-lived pairs that exert low intercellular forces compared with wild-type pairs, suggesting that the intracellular domains of TgrB1 and TgrC1 are essential to link the cells’ cytoskeletons and coordinate collective locomotion by bearing mechanical tension.
The intercellular tensional force could be responsible for the coordination of tandem cell migration, consistent with a mechanistic model in which the leading cell sets the period and the step length of the motility cycle. The observed ¾-cycle lag between the leading and trailing cells follows from the fact that the rear cell forms a new protrusion when the leading cell tugs on it during the leading cell’s retraction phase. In a very simplistic way, this could be viewed as the manner in which two individuals linked at their ankles through an elastic rope would synchronize their walk (Figure 8A).
We previously showed that polarized chemotaxing cells migrate in a quasiperiodic manner and advance an almost constant step length per motility cycle (del Álamo et al., 2007; Meili et al., 2010; Bastounis et al., 2011). Thus the trailing cell reusing the TAs of the leading cell observed in pairs (and also in longer streams of three and four cells) would be facilitated by the cells of the stream advancing a similar step length per cycle. However, in this scenario, the synchronization of the motility cycles of different cells is not purely geometrical and still requires transmission of mechanical forces. Otherwise, a stream would break in front of a cell with a slightly longer period or behind a cell with a slightly shorter period. Our finding that the formation of the frontal TA in trailing cells occurs during the retraction phase of leading cells without a further delay indicates that the cell–cell coordination is mediated by strong cell–cell adhesions that do not remodel during the motility cycle (high value of the spring constant and low value of the dashpot constant in Figure 8). Our observation that cells lacking TgrB1 and TgrC1 form transient, less-coordinated tandems compared with wild-type pairs further supports the idea that mechanical linkage is necessary and geometric effects alone cannot explain the reuse of the TAs.
Past studies suggested that certain molecules are externalized by the cells onto the substratum and that these molecules could act as signals for other cells to adhere at specific locations. One such molecule that is essential for streaming is DiscI, although direct evidence of its secretion by the cells is yet to be found (Springer et al., 1984; Barondes et al., 1985; Crowley et al., 1985; Alexander et al., 1992). To investigate whether the reuse of the TAs and the coordinated movements of tandems are enabled by DiscI secretion, we investigated wild-type and dscA− cells chemotaxing on collagen-coated and DiscI-coated substrata. Although we were unable to visualize DiscI externalization onto the substratum, we found that wild-type cells formed significantly fewer aligned pairs on DiscI-coated substrata than on collagen-coated ones. Similarly, dscA− cells formed fewer aligned pairs than wild-type cells on both collagen-coated and DiscI-coated substrata. Uchida and Yumura (1999) showed that cellular remnants are left behind migrating cells promoting cell aggregation, without identifying their molecular composition. Of interest, they showed that cells migrate faster and adhere less on substrata coated with these remnants, consistent with our findings showing that cells migrate faster on DiscI-coated substrata but exert very weak traction stresses. However, our measurements of the dynamics of traction stresses indicate that tandems lacking DiscI or migrating on DiscI-coated substrata still reuse the location of their TAs, particularly those where the cells exert large traction stresses. This could imply that TA reuse requires strong and stable cell–substratum adhesions. Indeed, when we placed cells on substrata of increased stiffness, on which cells exert stronger traction stresses, we observed that the vast majority of TAs were reused (Supplemental Figure S5). Thus we conclude that DiscI is implicated in cell–substratum adhesion and plays a role in streaming, but we have no evidence supporting the hypothesis that the coordination of the tandems is chemically driven via DiscI externalization. However, we cannot rule out that secretion of a different protein might contribute to the synchronization of the cell pairs. In fact, Kriebel et al. (2003, 2008) showed that localization of adenylyl cyclase (ACA; an enzyme that produces cAMP) at the posterior of cells is necessary for the formation of streams. Examining mixed pairs of aca− and wild-type cells cannot provide any additional insight because the wild type would be the leaders of the pairs. However, future studies should focus on examining cell pair dynamics of mutants overexpresssing or underexpressing ACA to study the potential role that ACA could play in the synchronization of the locomotion of the tandem pair described here.
