MEC-6 and POML-1 are similar proteins needed for touch sensitivity in Caenorhabditis elegans. These proteins reside primarily in the ER and affect the amount and localization of MEC-4, the DEG/ENaC mechanotransduction channel protein. MEC-6 also accelerates MEC-4 transport to the cell surface in vitro. Thus these proteins appear to act as MEC-4 chaperones.
Abstract
Caenorhabditis elegans senses gentle touch via a mechanotransduction channel formed from the DEG/ENaC proteins MEC-4 and MEC-10. An additional protein, the paraoxonase-like protein MEC-6, is essential for transduction, and previous work suggested that MEC-6 was part of the transduction complex. We found that MEC-6 and a similar protein, POML-1, reside primarily in the endoplasmic reticulum and do not colocalize with MEC-4 on the plasma membrane in vivo. As with MEC-6, POML-1 is needed for touch sensitivity, the neurodegeneration caused by the mec-4(d) mutation, and the expression and distribution of MEC-4 in vivo. Both proteins are likely needed for the proper folding or assembly of MEC-4 channels in vivo as measured by FRET. MEC-6 detectably increases the rate of MEC-4 accumulation on the Xenopus oocyte plasma membrane. These results suggest that MEC-6 and POML-1 interact with MEC-4 to facilitate expression and localization of MEC-4 on the cell surface. Thus MEC-6 and POML-1 act more like chaperones for MEC-4 than channel components.
INTRODUCTION
Gentle touch is sensed in the nematode Caenorhabditis elegans by six touch receptor neurons (TRNs; these cells are the 2 ALM, 2 PLM, 1 AVM, and 1 PVM neurons; Chalfie and Sulston, 1981). Touch is transduced in the TRNs by the activation of a trimeric channel formed by two degenerin/epithelial sodium channel (DEG/ENaC) proteins, MEC-4 and MEC-10 (O’Hagan et al., 2005; Árnadóttir et al., 2011; Chen et al., 2015). Previous work from our lab suggested that another protein, MEC-6, was also part of the mechanosensory channel complex in the TRNs, since it colocalized with MEC-4 in TRN neurites and coimmunoprecipitated with it in heterologous cells (Chelur et al., 2002).
The sequence of MEC-6 and several other predicted C. elegans proteins (Chelur et al., 2002) is similar to that of the three mammalian paraoxonases (PON1–PON3). Human PON1 and PON3 are serum proteins that contribute to high-density lipoprotein particles (Mackness and Walker, 1988; Reddy et al., 2001). In contrast, human PON2, which is ubiquitously expressed (Mochizuki et al., 1998), localizes to the endoplasmic reticulum (ER; Horke et al., 2007; Rothem et al., 2007). The exact function of these proteins is unclear, but they prevent lipid peroxidation (Aviram et al., 1998; Shih et al., 1998; Ng et al., 2001; Reddy et al., 2001; Besler et al., 2011; Devarajan et al., 2011; Huang et al., 2013), and PON1, but not PON2 or PON3, degrades the organophosphate paraoxon (Smolen et al., 1991; Davies et al., 1996; Stevens et al., 2008).
The localization and function of the mammalian paraoxonases suggest that MEC-6 may not be an integral component of the mechanosensory transduction complex but interacts with the channel-forming subunits elsewhere in the cell. Indeed, we recently found that MEC-6 does not colocalize with MEC-4 either on the plasma membrane of Xenopus oocytes or in TRN neurites (Chen et al., 2015). These results, when coupled to studies in Drosophila melanogaster, which has mechanosensory DEG/ENaC proteins (Liu et al., 2003; Zhong et al., 2010; Gorczyca et al., 2014; Guo et al., 2014; Mauthner et al., 2014) but no obvious MEC-6-like proteins (Hicks et al., 2011), led us to reinvestigate the role of MEC-6 in C. elegans touch sensitivity.
Here we show that MEC-6 and a second paraoxonase-like protein that is expressed in the TRNs, paraoxonase and MEC-6-like gene 1 (POML-1, previously named K11E4.3; Topalidou and Chalfie, 2011), primarily reside in the ER of the TRN cell body. Our results suggest that MEC-6 and POML-1 are important for MEC-4 production and localization.
RESULTS
MEC-6 and POML-1 localize to the TRN endoplasmic reticulum
The POML-1 sequence is 30% identical and 42% similar to that of MEC-6. Both MEC-6 (Chelur et al., 2002) and POML-1 contain a transmembrane domain and a nematode-specific region of 15 amino acids in the N-terminus (Supplemental Figure S1A). In contrast to MEC-6, which is expressed in many cells (Chelur et al., 2002), POML-1 was expressed in only a few neurons. In addition to the six TRNs (Topalidou and Chalfie, 2011), POML-1 was found in the IL1, AIM, ALN, and BDU neurons (Supplemental Figure S1B).
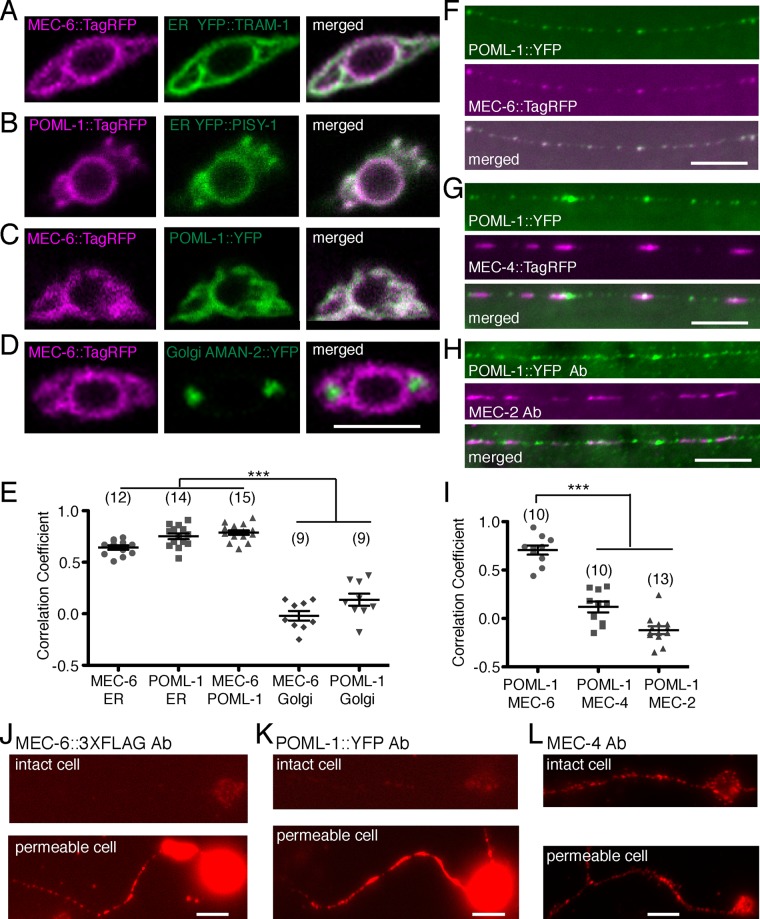
Rescuing and tagged translational fusions of MEC-6 and POML-1 are primarily expressed in the TRN cell body, with weak diffuse expression and puncta in the proximal TRN neurite (Supplemental Figure S1C; Chen et al., 2015). Using antibodies against tagged proteins, we usually saw POML-1 further along the TRN neurites (up to 100 μm of the ∼400-μm neurite) than MEC-6 (Supplemental Figure S1D). In the cell body, translational fusions for both proteins formed a perinuclear mesh-like structure that colocalized with markers for the ER (yellow fluorescent protein (YFP)::TRAM-1 and YFP::PISY-1, which label rough ER and general ER, respectively; Rolls et al., 2002) and each other (Figure 1, A–C), but not with a marker for the Golgi apparatus (AMAN-2::YFP; Figure 1, D and E, and Supplemental Figure S1E). POML-1 partially overlapped with the Golgi marker in 3 of 10 cells (Supplemental Figure S1E).
FIGURE 1:
TRN expression of MEC-6 and POML-1. Confocal sections of TRN cell bodies of (A) MEC-6::TagRFP and the ER marker YFP::TRAM-1, (B) POML-1::TagRFP and the ER marker YFP::PISY-1, (C) MEC-6::TagRFP and POML-1::YFP, and (D) MEC-6::TagRFP and the Golgi marker AMAN-2::YFP and their correlation coefficient (E). Scale bars, 5 μm (A–D, F–H, J–L). The number of examined TRNs is given in parentheses (E, I). Symbols for significance here and in all subsequent figures are described in Materials and Methods. Neurite expression of (F) POML-1::YFP and MEC-6::TagRFP, (G) POML-1::YFP and MEC-4::TagRFP, and (H) POML-1::YFP and MEC-2 and their correlation coefficient (I). Anti-GFP and anti-MEC-2 antibodies (Ab) were used to label the proteins in (H). (J) MEC-6::3XFLAG expression as detected by an anti-FLAG antibody in intact (top) and permeabilized (bottom) cultured TRNs. (K) POML-1::YFP expression as detected by an anti-GFP antibody in intact (top) and permeabilized (bottom) TRNs in culture. The faint immunofluorescence in intact cells (J, K) was not specific because it was often observed in cells that did not express MEC-6::3XFLAG or POML-1::YFP. Images in J and K are representative of 40 cells examined in two independent experiments. (L) MEC-4 expression detected with an anti-MEC-4 antibody that recognizes the extracellular domain in intact (top) and permeabilized (bottom) cultured TRNs. Images are representative of 20 cells examined in two independent experiments.
When seen, MEC-6 and POML-1 puncta colocalized in the proximal TRN neurite (Figure 1F). As with MEC-6 (Chen et al., 2015), POML-1 puncta differed from and did not colocalize with MEC-4::TagRFP or MEC-2 puncta in TRN neurites (Figure 1, G–I), although some overlap was observed. General ER but not rough ER is also present in TRN neurites (Supplemental Figure S1F; Rolls et al., 2002). In general, POML-1 puncta (of the cells represented in Figure 1, F and G) were smaller and closer together than the MEC-4 and MEC-2 puncta: POML-1 puncta were 0.90 ± 0.02 μm wide and separated by 1.17 ± 0.08 μm (25 PLM neurites), and MEC-2 puncta were 1.95 ± 0.05 μm wide and separated by 3.86 ± 0.13 μm (30 PLM neurites; p < 0.0001 for both puncta width and the distance between puncta for POML-1::YFP and MEC-2).
