Cell cycle and DNA repair involve steps requiring RanGTP-regulated nuclear-cytoplasmic transport (NCT). The activation of NCT via overexpression of RCC1 (the guanine nucleotide exchange factor for Ran) accelerated the cell cycle and DNA repair. Normal cells overexpressing RCC1 evaded DNA damage–induced senescence, mimicking cancer cells with high levels of RCC1.
Abstract
The coordination of cell cycle progression with the repair of DNA damage supports the genomic integrity of dividing cells. The function of many factors involved in DNA damage response (DDR) and the cell cycle depends on their Ran GTPase–regulated nuclear–cytoplasmic transport (NCT). The loading of Ran with GTP, which is mediated by RCC1, the guanine nucleotide exchange factor for Ran, is critical for NCT activity. However, the role of RCC1 or Ran⋅GTP in promoting cell proliferation or DDR is not clear. We show that RCC1 overexpression in normal cells increased cellular Ran⋅GTP levels and accelerated the cell cycle and DNA damage repair. As a result, normal cells overexpressing RCC1 evaded DNA damage–induced cell cycle arrest and senescence, mimicking colorectal carcinoma cells with high endogenous RCC1 levels. The RCC1-induced inhibition of senescence required Ran and exportin 1 and involved the activation of importin β–dependent nuclear import of 53BP1, a large NCT cargo. Our results indicate that changes in the activity of the Ran⋅GTP–regulated NCT modulate the rate of the cell cycle and the efficiency of DNA repair. Through the essential role of RCC1 in regulation of cellular Ran⋅GTP levels and NCT, RCC1 expression enables the proliferation of cells that sustain DNA damage.
INTRODUCTION
Regulation of the rate at which cells divide is critical to normal development and tissue homeostasis. Because many DNA-damaging events continuously challenge the genome integrity of dividing cells, conserved DNA damage response (DDR) signaling networks have evolved to coordinate DNA damage repair with the continuing cell cycle (Ciccia and Elledge, 2010; Smith et al., 2010). Central to DDR signaling is the activation of ATM (ataxia telangiectasia–mutated) and ATR (ataxia telangiectasia and Rad3–related) kinases. DNA damage checkpoints integrate the signaling of DNA damage, the cell cycle state, and other inputs to elicit a cell cycle pause, DNA repair or transcriptional changes, or if the damage is irreparable, programmed cell death or permanent exit from the cell cycle (Shaltiel et al., 2015). Because separate signaling networks control the termination of DDR, the cell cycle reentry in the presence of DNA damage is also a possible outcome (Kleiblova et al., 2013; Shaltiel et al., 2015).
The activities of many factors involved in the control of cell cycle and DDR depend on their Ran GTPase–regulated nuclear–cytoplasmic transport (NCT; Pemberton and Paschal, 2005; Thompson, 2010; Blackinton and Keene, 2014), suggesting that NCT activation could promote cell cycle and DDR kinetics.The driving force of NCT is the concentration gradient of Ran⋅GTP across the nuclear envelope: nuclei contain high Ran⋅GTP concentrations, and the cytoplasmic Ran is mostly GDP bound. This striking Ran⋅GTP compartmentalization depends on the nuclear localization of RCC1, the guanine nucleotide exchange factor for Ran, and cytoplasmic localization of RanGAP1, the Ran GTP-ase–activating protein. Ran⋅GTP interacts with ∼20 different nuclear transport receptors (NTRs), importins, and exportins (Pemberton and Paschal, 2005) and regulates either the loading or unloading of cargoes on NTRs. Cargoes are unloaded from importins that bind to Ran⋅GTP in the nucleus, while Ran⋅GTP is required for cargo loading onto exportins before their exit to the cytoplasm (Pemberton and Paschal, 2005). NTR cargoes include myriad proteins and several classes of nucleic acids, including the eIF4e-dependent mRNAs for many cyclins (Culjkovic et al., 2006; Muqbil et al., 2013). Following disassembly of the nuclear envelope in mitotic cells, mitotic chromosomes are surrounded by diffusional Ran⋅GTP gradients, which support the assembly and function of mitotic spindles through the continuing dynamics of Ran⋅GTP–NTR interactions (Kalab et al., 2002, 2006; Forbes et al., 2015). In addition to Ran⋅GTP-dependent NCT, Ran⋅GDP functions as the nuclear import receptor for proteins carrying the ankyrin repeat motif (Lu et al., 2014).
To date, the potential role of Ran activity in controlling the rate of the cell cycle or DNA repair is supported only by indirect or correlative evidence. Namely, since premature cell aging leads to decreased NCT activity (Snow et al., 2013), the reduced nuclear transport of various DDR factors could explain the decline in DDR function observed in aging cells (Klement and Goodarzi, 2014). The requirement of Ran for the continuation of the cell cycle is consistent with the evidence that Ran knockdown by RNA interference (RNAi) induces cell senescence in primary fibroblasts (Nagai and Yoneda, 2012). Furthermore, we previously showed that, compared with slow-growing cells, rapidly proliferating tissue culture cells expressed approximately four times higher levels of RCC1 (Hasegawa et al., 2013), the guanine nucleotide exchange factor for Ran, suggesting the potential role of RCC1 and Ran⋅GTP in accelerating the cell cycle.