In conclusion, our findings suggest that tandem migration of Dictyostelium pairs is a mechanically coordinated process requiring strong cell–cell adhesions but not secretion of DiscI, even if the potential concomitant role of a non-DiscI chemical secretion cannot be discarded yet. Although focused on only the early stages of stream formation, these observations might also provide insight and outline future directions in understanding the transition from single to collective cell migration as occurs in development or in cancer cell migration (Friedl et al., 2004; Roussos et al., 2011).
MATERIALS AND METHODS
In brief, for our experiments, we used the following Dictyostelium cell lines: wild-type cells (Ax2, obtained from the Dictyostelium Stock Center [dictybase.org/StockCenter/StockCenter.html]), dscA− cells (generated from Ax2 in our laboratory), and dscA−/dscE− double-knockout cells (generated from Ax2 in our laboratory). We also used wild-type cells Ax4 and tgrC1− and tgrB1− cells (a generous gift from G. Shaulsky (Baylor College of Medicine, Houston, TX). Before the time-lapse microscopy recordings, cells were starved 6 and 8 h for single chemotaxis and streaming assays, respectively. Cells were placed on a collagen I–coated polyacrylamide substratum that had embedded fluorescent beads on its upper layer. Using light or confocal microscopy to image the cells under study and the distribution of the beads embedded on the substratum, we acquired multichannel time-lapse images. Using correlation techniques and traction force microscopy, we calculated the stresses that cells or cell pairs apply to their substratum in order to drive their movement. Using our novel kymographic techniques and statistical methods, we categorized motility into distinct modes and characterized the mechanics of streaming cell pair migration. Full experimental details are provided in the Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Susan Lee and Christos Tzitzilonis for assistance. We are indebted to Shigenori Hirose, Gad Shaulsky, and Adam Kuspa (Baylor College of Medicine, Houston, TX) for advice and providing all of the Tgr cell lines used in this work. This work was funded by U.S. Public Health Service Grants R01 GM084227 and R01 GM037830.
Abbreviations used:
- dscA
discoidin I gene
- DiscI
discoidin I
- DOC
degree of cell–cell coupling
- TA
traction adhesion
- tgr
tiger gene.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0836) on February 24, 2016.
REFERENCES
- Alexander S, Sydow LM, Wessels D, Soll DR. Discoidin proteins of Dictyostelium are necessary for normal cytoskeletal organization and cellular morphology during aggregation. Differentiation. 1992;51:149–161. doi: 10.1111/j.1432-0436.1992.tb00691.x. [DOI] [PubMed] [Google Scholar]
- Álvarez-González B, Bastounis E, Meili R, del Álamo JC, Firtel R, Lasheras JC. Cytoskeletal mechanics regulating amoeboid cell locomotion. Appl Mech Rev. 2014;66:050804. doi: 10.1115/1.4026249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez B, Meili R, Bastounis E, Firtel RA, Lasheras JC, Del Alamo JC. Three-dimensional balance of cortical tension and axial contractility enables fast amoeboid migration. Biophys J. 2015;108:821–832. doi: 10.1016/j.bpj.2014.11.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagorda A, Mihaylov VA, Parent CA. Chemotaxis: moving forward and holding on to the past. Thromb Haemost. 2006;95:12–21. [PubMed] [Google Scholar]
- Barondes SH, Haywood-Reid PL, Cooper DN. Discoidin I, an endogenous lectin, is externalized from Dictyostelium discoideum in multilamellar bodies. J Cell Biol. 1985;100:1825–1833. doi: 10.1083/jcb.100.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E, Meili R, Alonso-Latorre B, del Álamo J, Lasheras J, Firtel R. The SCAR/WAVE complex is necessary for proper regulation of traction stresses during amoeboid motility. Mol Biol Cell. 2011;22:3995–4003. doi: 10.1091/mbc.E11-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E, Meili R, Álvarez-González B, Francois J, del Álamo JC, Firtel RA, Lasheras JC. Both contractile axial and lateral traction force dynamics drive amoeboid cell motility. J Cell Biol. 2014;204:1045–1061. doi: 10.1083/jcb.201307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wang J, Xu X, Wu X, Piao R, Siu CH. TgrC1 mediates cell-cell adhesion by interacting with TgrB1 via mutual IPT/TIG domains during development of Dictyostelium discoideum. Biochem J. 2013;452:259–269. doi: 10.1042/BJ20121674. [DOI] [PubMed] [Google Scholar]