Consistent with the ER localization of MEC-6 and POML-1, we found that both proteins were absent from the TRN surface. Previous work suggested that MEC-6 was a membrane protein that extended its C-terminus extracellularly (Chelur et al., 2002). Specifically, LacZ fused to the C-terminus of MEC-6 produced no β-galactosidase activity unless a synthetic transmembrane domain was inserted between MEC-6 and LacZ (LacZ produces β-galactosidase activity only intracellularly). We found that POML-1 acted similarly (Supplemental Figure S1G). This result suggests that the C-termini of MEC-6 and POML-1 are not located in the cytoplasm but are either outside the cell or in the lumen of an internal organelle. MEC-6 was glycosylated when expressed in CHO cells, and C-terminally hemagglutinin (HA)-tagged MEC-6 was detected by surface immunostaining against HA tags in CHO cells (Chelur et al., 2002). However, in cultured TRNs from wild-type embryos (using antibodies directed against C-terminal tags of MEC-6 and POML-1), we were able to detect the proteins only when the cells were permeabilized, indicating the absence of both MEC-6 and POML-1 expression on the cell surface (Figure 1, J and K; 40 cells for each). On the contrary, an antibody recognizing the extracellular region of MEC-4 detected clear expression of MEC-4 on the cell surface in intact cells (Figure 1L; 20 cells). Because MEC-6 and POML-1 could be expressed, albeit weakly, on the surface of Xenopus oocytes (Supplemental Figure S1H; Chen et al., 2015) and CHO cells in previous experiments (Chelur et al., 2002), either control over the subcellular localization is tighter in the TRNs or a small, undetected amount of MEC-6 and POML-1 goes to the TRN surface. Most of MEC-6 and POML-1, however, was detected in the TRN ER.
The foregoing fusion constructs, including poml-1::yfp, poml-1::tagrfp, mec-4::tagrfp, mec-6::3Xflag, and mec-6::tagrfp, produced functional products, since they could rescue poml-1, mec-4, and mec-6 null mutations (Supplemental Figure S1I; Chen et al., 2015).
POML-1 affects the function of MEC-4
The failure of POML-1 to colocalize with MEC-4 suggests that POML-1 may not function directly in transduction. To study the role of poml-1 gene, we first generated poml-1 null alleles. Using Mos1-mediated gene deletion (Frokjaer-Jensen et al., 2010; Frokjaer-Jensen et al., 2012), we obtained two null mutations, u881 and u882, which lacked the entire poml-1 coding sequence. We obtained four additional mutations using ethyl methanesulfonate mutagenesis: three splicing junction mutations (u851, u852, and u853) and one missense mutation (u854; Supplemental Figures S1A and S2A). Two poml-1 alleles (ok2266 and tm4234; Supplemental Figures S1A and S2A) with the gene partially deleted were previously known. For many of the experiments, we used the ok2266 allele, which acted as a null, since it produced the same phenotype as poml-1(u881; see later discussion).
None of the eight mutations produced touch insensitivity or any other obvious phenotype. The mutations, however, did render animals containing sensitizing mutations touch insensitive (Figure 2A and Supplemental Figure S2B). These sensitizing mutations were temperature-sensitive alleles of mec-4 and mec-6 (Gu et al., 1996), two hypomorphic alleles of mec-6 (u511(G235E) and u518(G213E); García-Añoveros, 1995), and null alleles of crt-1, which encodes the ER chaperone calreticulin (Park et al., 2001; Xu et al., 2001). Except for the crt-1 mutations, which lower MEC-4 protein level and cause slight touch defects (Xu et al., 2001), none of the sensitizing mutations produced touch insensitivity on their own. All of the mutations produced severe touch insensitivity in poml-1 null mutants. These synthetic phenotypes suggest a role for poml-1 in TRN touch sensitivity. The loss of MEC-6, POML-1, or CRT-1 did not affect the general physiology of the TRNs, since light activation of TRN-expressed channelrhodopsin-2 (Nagel et al., 2003) produced the same response in crt-1, mec-6; poml-1, and crt-1; poml-1 mutants as in wild type (Supplemental Figure S2C).
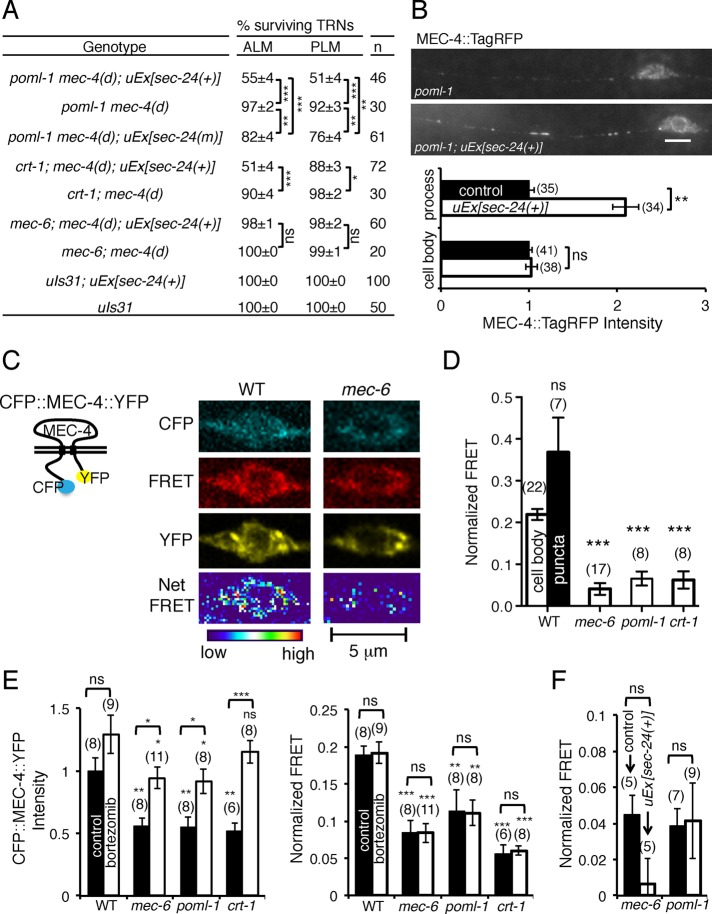
FIGURE 2:
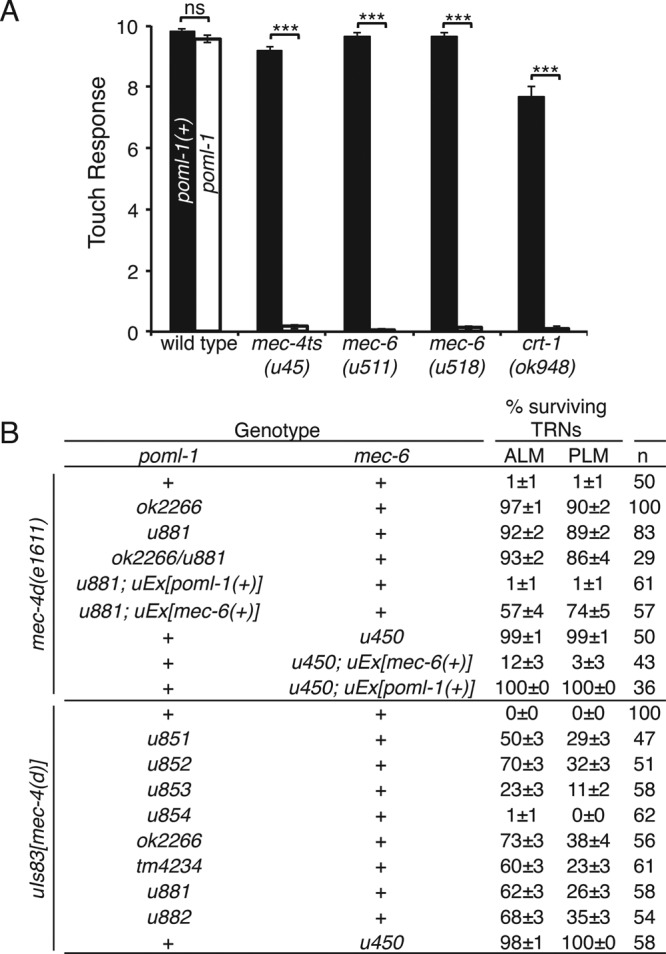
POML-1 is required for touch sensitivity and mec-4(d)–induced TRN degeneration. (A) poml-1(u882) reduced the touch response to 10 touches in sensitized backgrounds (30 animals were examined in three independent experiments). (B) poml-1 and mec-6 mutations suppress mec-4(d) degenerations. n, number of animals examined. TRNs labeled with GFP in L4 and young adult animals were scored as having survived. uIs83 is an integrated array that overexpresses mec-4(d). The rescue experiments used three to five stable lines.
Other evidence for a role of POML-1 in MEC-4 function comes from the suppression by poml-1 mutations of the TRN degeneration caused by the hyperactive channel encoded by the mec-4(d) gain-of-function mutation e1611 (Driscoll and Chalfie, 1991; Brown et al., 2007). mec-6 mutations, but not those of other touch sensitivity genes, suppressed these deaths (Chalfie and Wolinsky, 1990; Huang and Chalfie, 1994). In contrast to their weak effects on touch sensitivity, seven of the eight poml-1 alleles (all but the missense mutation u854) strongly suppressed mec-4(d) neuronal degeneration to different extents (Figure 2B) and did so on their own. The suppressed animals were all touch insensitive, suggesting that either insufficient MEC-4(d) is available for touch sensitivity or the mutant channels cannot transduce touch.
Overexpressing poml-1(+) rescued the poml-1 phenotype in poml-1 mec-4(d) animals (resulting in TRN degeneration) but not the mec-6 phenotype in mec-6 mec-4(d) animals (Figure 2B). Similarly, overexpressing mec-6(+) caused almost all of the TRNs to die in mec-6 mec-4(d) animals but only 35% of the TRNs in poml-1 mec-4(d) animals (Figure 2B). These results suggest that mec-6 and poml-1 have activities that cannot be replaced by the other gene.
Overexpressing mec-4(d) by the multiple-copy insertion uIs83 partially caused degeneration in poml-1 animals but not in animals with the mec-6(u450) mutation (Figure 2B), which deletes most of the mec-6 coding sequence and is considered to be a null allele (Chelur et al., 2002). Unlike mec-6 mutations (Chalfie and Wolinsky, 1990; Shreffler et al., 1995), poml-1 mutations did not suppress other DEG/ENaC gain-of-function mutations, including deg-1 (Supplemental Figure S2D) and unc-8 (20 of 20 animals were still Unc), even though POML-1 and DEG-1 are both expressed in IL1 neurons.