In this study, we set out to investigate the role of Ran activity in cell cycle and DDR regulation using RCC1 overexpression, RNAi, and chemical inhibitors. We manipulated the RCC1-dependent Ran⋅GTP levels in cells and the function of two major NTRs, importin β and exportin 1. Our results demonstrated that RCC1-dependent activation of Ran-regulated NCT accelerates cell cycle and DNA damage repair, leading to evasion of DNA damage–induced cell senescence.
RESULTS
RCC1 overexpression accelerates the cell cycle
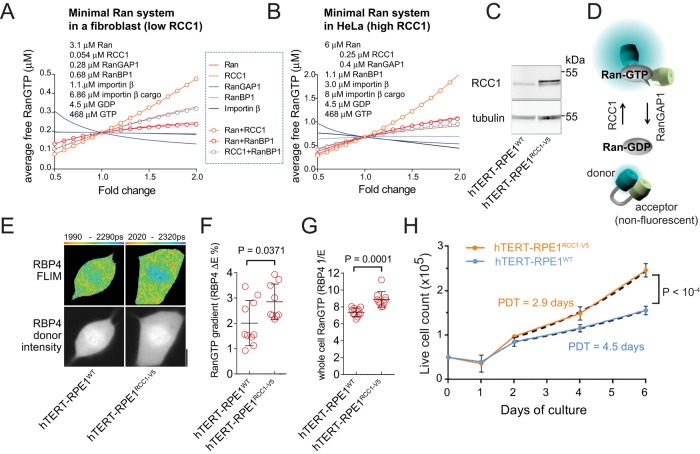
The cellular concentration of free Ran⋅GTP available for binding to NTRs is critical for most known Ran-regulated functions, including NCT. To determine changes in Ran regulation that could lead to increased Ran⋅GTP levels, we applied computational modeling of the minimal set of proteins controlling the GTP/GDP cycle on Ran. Within these models, we either increased or decreased the concentrations of their individual components and followed the changes of average cellular Ran⋅GTP concentration (Gorlich et al., 2003; Caudron et al., 2005; Kalab et al., 2006). The NCT rates for all Ran system components are not known. Therefore we used the previously published single-compartment models of mitotic HeLa cells (Caudron et al., 2005; Kalab et al., 2006) to simulate the Ran system in rapidly dividing mitotic cells with high Ran⋅GTP levels (Hasegawa et al., 2013; Figure 1B). From the relative expression in HeLa and human fibroblasts (HFF-1; Supplemental Figure S1), we deduced the composition of the Ran system in slowly dividing cells (Hasegawa et al., 2013; Figure 1A, Supplemental Figure S1, Supplemental Text S1, and Supplemental Tables S1 and S2). In both models, Ran⋅GTP levels were most responsive to simultaneous changes in Ran and RCC1 concentrations and reduced RanGAP1 expression (Figure 1, A and B). When overexpression of individual factors was considered, Ran⋅GTP was most sensitive to RCC1 concentration in the fibroblast model (Figure 1A) and to changes in Ran in the HeLa model (Figure 1B). Therefore, to examine how increased Ran⋅GTP levels affect the cell cycle in relatively slowly dividing cells, we tested RCC1 overexpression.
FIGURE 1:
RCC1-dependent increase in Ran⋅GTP accelerates the cell cycle. (A and B) Changes in cellular Ran⋅GTP concentration analyzed by computational models of the minimal Ran system in a fibroblast-like cell (A) and a HeLa-like cell (B). (C) Immunoblots of RCC1 in the hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cell lysates. (D) Schematic of the RBP-4 FRET sensor for Ran⋅GTP detection with FLIM. The binding of Ran⋅GTP to RBP-4 increases the donor–acceptor distance, resulting in a longer donor fluorescence lifetime, τdonor. (E) Detection of mitotic Ran⋅GTP gradients in hTERT-RPE1WT and hTERT-ΔRPE1RCC1-V5 cells, using FLIM with RBP-4. The top row shows the donor intensity images and the bottom row shows the pseudocolor FLIM images. The range of the displayed values (corresponding to τdonor values) is indicated beneath the FLIM panels. Scale bar: 10 μm. (F) Scatter plot of the mitotic Ran⋅GTP gradients quantified as the difference between the cytoplasmic and chromatin E in each cell (ΔE; single-cell data; means ± SD; t test). (G) Scatter plot of the inverse of the average cellular RBP-4 E, which is proportional to Ran⋅GTP concentration (E−1; single-cell data; means ± SD; t test). (H) Cell number in cultures of hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells grown in parallel. Means ± SD from two experiments performed in triplicate were fitted with exponential growth equations after 2 d from the start of culture (dashed lines) to calculate the PDT. The null hypothesis was tested: one curve for both data sets.