- Coates JC, Harwood AJ. Cell-cell adhesion and signal transduction during Dictyostelium development. J Cell Sci. 2001;114:4349–4358. doi: 10.1242/jcs.114.24.4349. [DOI] [PubMed] [Google Scholar]
- Crowley TE, Nellen W, Gomer RH, Firtel RA. Phenocopy of discoidin I-minus mutants by antisense transformation in Dictyostelium. Cell. 1985;43:633–641. doi: 10.1016/0092-8674(85)90235-1. [DOI] [PubMed] [Google Scholar]
- del Álamo JC, Meili R, Alonso-Latorre B, Rodríguez-Rodríguez J, Aliseda A, Firtel RA, Lasheras JC. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc Natl Acad Sci USA. 2007;104:13343–13348. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll MK, McCann C, Kopace R, Homan T, Fourkas JT, Parent C, Losert W. Cell shape dynamics: from waves to migration. PLoS Comput Biol. 2012;8:e1002392. doi: 10.1371/journal.pcbi.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Clark AM, Shaulsky G, Kuspa A, Loomis WF, Firtel RA. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- Fey P, Stephens S, Titus MA, Chisholm RL. SadA, a novel adhesion receptor in Dictyostelium. J Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Borgmann S, Brocker E. Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J Leukoc Biol. 2001;70:491–509. [PubMed] [Google Scholar]
- Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- Harloff C, Gerisch G, Noegel AA. Selective elimination of the contact site A protein of Dictyostelium discoideum by gene disruption. Genes Dev. 1989;3:2011–2019. doi: 10.1101/gad.3.12a.2011. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Awrey DE, Cox BJ, Ravandi A, Tsang A, Siu CH. Involvement of a triton-insoluble floating fraction in Dictyostelium cell-cell adhesion. J Biol Chem. 2001;276:18640–18648. doi: 10.1074/jbc.M010016200. [DOI] [PubMed] [Google Scholar]
- Hirose S, Benabentos R, Ho H-I, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science. 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebel PW, Barr VA, Parent CA. Adenylyl cyclase localization regulates streaming during chemotaxis. Cell. 2003;112:549–560. doi: 10.1016/s0092-8674(03)00081-3. [DOI] [PubMed] [Google Scholar]
- Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–961. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lee HH, Lee HC, Chou CC, Hur SS, Osterday K, del Alamo JC, Lasheras JC, Chien S. Shp2 plays a crucial role in cell structural orientation and force polarity in response to matrix rigidity. Proc Natl Acad Sci USA. 2013;110:2840–2845. doi: 10.1073/pnas.1222164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF, Fuller D, Gutierrez E, Groisman A, Rappel W-J. Innate non-specific cell substratum adhesion. PLoS One. 2012;7:e42033. doi: 10.1371/journal.pone.0042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu SV, Aragão KS, Imberty A, Varrot A. Discoidin I from Dictyostelium discoideum and interactions with oligosaccharides: specificity, affinity, crystal structures and comparison with Discoidin II. J Mol Biol. 2010;400:540–554. doi: 10.1016/j.jmb.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Alonso-Latorre B, Álamo del JC, Firtel RA, Lasheras JC. Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol Biol Cell. 2010;21:405–417. doi: 10.1091/mbc.E09-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer WR, Cooper DNW, Barondes SH. Discoidin I is implicated in cell-substratum attachment and ordered cell migration of Dictyostelium discoideum and resembles fibronectin. Cell. 1984;39:557–564. doi: 10.1016/0092-8674(84)90462-8. [DOI] [PubMed] [Google Scholar]
- Uchida K, Yumura S. Novel cellular tracks of migrating Dictyostelium cells. Eur J Cell Biol. 1999;78:757–766. doi: 10.1016/S0171-9335(99)80044-2. [DOI] [PubMed] [Google Scholar]
- Weber I, Wallraff E, Albrecht R, Gerisch G. Motility and substratum adhesion of Dictyostelium wild-type and cytoskeletal mutant cells: a study by RICM/bright-field double-view image analysis. J Cell Sci. 1995;108:1519–1530. doi: 10.1242/jcs.108.4.1519. [DOI] [PubMed] [Google Scholar]
- Wong E, Yang C, Wang J, Fuller D, Loomis WF, Siu CH. Disruption of the gene encoding the cell adhesion molecule DdCAD-1 leads to aberrant cell sorting and cell-type proportioning during Dictyostelium development. Development. 2002;129:3839–3850. doi: 10.1242/dev.129.16.3839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.