POML-1 modulates MEC-4(d) channel activity in Xenopus oocytes
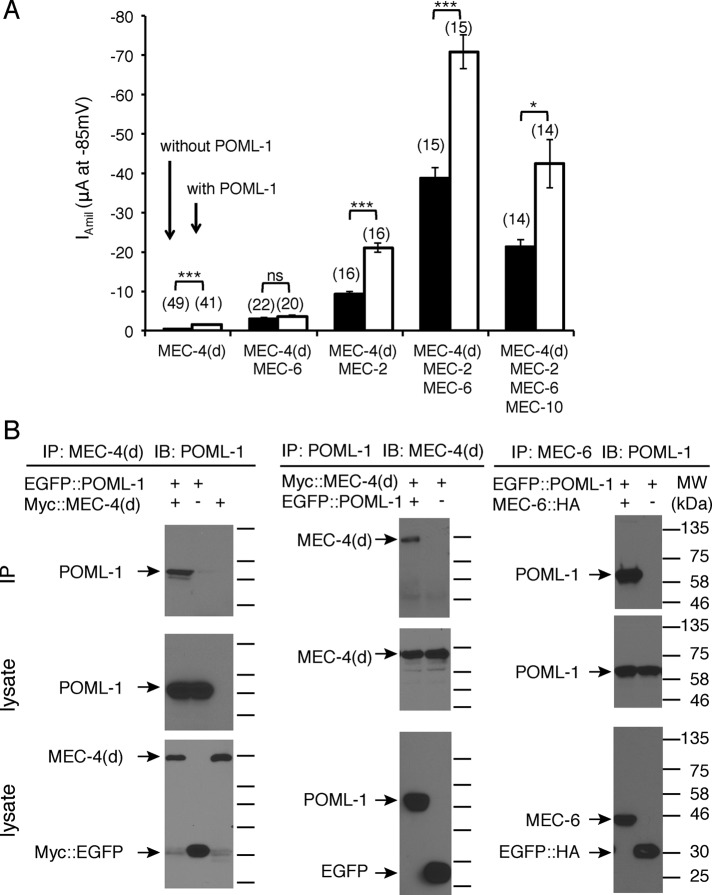
As with MEC-6 (Chelur et al., 2002), POML-1 increased the activity of the MEC-4(d) channel in Xenopus oocytes (Figure 3A and Supplemental Figure S3A). POML-1 increased the amiloride-sensitive Na+ current by threefold, a smaller effect than the 10-fold increase produced by MEC-6. In addition, N-terminally enhanced green fluorescent protein (EGFP)–tagged POML-1 and N-terminally Myc-tagged MEC-4(d) immunoprecipitated each other in oocytes (Figure 3B). Moreover, POML-1 also immunoprecipitated C-terminally HA-tagged MEC-6 (Figure 3B), which is consistent with their colocalization in the ER and suggests that they physically interact. Like MEC-6, POML-1 worked synergistically with MEC-2, but not with MEC-6, to increase channel activity ∼40-fold (Figure 3A). MEC-2, a stomatin-like protein that binds cholesterol, increases MEC-4 channel activity both in vivo and in vitro, perhaps through modulating the lipid environment surrounding the channel (Goodman et al., 2002; O’Hagan et al., 2005; Huber et al., 2006). At higher concentrations, POML-1 on its own, like MEC-6 (Chelur et al., 2002), produced an amiloride-resistant current in oocytes (Supplemental Figure S3, B and C).
FIGURE 3:
The effect of POML-1 on MEC-4(d) channel activity and their physical interaction in Xenopus oocytes. (A) Effect of POML-1 (white bars) on the MEC-4(d) amiloride-sensitive current at −85 mV in oocytes. The number of oocytes tested is given in parentheses. The oocytes were from at least two frogs. (B) Immunoprecipitation (IP) of POML-1 with MEC-4(d) and MEC-6 expressed in oocytes. IB, immunoblot probe. Images are representative of two or three independent experiments. Molecular weights (kilodaltons) of the protein markers are indicated on the right. EGFP::POML-1 is functional, since its coexpression with MEC-4(d) generated amiloride-sensitive currents (EGFP::POML-1 and Myc::MEC-4(d), Iamil = −1.4 ± 0.3 μA, n = 5) that were similar to those of coexpressing untagged POML-1 and MEC-4(d) (Iamil = −1.6 ± 0.6 μA, n = 5) 6 d after cRNA injection. The negative control (–) is EGFP with the same tag.
POML-1 and MEC-6 affect the amount and distribution of MEC-4
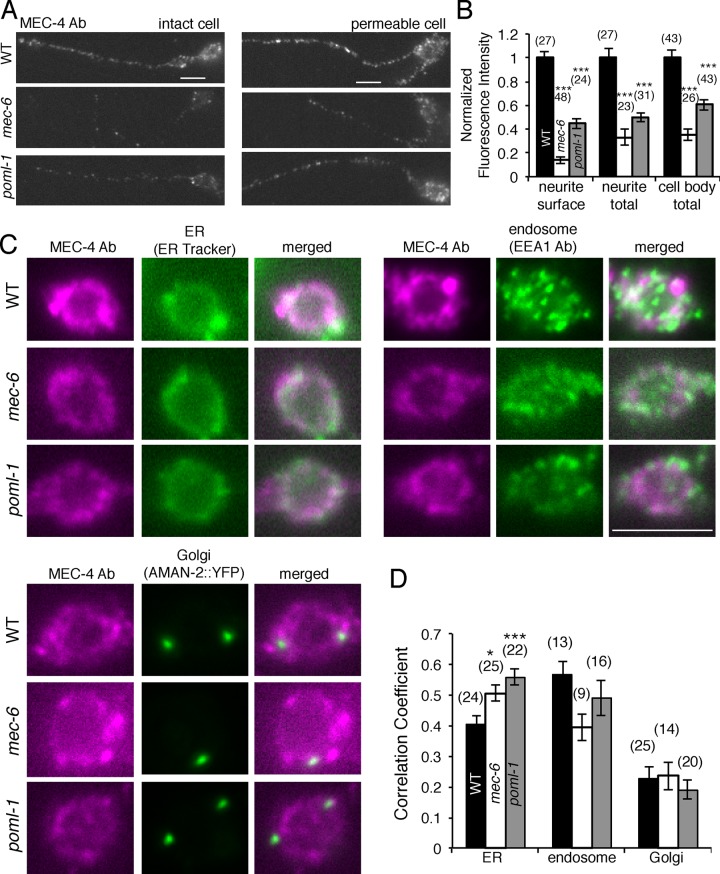
Because POML-1 and MEC-6 affected MEC-4 channel function in vivo and channel activity in oocytes (Figures 2 and 3), we next tested their effect on the production and distribution of MEC-4. We examined wild-type MEC-4 expression in cultured TRN cells with anti-MEC-4 antibodies. Both total expression (detected in permeable cells) and surface expression (detected in intact cells) of MEC-4 were substantially reduced by mec-6 and poml-1 mutations. MEC-4 surface expression was barely detected in cells with a mec-6 null mutation and was reduced by 50% in cells with a poml-1 null mutation (Figure 4, A and B). MEC-4 partially colocalized with ER and endosome markers in wild-type TRN cell bodies but not with Golgi markers (Figure 4C). Less MEC-4 protein appeared in the TRN cell bodies in mec-6 and poml-1 mutants, but the localization of MEC-4 vis-à-vis the ER, endosome, and Golgi markers did not dramatically change (Figure 4, C and D).
FIGURE 4:
MEC-4 expression in cultured TRNs with mec-6(u450) or poml-1(ok2266) mutation. Images (A) and quantification (B) of MEC-4 expression as detected by an anti–MEC-4 antibody in intact (left) and permeabilized (right) TRNs in culture. MEC-4 immunofluorescence intensity was normalized to that of wild-type (WT) TRNs. Scale bars, 5 μm (A, C). Because most cell bodies leaked after immunostaining for intact cells (as evident by the staining for MEC-18, a cytoplasmic protein), immunofluorescence was only measured for MEC-4 surface expression in intact neurites (which did not show MEC-18 staining; Materials and Methods). Statistical significance is indicated for comparison with the WT cells by one-way ANOVA with Tukey post hoc (B, D). The number of cell bodies tested from two or three experiments is given in parentheses (B, D). The subcellular localization of MEC-4 and markers for the ER, endosome, and Golgi (C) in cultured TRNs and their correlation coefficient (D).
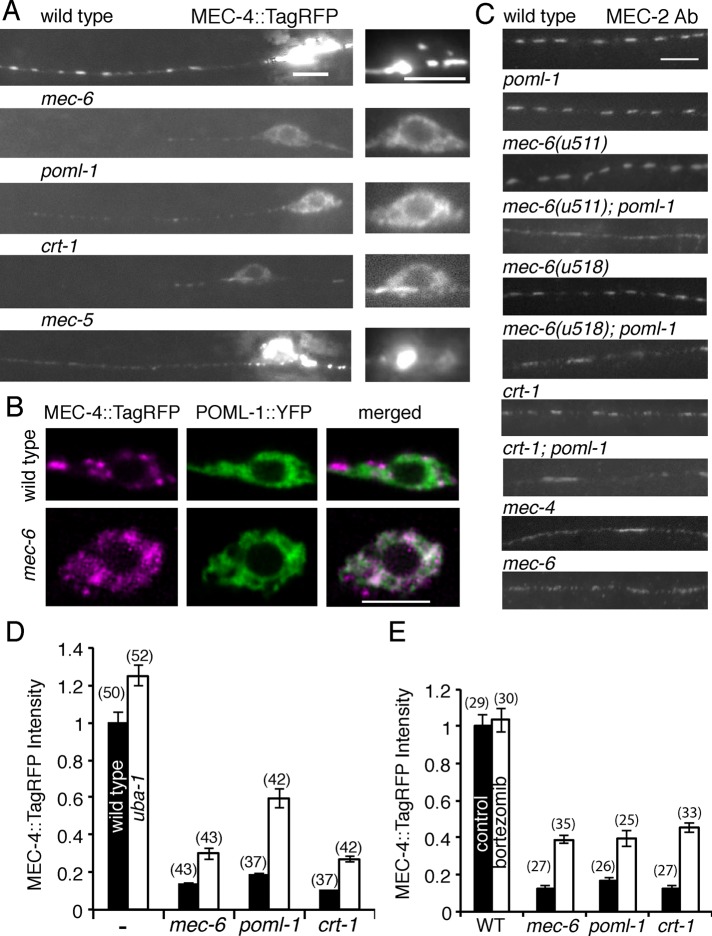
We also examined MEC-4 expression in vivo using a fluorescent protein tag and an anti–MEC-4 antibody. Tagged MEC-4, which produced very strong fluorescence, appeared as large spots in the TRN cell body and smaller puncta in the neurite (Figure 5A; in some cases, the spots were less prominent in the cell body, and a meshwork was seen). Such MEC-4 aggregates were also detected by the anti-MEC-4 antibody in cell bodies, although this fluorescence was much less bright (under these conditions, the diffuse expression was more obvious). In the cell body, MEC-4::TagRFP spots partially colocalized with POML-1 (Figure 5B) and the ER (YFP::TRAM-1), but some were always adjacent to the Golgi (AMAN-2::YFP; 20 TRNs) and the ER exit site (SEC-23::GFP; 10 TRNs; Supplemental Figure S4A). Thus MEC-4 may reside, at least for some time, in ER–Golgi intermediate compartment (Appenzeller-Herzog and Hauri, 2006) or trans-Golgi network (Traub and Kornfeld, 1997).
FIGURE 5:
Effect of mec-6, poml-1, and crt-1 mutations on MEC-4 and MEC-2 expression in the TRNs. Unless noted, the following mutations were used: crt-1(ok948), mec-4(u253), mec-5(u444), mec-6(u450), uba-1(it129), and poml-1(ok2266). (A) MEC-4::TagRFP expression. Left, merged images of expression at 10 focal planes; right, images of the single plane showing the best-focused image of the same cell body. Scale bar, 5 μm (A–C). (B) Confocal images of MEC-4::TagRFP and POML-1::YFP in the TRN cell body of WT animals (top) and mec-6(u450) mutants (bottom). Their correlation coefficient is 0.2 ± 0.04 and 0.7 ± 0.04 in WT and mec-6 mutants (10 TRNs), respectively. (C) MEC-2 expression in the TRN neurite. Representative of 20–30 TRNs examined in two experiments. (D) MEC-4::TagRFP fluorescence intensity (normalized to WT) in PLM cell bodies of L4 larvae and young adults of controls and mec-6(u450), poml-1, and crt-1 without (black) or with (white) a uba-1 mutation. For each pair of white and black bars, the effect of the uba-1 mutation was significant at p < 0.001, except for WT, for which it was at p < 0.01. The comparison to the control within each group (with or without uba-1) was significant at p < 0.001 by two-way ANOVA with Bonferroni posttest. The number of examined PLM cells collected from three independent experiments is given in parentheses (D, E). (E) Fluorescence intensity of MEC-4::TagRFP (normalized to WT control) in PLM cell bodies of WT and mutants either untreated (black) or treated (white) with 50 μM bortezomib for 8 h. Statistical comparisons are as in D, with the exception that the difference between treated and untreated WT animals was not significant.
Consistent with the results seen in cultured TRNs, mec-6 and poml-1 mutations reduced MEC-4 protein levels in vivo as seen with fusion proteins (Figure 5A) and an anti-MEC-4 antibody (Supplemental Figure S4B). As in cultured TRNs (Figure 4, A–B), the reduction was stronger with mec-6 in vivo (Supplemental Figure S4, B and C) and thus correlates with the stronger phenotype of mec-6 with regard to touch sensitivity and mec-4(d) degeneration. Proteins were found mainly in the perinuclear mesh-like structure in the TRN cell body, indicating the ER, and puncta were not seen in the neurites. Indeed, in the TRN cell body with mec-6 mutations, MEC-4 fusion proteins colocalized with POML-1 in the ER (Figure 5B). In addition, the half-maximal pressure (P1//2) for the mechanoreceptor current (MRC) differed slightly between poml-1 and wild-type TRNs (poml-1 P1/2 = 7.7 ± 0.8 nN/μm2 vs. wild-type P1/2 = 4.5 ± 0.7 nN/μm2, mean ± SD; Supplemental Figure S4D), but poml-1 mutations did not affect MRC peak amplitude or kinetics (Supplemental Figure S4E). The effects of mec-6 and poml-1 mutations on MEC-4 appeared to be specific because they did not affect the expression of each other (Supplemental Figure S5A) or of MEC-18 (Supplemental Figure S4B).
The effect of mec-6 and poml-1 mutations was similar to that of a crt-1 null mutation, which also reduced the amount of MEC-4 and its appearance as puncta in TRN cell bodies and neurites (Figure 5A). We found that MEC-6, POML-1, and CRT-1 affected the expression and distribution of MEC-4 protein rather than the amount of mec-4 mRNA as detected by single-molecule fluorescence in situ hybridization (the crt-1 mutation slightly increased the number of mec-4 mRNA molecules in TRN; Supplemental Figure S5B).
The effects of mec-6 and poml-1 mutations on MEC-4 protein levels and distribution suggest that MEC-6 and POML-1 act early in MEC-4 production and/or transport. Consistent with this hypothesis, mutations that presumably affect touch sensitivity after MEC-4 is made (e.g., null mutation of mec-5, a gene encoding an extracellular matrix collagen needed for touch sensitivity; Emtage et al., 2004) disrupted the neurite localization of MEC-4 without affecting the level of MEC-4 protein (normalized intensity of MEC-4::TagRFP in the cell body: wild-type 1 ± 0.06 vs. mec-5(u444) 0.92 ± 0.05; 29 PLM cells; not significant by the Student’s t test) or its distribution in the cell body (Figure 5A).
Because the production of MEC-2 puncta requires MEC-4 (Emtage et al., 2004; Zhang et al., 2004), we also tested the role of POML-1 on MEC-2 distribution using an anti–MEC-2 antibody in poml-1 mutants, the two mec-6 hypomorphic mutants u511(G235E) and u518(G213E), crt-1 mutants, and mec-6 (u511); poml-1, mec-6 (u518); poml-1 and crt-1; poml-1 double mutants. No single mutation caused obvious defects in MEC-2 puncta. In contrast, the double mutants, as in the null mutants of mec-4 and mec-6, had disrupted MEC-2 puncta (Figure 5C), a result that is consistent with the need for the double mutations to cause touch insensitivity.
MEC-6 and POML-1 likely act as chaperones
The similarity of the phenotypes of crt-1, mec-6, and poml-1 mutants, the expression of all three proteins primarily in the ER, and the additivity of their phenotypes with regard to touch sensitivity suggest that MEC-6 and POML-1, like CRT-1, may, at least in part, act as chaperones. If these proteins facilitate the folding/assembly of MEC-4 in the ER, the reduction in MEC-4 protein in these mutants could be a consequence of increased protein degradation. Indeed, the loss of CRT-1, MEC-6, or POML-1 caused an increase in MEC-4 degradation. Mutation of the ubiquitin-activating (E1) enzyme gene uba-1 (Jones et al., 2002) or treatment of animals with the proteasome inhibitor bortezomib increased MEC-4 levels twofold to threefold in mec-6, poml-1, and crt-1 mutants (Figure 5, D and E). uba-1 mutation, but not bortezomib, affected the removal of wild-type MEC-4 from the cell surface (Chen and Chalfie, 2015). The effects of bortezomib on MEC-4 levels in the mec-6, poml-1, and crt-1 mutants thus reveal a different process, perhaps a consequence of the accumulation of misfolded protein.
This increase, however, did not restore MEC-4 levels to those seen in wild type, in part, perhaps, because less MEC-4 protein was produced in mec-6, poml-1, and crt-1 mutants even when the degradation pathway was blocked or the uba-1 mutation and bortezomib treatment only partially suppressed the degradation pathway. Although these treatments increased the amount of MEC-4, they did not change its distribution. MEC-4 was still largely restricted to the same mesh-like structure at perinuclear sites in the cell body seen in the mutants without the uba-1 mutation or bortezomib treatment (Figure 5A and Supplemental Figure S5C). Moreover, the MEC-4 puncta were not restored in TRN neurites (Supplemental Figure S5C). In addition, because uba-1 mutation did not restore touch sensitivity to mec-6 null mutants or crt-1; poml-1 double mutants (Supplemental Figure S5D) and resulted in only a modest increase (16% in ALM for uba-1) of touch cell deaths in poml-1 mec-4(d) animals but not in mec-6; mec-4(d) animals (Supplemental Figure S5E), the increased MEC-4 does not function, perhaps because it is misfolded or not properly localized.
Because overexpression of the ER transport protein SEC-24 rescued the trafficking defects caused by the loss of a putative ER chaperone/cargo receptor but not the folding defects in yeast (Pagant et al., 2015), we tested whether overexpression of C. elegans sec-24 genes (sec-24.1 and sec-24.2) in the TRNs could similarly suppress the effects of crt-1, mec-6, and poml-1 mutations (CRT-1 is also required for the degeneration caused by hyperactive MEC-4(d) channels; Xu et al., 2001). The effects were partial: ∼50% of the TRNs died in poml-1 mec-4(d) animals and 30% in crt-1; mec-4(d) animals, but no TRNs died in mec-6; mec-4(d) animals overexpressing the sec-24 genes (Figure 6A). We also noticed that overexpression of sec-24(+) caused morphological defects: wavy neurites, extra neurites, and misplaced cell bodies; these defects were rarely seen in animals that did not overexpress sec-24(+) (Supplemental Figure S5F). In addition, the overexpression of the C. elegans sec-24 genes with mutated potential cargo-binding sites (corresponding to yeast SEC-24 R230A, R235A, L616W; Miller et al., 2003) reduced but did not eliminate the effect; 20% of the TRNs died in poml-1 mec-4(d) animals (Figure 6A). Although overexpressing SEC-24 in poml-1 mutants doubled the amount of MEC-4::TagRFP in proximal TRN neurites but not in the cell bodies (Figure 6B), it did not restore touch sensitivity to mec-6(u511); poml-1 animals (0.21 ± 0.09 response to 10 touches; 20 animals).
FIGURE 6:
MEC-6, POML-1, and CRT-1 may function as chaperones. The mutations mec-6(u450), poml-1(ok2266), and crt-1(ok948) were used. (A) The effect of sec-24.1 and sec-24.2 overexpression, uEx[sec-24(+)], on the suppression of mec-4(d) deaths by poml-1, crt-1, and mec-6 mutations. In some strains, the cargo-binding sites of sec-24.1 and sec-24.2 were mutated (sec-24(m)). n, number of animals examined. The results with uEx[sec-24(+)] and uEx[sec-24(m)] were collected from two to five stable lines. (B) Effect of overexpressing sec-24(+) on MEC-4::TagRFP fluorescence intensity in the PLM cell bodies and proximal neurites of L4 larvae and young adults with the poml-1 mutation. Data for uEx[sec-24(+)] were collected from two stable lines. The number of PLM cells examined is given in parentheses. Fluorescence intensity was normalized to that of PLM in poml-1 mutants (control). Scale bar, 5 μm. (C) Schematic of CFP::MEC-4::YFP protein (left) and images of CFP::MEC-4::YFP in the TRN cell body taken with the CFP (blue), FRET (red), and YFP (yellow) channels, respectively (right). The Net FRET signal is given by a pseudocolored image to show the relative intensity. (D) The normalized FRET signal (see Materials and Methods) of CFP::MEC-4::YFP either in the TRN cell bodies (white bars) of WT animals and mutants or in the puncta of WT animals (black bar). The number of cell bodies or strongly fluorescent puncta tested (from two or three stable lines with extrachromosomal arrays collected from three of four experiments) is given in parentheses. Statistical significance is indicated for comparison with the FRET signal in the WT cell body. (E) CFP::MEC-4::YFP intensity (measured in the YFP channel and normalized to that WT controls) and FRET signals in the TRN cell body of WT, mec-6, poml-1, and crt-1 animals treated with bortezomib. The number of examined cells bodies here and in F is indicated in parentheses. These experiments used cells from an integrated line, which produced similar FRET signals to the stable lines with extrachromosomal arrays used in D. Bortezomib treatment had a significant effect on CFP::MEC-4::YFP intensity (left, F(1, 58) = 33.56, p < 0.0001) but no effect on the FRET signal (right, F(1, 58) = 0.01, p = 0.9162, by two-way ANOVA with Bonferroni posttests). The value above the bracket is that of the pairwise comparison. The values below the bracket are for the comparison to the WT of each untreated (black bars) or treated group (white bars) by two-way ANOVA with Bonferroni posttests. The difference in CFP::MEC-4::YFP fluorescence intensity (left) between control WT and bortezomib-treated mec-6, poml-1, and crt-1 mutants was not significant. (F) FRET signals in mec-6 and poml-1 animals overexpressing sec-24(+) in TRNs.
These results can be explained if little, if any, MEC-4 folds in mec-6 mutants, but some MEC-4 folds but is not transported to the surface in poml-1 and crt-1 mutants. Thus, by increasing transport to the surface, SEC-24 could cause more mec-4(d)-induced deaths in the poml-1 and crt-1 mutants but not in the mec-6 mutants because they contain no functioning protein. These results suggest an absolute need for MEC-6 in MEC-4 expression and distribution and are consistent with the need for MEC-6 (Chalfie and Sulston, 1981), but not of POML-1 and CRT-1, in touch sensitivity in wild-type animals (Figure 2A).
CRT-1 can bind Ca2+ and regulate Ca2+ homeostasis in the ER (Michalak et al., 2009). Xu et al. (2001) suggested that crt-1 suppression of MEC-4(d)–induced cell death can be attributed to the Ca2+-binding capacity of CRT-1 in the ER and partially reversed by the release of ER Ca2+ induced by thapsigargin. We tested whether poml-1 and mec-6 suppress MEC-4(d) through a similar mechanism and found that, in contrast to crt-1, the effect of poml-1 and mec-6 on cell death was not affected by thapsigargin treatment (Supplemental Figure S5G). Thus poml-1 and mec-6 mutations are less likely to suppress MEC-4(d)–induced cell death by affecting the subcellular Ca2+ level. Moreover, manipulating the subcellular Ca2+ level has no effect on MEC-4 expression (Xu et al., 2001). Therefore mec-6 and poml-1 suppression of MEC-4(d) is primarily due to their effect on MEC-4 expression and distribution.
Because Förster resonance energy transfer (FRET) can be used to monitor protein folding (Philipps et al., 2003), we used a cyan fluorescent protein (CFP)::MEC-4::YFP fusion to examine whether MEC-6, POML-1, and CRT-1 affect the MEC-4 protein folding. This fusion was expressed in the TRNs: as with MEC-4::GFP and MEC-4::TagRFP, the protein formed regular puncta in the neurite (Supplemental Figure S5H) and a mesh-like structure and spots in the cell body, although the spots in the cell body were smaller and dimmer than with MEC-4::TagRFP (Figure 6C). In mec-6, poml-1, or crt-1 mutants, CFP::MEC-4::YFP was restricted to the cell body, where the fluorescence intensity was reduced by nearly 50% (Figure 6C). CFP::MEC-4::YFP produced an efficient FRET signal in wild-type animals (Figure 6, C and D), suggesting that CFP and YFP were close to each other.
In contrast to the FRET signal in wild type, FRET from CFP::MEC-4::YFP was reduced by 70–80% in mec-6, poml-1, or crt-1 mutants (Figure 6, C and D). The reduction of FRET in these mutants was not due to reduced CFP::MEC-4::YFP expression, because the FRET signal was normalized to the CFP and YFP intensities (see Materials and Methods), and wild-type animals expressing similar level of CFP::MEC-4::YFP to these mutants (from uIs190(mec-4p::cfp::mec-4::yfp)/+ animals) produced robust FRET signals (normalized FRET: 0.24 ± 0.06, n = 5). Moreover, bortezomib treatment increased CFP::MEC-4::YFP expression in mec-6, poml-1, and crt-1 mutants to a level similar to that in wild-type untreated animals but had no effect on the FRET signal (Figure 6E). These data suggest that the folding and/or assembly of MEC-4 is compromised in these mutants. Overexpression of SEC-24 also failed to increase the FRET signals in mec-6 and poml-1 mutants (Figure 6F), a result that is consistent with the role for SEC-24 in protein transport but not in protein folding.
Given the localization of MEC-6 and POML-1 in the ER and their potential effects on folding, we wondered whether mec-6 or poml-1 mutations induced a general ER stress response. These mutations, however, did not significantly change the expression of xbp-1b::gfp (Supplemental Figure S5I), which produces a GFP translation product only in response to ER stress (Shim et al., 2004). Wild-type and the mutant strains showed a similar ER stress response when proteasomes were inhibited by bortezomib (Supplemental Figure S5I).
MEC-6 accelerates the surface expression of MEC-4 in Xenopus oocytes
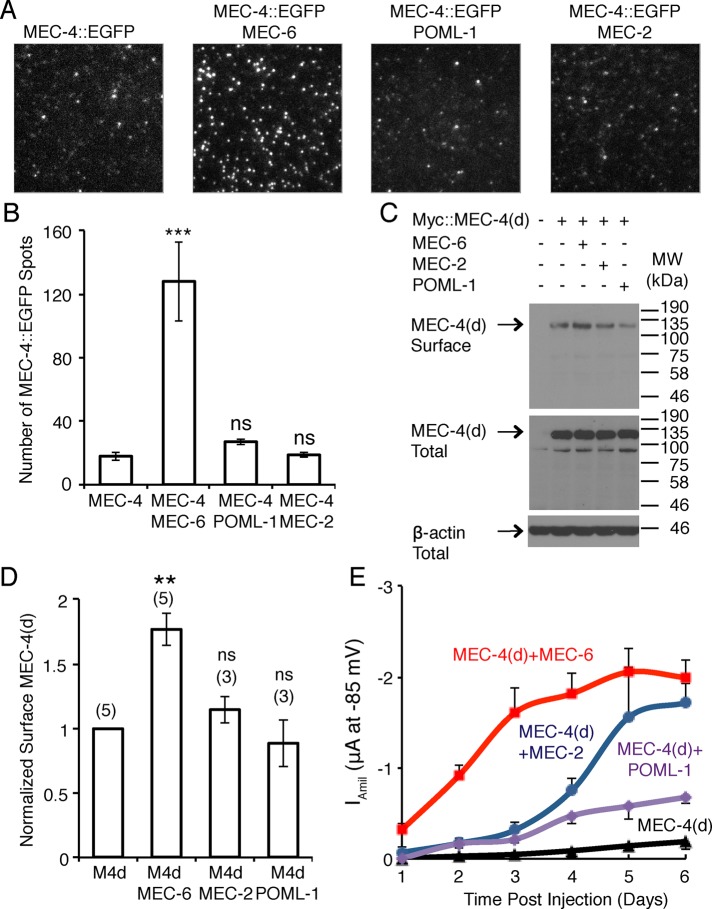
Consistent with its role as a chaperone, we found that MEC-6 greatly increased the amount of surface expression of MEC-4 in Xenopus oocytes, using total internal reflection fluorescence (TIRF) microscopy and biotinylation (Figure 7, A–D) 2 d after complementary RNA (cRNA) injection. In contrast, MEC-2 and POML-1 did not increase the surface expression of MEC-4 (Figure 7, A–D). Although TIRF microscopy cannot be used to determine the position of the protein on the plasma membrane, the biotinylation experiments suggest that MEC-6 affects MEC-4 surface expression. These results support the hypothesis that MEC-6 assists the maturation of MEC-4 channels and/or its transport to the plasma membrane.
FIGURE 7:
Effect of MEC-6 and POML-1 on MEC-4 surface expression in Xenopus oocytes. (A) Images and (B) quantification of MEC-4::EGFP fluorescent spots on the oocyte surface visualized by TIRF imaging (19–29 patches from 14–16 cells from two different batches) 2 d after cRNA injection. Values are compared with the expression of MEC-4::EGFP alone using the Mann–Whitney test. The field dimensions are 13 μm × 13 μm. (C) Western blot of Myc::MEC-4(d) on the surface of oocytes as detected by biotinylation (top) and the expression of Myc::MEC-4(d) in total lysate of oocytes (middle) at 2 d after cRNA injection. β-Actin detected in total lysate was used as an input control (bottom). Molecular weights (kilodaltons) of the protein markers are indicated on the right. (D) Quantification of changes in surface Myc::MEC-4(d) detected by biotinylation at 2 d after cRNA injection (the number of independent experiments is given in parentheses). All data are normalized and compared with Myc::MEC-4(d) expression alone by the one-sample t test. MEC-6, MEC-2, and POML-1 did not affect Myc::MEC-4(d) levels in total lysates at 2 d after injection (MEC-6, 1.0 ± 0.1; MEC-2, 1.0 ± 0.1; POML-1, 0.9 ± 0.1; four or five independent experiments, normalized and compared with the expression of Myc::MEC-4(d) alone; not significant by one-sample t test). The normalized amount of total Myc::MEC-4(d) differed by no more than 25% in any of the experiments. (E) The amiloride-sensitive MEC-4(d) current at −85 mV (12–18 oocytes [2 d after cRNA injection] or 6–12 oocytes [other times] of three batches) on its own and in the presence of MEC-2, MEC-6, and POML-1 at various times after cRNA injection. p < 0.001 for Iamil at −85 mV between oocytes 2 d after injected with MEC-4(d) and MEC-6 vs. MEC-4(d) alone, MEC-4(d) and MEC-2, or MEC-4(d) and POML-1; no statistically significance was found between oocytes 2 d after injection with MEC-4(d) alone vs. MEC-4(d) and MEC-2, or MEC-4(d) and POML-1. p < 0.001 for Iamil at −85 mV between oocytes 1 d after injected with MEC-4(d) and MEC-6 vs. MEC-4(d) alone; p < 0.01 between MEC-4(d) and MEC-6 vs. MEC-4(d) and MEC-2, or MEC-4(d) and POML-1. One-way ANOVA with Tukey post hoc.
The effect of MEC-6 on MEC-4(d) surface expression in oocytes was not seen 5 d after injection by biotinylation (Supplemental Figure S6A), which is consistent with previous experiments (Chelur et al., 2002; Brown et al., 2008). Presumably, the maximum steady-state amount of MEC-4 is found on the surface with or without MEC-6 by 5 d. The lack of an effect in those previous experiments is due, at least in part, to the longer period of expression and presumably to the accumulation of more inactive MEC-4 in the absence of MEC-6.
We also tested whether the early, MEC-6-induced change in membrane-associated MEC-4 affected the MEC-4(d) current in oocytes. Indeed, MEC-6, but not MEC-2 or POML-1, increased the MEC-4(d) current >30-fold to nearly 50% of the maximum current 2 d after injection (Figure 7E; the fold difference is greater here than before [ Figure 3A] because these oocytes had been injected with the lesser amount of mec-4 cRNA, so the MEC-4(d) current was lower). The early effect of MEC-6 compared with POML-1 and MEC-2 on MEC-4(d) activity 2 d after injection was due to different amounts of surface MEC-4(d) rather than of total MEC-4(d) (Figure 7, C and D). Differences in the timing of MEC-6, MEC-2, and POML-1 expression were unlikely to cause these differences in the MEC-4(d) current. MEC-6, MEC-2, and POML-1 were all expressed well 2 d after injection, and all had higher expression level 5 d after injection (Supplemental Figure S6, B–D). In addition, although injecting oocytes with greater amounts of mec-2 and poml-1 cRNA increased the levels of total MEC-2 and POML-1 at 2 d after injection to the levels normally seen at 5 d after injection (Supplemental Figure S6, C and D), MEC-4(d) currents 2 d after injection were not increased (Supplemental Figure S6, E and F).
In addition to MEC-6 increasing MEC-4 membrane expression, it also increased, albeit weakly, the membrane expression of MEC-2 in oocytes (Supplemental Figure S6, G and H). In contrast, although coexpression of POML-1 with MEC-2 doubled MEC-4(d) channel activity over that in oocytes without POML-1 (Figure 3A), POML-1 did not affect MEC-2 surface expression (Supplemental Figure S6, G and H). In addition, neither POML-1 nor MEC-6 changed total MEC-2 expression in oocytes (Supplemental Figure S6I), stabilized MEC-2 (which moves on the surface of oocytes; Chen et al., 2015), or caused it to colocalize with MEC-4 (Supplemental Video S1).
DISCUSSION
MEC-6 is essential for TRN touch sensitivity (Chalfie and Sulston, 1981; Chelur et al., 2002). Previously the interaction of MEC-4 and MEC-6 (Chelur et al., 2002) led us to conclude that MEC-6 was a component of the transduction channel. The findings that MEC-4 and MEC-6 do not colocalize on the plasma membrane of Xenopus oocytes (Chen et al., 2015) and that MEC-6 and the related protein POML-1 fail to localize with MEC-4 and MEC-2 in TRN neurites (this work) argue against MEC-6 and POML-1 being components of the transduction complex. The primary localization of these proteins in the ER, their colocalization with MEC-4 in the ER, and their effect on the expression and localization of MEC-4 suggest that MEC-6 and POML-1 act instead as ER-resident chaperones. Thus MEC-6 and POML-1 represent a new class of chaperones. The effects on functional expression of MEC-6 and POML-1 (and, in the case of MEC-6, on transport) may underlie the increases in MEC-4(d) channel currents that we saw in oocytes. MEC-6 and POML-1, however, are likely to affect relatively few proteins, since general ER stress was not induced in mec-6 or poml-1 mutants.
Although both MEC-6 and POML-1 are required for MEC-4 expression and localization, they do not act identically. Specifically, MEC-6, but not POML-1, accelerated the appearance of MEC-4 on the oocyte surface, and SEC-24 proteins partially suppressed the inhibition of mec-4(d)–induced TRN cell death caused by poml-1 but not mec-6 mutations. These results and the finding that overexpression of mec-6 or poml-1 did not rescue mutations in the other gene argue that MEC-6 and POML-1 may have distinct but overlapping roles in MEC-4 channel maturation. In contrast to mec-6, poml-1 mutants still have nearly normal touch sensitivity and MEC-2 puncta, indicating that these animals have a reduced but still sufficient amount of functional MEC-4 in the TRN neurites. This MEC-4 can be detected on the surface of the cultured TRNs but does not form visible puncta in vivo, although they presumably are sufficient enough to allow for the formation of MEC-2 puncta, which depend on MEC-4 (Zhang et al., 2004). This result agrees with stoichiometry data suggesting that visible MEC-4 puncta are not necessary for the channel function (Chen et al., 2015).
In contrast to CRT-1, which has more general functions in cells—for example, facilitating glycoprotein folding and regulating Ca2+ homeostasis in the ER (Michalak et al., 2009)—the action of MEC-6 and POML-1 is more restricted. Unlike calreticulin, which can bind up to 25 Ca2+ ions (Baksh and Michalak, 1991; Michalak et al., 2009), MEC-6 and POML-1 lack a similar Ca2+-binding domain or even the two Ca2+-binding residues in mammalian PONs (Harel et al., 2004) and thus are unlikely to play a direct role in regulating subcellular Ca2+ content. Consistent with the absence of Ca2+-binding domains in MEC-6 and POML-1, induction of ER Ca2+ release by thapsigargin had no effect on the suppression of MEC-4(d) by mec-6 and poml-1 (such treatment does affect the suppression by crt-1; Xu et al., 2001). Moreover, genetic screens for mutations that reverse poml-1 suppression of MEC-4(d) identified genes that encode MEC-10 and MEC-19, which normally inhibit MEC-4(d) channel activity and surface expression (Chen et al., 2016). Therefore the effect of poml-1, and perhaps mec-6, on MEC-4(d) is largely due to effects on MEC-4 expression and production rather than modulation of cellular Ca2+.
Both MEC-6 and POML-1 are needed for the maturation of DEG/ENaC proteins. MEC-6, which is expressed in many neurons and muscles, is needed for the action of gain-of-function (d) mutations affecting several DEG-ENaC proteins (DEG-1 and UNC-8) and ectopically expressed MEC-4(d) but not a gain-of-function, degeneration-causing mutation affecting the nicotinic acetylcholine receptor protein DEG-3 (Chalfie and Wolinsky, 1990; García-Añoveros, 1995; Shreffler et al., 1995; Harbinder et al., 1997).
The action of MEC-6 and POML-1, however, may not be restricted to only DEG/ENaC proteins, proteins that form amiloride-sensitive channels. That both MEC-6 (Chelur et al., 2002) and POML-1 (this work) can produce an amiloride-resistant current in oocytes hints that they may act on other proteins. In addition, we found that MEC-6, but not POML-1, increased MEC-2 surface expression by ∼60% in oocytes. POML-1 might affect MEC-2 activity, since POML-1 increased MEC-4(d) channel activity in oocytes with MEC-2 or with MEC-2 and MEC-6 together but not with MEC-6 on its own. Because POML-1 did not change MEC-2 expression in oocytes or in the TRNs, we do not know whether this enhancement was direct—for example, by altering MEC-2 conformation—or indirect through changes in MEC-4.
MEC-6 and POML-1 and the human PONs are ∼27% identical (over the C-terminal 260 residues), and all have an N-terminal hydrophobic region (Sorenson et al., 1999). Of interest, two of the three mammalian PONs—PON2 and PON3—are found in the ER and can reduce the cell death induced by the unfolded protein response (Horke et al., 2007; Schweikert et al., 2012). The characterization of MEC-6 and POML-1 in C. elegans suggests a novel function of this protein family: ER chaperones that facilitate the maturation and transport of DEG/ENaC and, perhaps in vertebrates, other proteins.
MATERIALS AND METHODS
C. elegans procedures
Unless otherwise indicated, strains were maintained and studied at 20°C on the OP50 strain of Escherichia coli according to Brenner (1974). All of the translational fusions were based on pPD95.75 (www.addgene.org/static/cms/files/Vec95.pdf). Transgenic animals were prepared by microinjection, and integrated transgenes were generated by ultraviolet irradiation (Chelur and Chalfie, 2007). Details about strains, plasmids, and cDNAs are given in the Supplemental Materials. Ethyl methanesulfonate mutagenesis was performed according to Brenner (1974) to obtain additional alleles of poml-1 (see the Supplemental Materials).
We studied gentle touch sensitivity in blind tests as described (Chalfie and Sulston, 1981) and quantified the response by counting the number of responses to 10 touches delivered alternately near the head and tail in 30 animals. We performed in vivo electrophysiology as previously described (O’Hagan et al., 2005) and also used blue light and channelrhodopsis-2 expressed in the TRNs to stimulate these cells as previously described (Chen and Chalfie, 2014). We performed single-molecule fluorescence in situ hybridization to count mec-4 mRNA as previously described (Topalidou et al., 2011).
Bortezomib (Selleckchem, Houston, TX) was dissolved in dimethyl sulfoxide (DMSO) to make 130 mM stock and added to nematode growth medium (NGM) to make plates containing 50 μM bortezomib. L3 to L4 larvae were transformed onto NGM plates with bortezomib and grown for 8 h. Animals get sick if treated for longer times. In the control group, animals of the same age were transferred to NGM plates containing the same volume of DMSO without bortezomib.
C. elegans microscopy and immunofluorescence
Confocal images were acquired using a 63×/numerical aperture 1.40 oil immersion objective on a Zeiss LSM700 confocal microscope (Zeiss, Jena, Germany). Colocalization of MEC-6 and POML-1 with each other and with ER and Golgi markers in the cell body was quantified by the colocalization function in ZEN 2010 software and is represented by Pearson’s R value. Live animals were anesthetized using 100 mM 2-3-butanedione monoxime in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4.
Fluorescence was observed using a Zeiss Axio Observer Z1 inverted microscope equipped with a Photometrics CoolSnap HQ2 camera (Photometrics, Tucson, AZ) and a Zeiss Axioskop II equipped with a SPOT 2 slider camera (SPOT Imaging Solutions, Sterling Heights, MI).
Immunostaining of larvae and adults was performed according to Miller and Shakes (1995), except for MEC-4 immunostaining, which used a mouse anti–MEC-4 antibody (ab22184; Abcam, Cambridge, MA) and was performed according to Bellanger et al. (2012). The following antibodies were used for immunostaining of C. elegans: anti–MEC-18 (Zhang, 2004), anti–MEC-2 N-terminus (Zhang, 2004), anti–MEC-4 (mouse, ab22184; Abcam), anti-FLAG (mouse, F1804; Sigma, St. Louis, MO), and anti-GFP (rabbit polyclonal A11122 and mouse monoclonal 3E6; Life Technologies, Carlsbad, CA) diluted 1:200, Rhodamine Red-X–conjugated goat anti-rabbit immunoglobulin G (IgG), Alexa Fluor 488–conjugated goat anti-rabbit IgG, and Alexa Fluor 647–conjugated goat anti-rabbit/mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), and Alexa Fluor 488/555–conjugated goat anti-mouse (Life Technologies) diluted 1:700.
MEC-2 and POML-1 immunofluorescence puncta were analyzed over ~50-μm length of TRN neurites with regular puncta using ImageJ (rsbweb.nih.gov/ij/) and the Puncta Analysis Toolkit beta developed by Mei Zhen (Samuel Lunenfeld Research Institute, Toronto, Canada). The width of puncta was the length at half-maximum of each peak, and the average distance between puncta was calculated as 1/(number of puncta per micrometer). Colocalization of POML-1, MEC-4, MEC-2, and MEC-6 puncta in the TRN neurite was analyzed by ImageJ plug-in Coloc 2 (http://fiji.sc/Coloc2) as described in Chen et al. (2015).
We measured fluorescence intensity in the cell body by selecting the cell body area (20–30 μm2) and measuring the mean intensity subtracted from the background of the same-size area by ImageJ. The intensity of the MEC-4::TagRFP puncta intensity was measured in the best- focused image of six images taken at different z-planes using the Puncta Analysis Toolkit beta. Puncta were examined over a region approximately equivalent to 10 cell body lengths starting near the cell body.
Isolated, embryonic TRNs that had been cultured for 15–24 h (Zhang et al., 2002) were fixed in 4% paraformaldehyde, blocked in phosphate-buffered saline (PBS) with 1% bovine serum albumin (in some experiments, 0.2% Triton-X-100 was added to permeabilize the plasma membrane), incubated with primary antibodies (as indicated) at 4°C for 2 h, washed three times in PBS, incubated with secondary antibodies (Rhodamine Red-X–conjugated goat anti-rabbit IgG and Alexa Fluor 647–conjugated goat anti-mouse IgG diluted 1:2000) at room temperature for 30 min, and washed three times in PBS. Immunofluorescence of MEC-18, a cytoplasmic TRN protein, was used as an internal control for nonpermeabilized immunostaining (Chen and Chalfie, 2015). Most cell bodies and some neurites became leaky during immunostaining and displayed clear immunofluorescence signal of MEC-18. Only the fluorescence in intact neurites was measured. We quantified the mean immunofluorescence intensity of MEC-4 in the cell body and the neurite by ImageJ.
After immunostaining for MEC-4, cultured TRNs were incubated with ER tracker Blue-White DPX (E12353; Life Technologies) diluted 1:1000 in PBS at room temperature for 30 min and washed three times in PBS before imaging. An anti-EEA1 antibody (ab2900; Abcam) diluted 1:400 was used to label endosome in cultured TRNs. Correlation coefficients of MEC-4 with markers for ER, endosome, and Golgi were analyzed using ImageJ plug-in Coloc 2 as described earlier.
FRET
FRET was performed on L4 to young adult animals glued with Dermabond (Ethicon, Somerville, NJ) onto 2% agarose pads according to Youvan et al. (1997), using a Zeiss LSM700 confocal microscope. Mean fluorescence intensity minus background was determined in the cell body (and in the puncta for wild type) for three channels: CFP (CFPexcitation 405 nm – CFPemission 420–475 nm), YFP (YFPexcitation 488 nm – YFPemission ≥ 520 nm), and FRET (CFPexcitation 405 nm – YFPemission ≥ 520 nm). The cross-talk of CFP into the FRET channel (Df = FRET/CFP = 84%) was determined in animals expressing mec-4p::cfp::mec-4. Similarly, the cross-talk coefficient of YFP to the FRET channel is determined by expressing mec-4p::mec-4::yfp only and calculated as Af = FRET/YFP = 1.4%. Net FRET was calculated as FRET − 0.84 × CFP − 0.014 × YFP (Youvan et al., 1997). Normalized FRET was calculated as Net FRET/(CFP × YFP)1/2 (Xia and Liu, 2001).
Electrophysiology, biochemistry, and single-molecule imaging in Xenopus ooctyes
A 1050–base pair poml-1 cDNA coding sequence was generated by RACE PCR using the FirstChoice RLM-RACE kit (Ambion, Grand Island, NY) with mRNA from wild-type animals and cloned in pGEM-HE (Liman et al., 1992). cDNA of POML-1 was cloned into pGEMHE-EGFP-X (Ulbrich and Isacoff, 2007) to generate proteins tagged with EGFP at their N-termini.
cRNA expression and electrophysiology in Xenopus laevis oocytes (Xenopus I, Dexter, MI; Nasco, Fort Atkinson, WI; Ecocyte, Austin, TX) followed the procedures and used the plasmids previously described (Goodman et al., 2002). In the experiments described in Figure 3, 10 ng of cRNA of MEC-4, MEC-2, and MEC-10, 1 ng of MEC-6 cRNA, and/or 5 ng of cRNA of POML-1 were injected into oocytes unless noted otherwise. Oocytes were maintained as previously described (Árnadóttir et al., 2011). Membrane current was measured 4–6 d after cRNA injection using a two-electrode voltage clamp as previously described (Goodman et al., 2002). In the experiments described in Figure 7, A and B, oocytes were injected with 3.75 ng of MEC-4::EGFP cRNA, 1 ng of MEC-6 cRNA, 3.75 or 7.5 ng of MEC-2 cRNA, and 3.75 or 5 ng of cRNA of POML-1 (no difference was seen for the different MEC-2 and POML-1 injections, and data were pooled). For the remainder of the experiments in Figure 7 and Supplemental Figure S6, oocytes were injected with 3.75 ng of Myc::MEC-4(d) cRNA, 1 ng of MEC-6::HA cRNA, 10 ng of MEC-2 cRNA, and 7.5 ng of cRNA of EGFP::POML-1 unless otherwise noted. Coexpression of Myc::MEC-4(d) and tagged MEC-6 and POML-1 produced amiloride-sensitive currents that were similar to those of the coexpressed untagged proteins at different time points (for MEC-4(d) and MEC-6 vs. Myc::MEC-4(d) and MEC-6::HA at 2 d, −0.8 ± 0.1 vs. −0.7 ± 0.1, and at 5–6 d, −1.7 ± 0.3 vs. −1.7 ± 0.3; for MEC-4(d) and POML-1 vs. Myc::MEC-4(d) and EGFP::POML-1 at 2 d, −0.1 ± 0 vs. −0.2 ± 0.1, and at 5–6 d, −0.7 ± 0.2 vs. −0.8 ± 0.1; five oocytes).
Immunoprecipitation was performed 5–6 d after cRNA injection as previously described (Goodman et al., 2002). Protein complex was precipitated by using the following antibodies conjugated to Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Dallas, TX): antibodies against GFP (rabbit polyclonal sc-8334; Santa Cruz Biotechnology), Myc (rabbit polyclonal sc-789; Santa Cruz Biotechnology), and HA tags (rabbit polyclonal, sc-805; Santa Cruz Biotechnology). Protein samples were subjected to SDS–PAGE and Western blot. Four to eight oocyte equivalents were loaded per lane for immunoprecipitation, and one oocyte equivalent was loaded per lane for total lysate. The specificity of the immunoprecipitation was confirmed in two ways. First, 1 ng of cRNA encoding Myc::EGFP (for Myc::MEC-4(d) immunoprecipitation of EGFP::POML-1), EGFP (for EGFP::POML-1 immunoprecipitation of Myc::MEC-4(d)), and EGFP::HA (for MEC-6::HA immunoprecipitation of EGFP::POML-1) were used as negative controls; none of the proteins was immunoprecipitated. Second, we probed the immunocomplexes for a Xenopus oocyte membrane protein, β-integrin, by using a monoclonal antibody (8C8; Developmental Studies Hybridoma Bank, Iowa City, IA) and did not detect the β-integrin.
Protein was detected by Western blot using antibodies against GFP (mouse monoclonal, sc-9996; Santa Cruz Biotechnology), Myc (mouse monoclonal 9E10; Sigma-Aldrich), the HA tags (mouse monoclonal, sc-7392; Santa Cruz Biotechnology), MEC-2 N-terminus (rabbit polyclonal; Zhang, 2004), or actin (rabbit polyclonal, A2066; Sigma-Aldrich) and horseradish peroxidase (HRP)–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Horseradish peroxidase was detected using the ECL Western Blotting reagent (Amersham, Little Chalfont, United Kingdom).
Biotinylation assays to detect the surface expression of MEC-4 generally followed the methods described previously (Goodman et al., 2002). Surface protein was labeled and isolated using the membrane-impermeable EZ-Link Sulfo-NHS-SS-Biotin and NeutrAvidin agarose provided in the Pierce Cell Surface Protein Isolation Kit (Thermo Scientific, Waltham, MA). Samples collected from 30 oocytes from each group were loaded per lane in SDS–PAGE and detected by Western blotting using a primary monoclonal Myc antibody (clone 9E10; Sigma-Aldrich) and HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). The total lysate of one oocyte equivalent was loaded as input. β-Actin was detected in total lysate as an input control by Western blotting using a rabbit polyclonal antibody against actin (A2066; Sigma-Aldrich). Cytoplasmic EGFP was detected in the supernatants but not in the avidin precipitates. Band density was measured from the autoradiography films using ImageJ (National Institutes of Health, Bethesda, MD).
TIRF imaging of oocytes was performed as described in Chen et al. (2015).
Statistics
Data are presented as mean ± SEM unless noted otherwise. Statistical significance was determined using GraphPad Prism5 software (GraphPad, La Jolla, CA). We used the Student’s t test (with Welch’s correction when data being compared did not have equal variances) for most experiments. For the quantification of MEC-4::EGFP spots on the surface of Xenopus oocytes, we used the Mann–Whitney test. For the quantification of Western blot, we used the one-sample t test. All p values from the Student’s t, Mann–Whitney, and one-sample t test were adjusted with a Bonferroni correction. When three or more groups were compared, we used one-way analysis of variance (ANOVA) with Tukey post hoc or two-way ANOVA with Bonferroni posttests. In the figures, *p < 0.05, **p < 0.01, and ***p < 0.001 are corrected p values; ns indicates not significant. All values were determined with the Student’s t test unless noted otherwise.
Supplementary Material
Acknowledgments
We thank Jian Yang and Yong Yu for providing the Xenopus laevis oocytes, Mei Zhen for the puncta analysis software, and Oliver Hobert, Elizabeth Miller, and members of the Chalfie lab for discussion. This work was supported by Grants GM30997 to M.C. and NS35549 to E.Y.I. from the National Institutes of Health. R.O. was supported by NJCSCR Postdoctoral Fellowship 10-2951-SCR-E-0.
Abbreviations used:
- ER
endoplasmic reticulum
- POML-1
paraoxonase and MEC-6-like gene 1
- PON
paraoxonase
- TRN
touch receptor neuron.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0561) on March 3, 2016.
REFERENCES
- Appenzeller-Herzog C, Hauri HP. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Árnadóttir J, O’Hagan R, Chen Y, Goodman MB, Chalfie M. The DEG/ENaC protein MEC-10 regulates the transduction channel complex in Caenorhabditis elegans touch receptor neurons. J Neurosci. 2011;31:12695–12704. doi: 10.1523/JNEUROSCI.4580-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J Clin Invest. 1998;101:1581–1590. doi: 10.1172/JCI1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266:21458–21465. [PubMed] [Google Scholar]
- Bellanger JM, Cueva JG, Baran R, Tang G, Goodman MB, Debant A. The doublecortin-related gene zyg-8 is a microtubule organizer in Caenorhabditis elegans neurons. J Cell Sci. 2012;125:5417–5427. doi: 10.1242/jcs.108381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besler C, Heinrich K, Rohrer L, Doerries C, Riwanto M, Shih DM, Chroni A, Yonekawa K, Stein S, Schaefer N, et al. Mechanisms underlying adverse effects of HDL on eNOS-activating pathways in patients with coronary artery disease. J Clin Invest. 2011;121:2693–2708. doi: 10.1172/JCI42946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Fernandez-Illescas SM, Liao Z, Goodman MB. Gain-of-function mutations in the MEC-4 DEG/ENaC sensory mechanotransduction channel alter gating and drug blockade. J Gen Physiol. 2007;129:161–173. doi: 10.1085/jgp.200609672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Liao Z, Goodman MB. MEC-2 and MEC-6 in the Caenorhabditis elegans sensory mechanotransduction complex: auxiliary subunits that enable channel activity. J Gen Physiol. 2008;131:605–616. doi: 10.1085/jgp.200709910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Wolinsky E. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans. Nature. 1990;345:410–416. doi: 10.1038/345410a0. [DOI] [PubMed] [Google Scholar]
- Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proc Natl Acad Sci USA. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, R OH, Chalfie M. The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature. 2002;420:669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bharill S, Isacoff EY, Chalfie M. Subunit composition of a DEG/ENaC mechanosensory channel of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2015;112:11690–11695. doi: 10.1073/pnas.1515968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bharill S, O’Hagan R, Isacoff EY, Chalfie M. MEC-10 and MEC-19 reduce the neurotoxicity of the MEC-4(d) DEG/ENaC channel in Caenorhabditis elegans. G3 (Bethesda) 2016;6:1121–1130. doi: 10.1534/g3.115.023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chalfie M. Modulation of C. elegans touch sensitivity is integrated at multiple levels. J Neurosci. 2014;34:6522–6536. doi: 10.1523/JNEUROSCI.0022-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chalfie M. Regulation of mechanosensation in C. elegans through ubiquitination of the MEC-4 mechanotransduction channel. J Neurosci. 2015;35:2200–2212. doi: 10.1523/JNEUROSCI.4082-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- Devarajan A, Bourquard N, Hama S, Navab M, Grijalva VR, Morvardi S, Clarke CF, Vergnes L, Reue K, Teiber JF, et al. Paraoxonase 2 deficiency alters mitochondrial function and exacerbates the development of atherosclerosis. Antioxid Redox Signal. 2011;14:341–351. doi: 10.1089/ars.2010.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- Emtage L, Gu G, Hartwieg E, Chalfie M. Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron. 2004;44:795–807. doi: 10.1016/j.neuron.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hollopeter G, Taylor J, Harris TW, Nix P, Lofgren R, Prestgard-Duke M, Bastiani M, Moerman DG, et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Añoveros J. Genetic Analysis of Degenerin Proteins in Caenorhabditis elegans. New York, NY: Columbia University; 1995. PhD Thesis. [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O’Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- Gorczyca DA, Younger S, Meltzer S, Kim SE, Cheng L, Song W, Lee HY, Jan LY, Jan YN. Identification of Ppk26, a DEG/ENaC channel functioning with Ppk1 in a mutually dependent manner to guide locomotion behavior in Drosophila. Cell Rep. 2014;9:1446–1458. doi: 10.1016/j.celrep.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Caldwell GA, Chalfie M. Genetic interactions affecting touch sensitivity in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1996;93:6577–6582. doi: 10.1073/pnas.93.13.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Wang Q, Wang Z. The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep. 2014;9:1183–1190. doi: 10.1016/j.celrep.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Harbinder S, Tavernarakis N, Herndon LA, Kinnell M, Xu SQ, Fire A, Driscoll M. Genetically targeted cell disruption in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:13128–13133. doi: 10.1073/pnas.94.24.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- Hicks MA, Barber AE, 2nd, Giddings LA, Caldwell J, O’Connor SE, Babbitt PC. The evolution of function in strictosidine synthase-like proteins. Proteins. 2011;79:3082–3098. doi: 10.1002/prot.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horke S, Witte I, Wilgenbus P, Kruger M, Strand D, Forstermann U. Paraoxonase-2 reduces oxidative stress in vascular cells and decreases endoplasmic reticulum stress-induced caspase activation. Circulation. 2007;115:2055–2064. doi: 10.1161/CIRCULATIONAHA.106.681700. [DOI] [PubMed] [Google Scholar]
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wu Z, Riwanto M, Gao S, Levison BS, Gu X, Fu X, Wagner MA, Besler C, Gerstenecker G, et al. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J Clin Invest. 2013;123:3815–3828. doi: 10.1172/JCI67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TB, Schermer B, Muller RU, Hohne M, Bartram M, Calixto A, Hagmann H, Reinhardt C, Koos F, Kunzelmann K, et al. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc Natl Acad Sci USA. 2006;103:17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Crowe E, Stevens TA, Candido EP. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Liu L, Leonard AS, Motto DG, Feller MA, Price MP, Johnson WA, Welsh MJ. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003;39:133–146. doi: 10.1016/s0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- Mackness MI, Walker CH. Multiple forms of sheep serum A-esterase activity associated with the high-density lipoprotein. Biochem J. 1988;250:539–545. doi: 10.1042/bj2500539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthner SE, Hwang RY, Lewis AH, Xiao Q, Tsubouchi A, Wang Y, Honjo K, Skene JH, Grandl J, Tracey WD., Jr Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr Biol. 2014;24:2920–2925. doi: 10.1016/j.cub.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417:651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Mochizuki H, Scherer SW, Xi T, Nickle DC, Majer M, Huizenga JJ, Tsui LC, Prochazka M. Human PON2 gene at 7q21.3: cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene. 1998;213:149–157. doi: 10.1016/s0378-1119(98)00193-0. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CJ, Wadleigh DJ, Gangopadhyay A, Hama S, Grijalva VR, Navab M, Fogelman AM, Reddy ST. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J Biol Chem. 2001;276:44444–44449. doi: 10.1074/jbc.M105660200. [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- Pagant S, Wu A, Edwards S, Diehl F, Miller EA. Sec24 is a coincidence detector that simultaneously binds two signals to drive ER export. Curr Biol. 2015;25:403–412. doi: 10.1016/j.cub.2014.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Lee DG, Yu JR, Jung SK, Choi K, Lee J, Lee J, Kim YS, Lee JI, Kwon JY, et al. Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2835–2845. doi: 10.1091/mbc.12.9.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps B, Hennecke J, Glockshuber R. FRET-based in vivo screening for protein folding and increased protein stability. J Mol Biol. 2003;327:239–249. doi: 10.1016/s0022-2836(03)00077-9. [DOI] [PubMed] [Google Scholar]
- Reddy ST, Wadleigh DJ, Grijalva V, Ng C, Hama S, Gangopadhyay A, Shih DM, Lusis AJ, Navab M, Fogelman AM. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arterioscler Thromb Vasc Biol. 2001;21:542–547. doi: 10.1161/01.atv.21.4.542. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Hall DH, Victor M, Stelzer EH, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothem L, Hartman C, Dahan A, Lachter J, Eliakim R, Shamir R. Paraoxonases are associated with intestinal inflammatory diseases and intracellularly localized to the endoplasmic reticulum. Free Radic Biol Med. 2007;43:730–739. doi: 10.1016/j.freeradbiomed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Schweikert EM, Devarajan A, Witte I, Wilgenbus P, Amort J, Forstermann U, Shabazian A, Grijalva V, Shih DM, Farias-Eisner R, et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012;19:1549–1560. doi: 10.1038/cdd.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- Shim J, Umemura T, Nothstein E, Rongo C. The unfolded protein response regulates glutamate receptor export from the endoplasmic reticulum. Mol Biol Cell. 2004;15:4818–4828. doi: 10.1091/mbc.E04-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreffler W, Magardino T, Shekdar K, Wolinsky E. The unc-8 and sup-40 genes regulate ion channel function in Caenorhabditis elegans motorneurons. Genetics. 1995;139:1261–1272. doi: 10.1093/genetics/139.3.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen A, Eckerson HW, Gan KN, Hailat N, La Du BN. Characteristics of the genetically determined allozymic forms of human serum paraoxonase/arylesterase. Drug Metab Dispos. 1991;19:107–112. [PubMed] [Google Scholar]
- Sorenson RC, Bisgaier CL, Aviram M, Hsu C, Billecke S, La Du BN. Human serum Paraoxonase/Arylesterase’s retained hydrophobic N-terminal leader sequence associates with HDLs by binding phospholipids: apolipoprotein A-I stabilizes activity. Arterioscler Thromb Vasc Biol. 1999;19:2214–2225. doi: 10.1161/01.atv.19.9.2214. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Suzuki SM, Cole TB, Park SS, Richter RJ, Furlong CE. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc Natl Acad Sci USA. 2008;105:12780–12784. doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Chalfie M. Shared gene expression in distinct neurons expressing common selector genes. Proc Natl Acad Sci USA. 2011;108:19258–19263. doi: 10.1073/pnas.1111684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, van Oudenaarden A, Chalfie M. Caenorhabditis elegans aristaless/Arx gene alr-1 restricts variable gene expression. Proc Natl Acad Sci USA. 2011;108:4063–4068. doi: 10.1073/pnas.1101329108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Kornfeld S. The trans-Golgi network: a late secretory sorting station. Curr Opin Cell Biol. 1997;9:527–533. doi: 10.1016/s0955-0674(97)80029-4. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Tavernarakis N, Driscoll M. Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca2+ release from the endoplasmic reticulum. Neuron. 2001;31:957–971. doi: 10.1016/s0896-6273(01)00432-9. [DOI] [PubMed] [Google Scholar]
- Youvan DC, Silva CM, Bylina EJ, Coleman WJ, Dilworth MR, Yang MM. Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel chelating beads. Biotechnology. 1997;3:1–18. [Google Scholar]
- Zhang S. Stomatin Gene Family in Caenorhabditis elegans. New York, NY: Columbia University; 2004. PhD Thesis. [Google Scholar]
- Zhang S, Árnadóttir J, Keller C, Caldwell GA, Yao CA, Chalfie M. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.