We chose the telomerase-immortalized normal epithelial RPE1 cells (hTERT-RPE1WT) as a model, because these cells display intermediate mitotic Ran⋅GTP gradients and Ran⋅GTP levels (Hasegawa et al., 2013). Because the models predicted that the effect of RCC1 on Ran⋅GTP would saturate at low micromolar RCC1 concentrations (Supplemental Figure S1), we selected human phosphoglycerate kinase (PGK) promoter to drive moderate overexpression of RCC1 with a C-terminal V5 tag in a stable hTERT-RPE1RCC1-V5 cell line. From immunoblots (Figure 1C), we estimated that the total RCC1 concentration in the hTERT-RPE1RCC1-V5 cells was four to six times higher than in the hTERT-RPE1WT cells. Such an increase in RCC1 levels was similar to the difference between HFF-1 and HeLa cells (Figure 1, A and B, and Supplemental Figure S1), suggesting that the RCC1 overexpression mimicked the physiologically relevant range. We then verified the effect of RCC1 overexpression using RBP-4, the Förster resonance energy transfer (FRET) biosensor for Ran⋅GTP in mitotic cells (Hasegawa et al., 2013). RBP-4 consists of a Ran⋅GTP-binding domain flanked by the cyan mTFP-1 donor and a nonfluorescent dsREACh acceptor (Figure 1D; Hasegawa et al., 2013). RBP-4 binding to Ran⋅GTP extends the donor from the acceptor, leading to decreased FRET efficiency (ERBP-4) and increased donor fluorescence lifetime (τdonor). Because the RBP-4 FRET activity decreases with Ran⋅GTP binding, the average (ERBP-4)−1 value could be used to compare the Ran⋅GTP concentration between different mitotic cells quantitatively (Hasegawa et al., 2013). We applied fluorescence lifetime imaging microscopy (FLIM) to detect the changes of ERBP-4 in live mitotic cells transiently expressing the RBP-4 sensor. The mitotic hTERT-RPE1RCC1-V5 cells displayed larger differences in ERBP-4 between the chromatin and cytoplasmic areas (Figure 1F) and higher (ERBP-4)−1 values (Figure 1G), confirming the expected increased mitotic Ran⋅GTP gradients and elevated Ran⋅GTP levels. The compartmentalization of Ran and its regulators RCC1 and RanGAP1 between the nucleus and cytoplasm precludes the direct measurements of Ran⋅GTP with RBP-4 FRET in interphase cells. However, previous studies showed that the expression of Ran, RCC1, and RanGAP1 remained stable during the exit from mitosis (Ciciarello et al., 2010), indicating that the mitotic Ran⋅GTP levels correspond to the Ran⋅GTP-generating activity in the interphase cells as well. Fluorescence-activated cell sorting (FACS) analysis showed that the fraction of cells in different cell cycle phases was not appreciably affected by RCC1 overexpression (Supplemental Figure S1). However, consistent with our initial hypothesis, the hTERT-RPE1RCC1-V5 cells proliferated significantly faster than the WT cells (Figure 1H; population doubling time [PDT] = 2.9 vs. 4.5 d).
Ran⋅GTP levels decline in nondividing cells
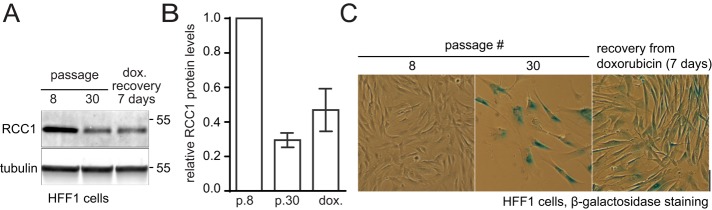
If the RCC1-dependent rise of Ran⋅GTP activates cell cycle progression, stimuli inducing cell cycle arrest should oppose RCC1 by lowering its activity or expression. To test this scenario, we induced cell cycle arrest in HFF-1 fibroblasts by a long-term in vitro culture or by inducing DNA damage through treatment with doxorubicin (Chang et al., 2002; Sliwinska et al., 2009). Both replicative exhaustion and doxorubicin treatment induced the onset of cell senescence, as indicated by the appearance of the senescence-associated β-galactosidase (SABG) staining (Debacq-Chainiaux et al., 2009; Figure 2C) and led to a strong decline in RCC1 expression (Figure 2B). We also observed lower Ran and RanBP1 levels in senescent HFF-1 cells and a decrease in RCC1 levels in doxorubicin-treated Wi-38 and CRL-1474 primary human fibroblasts (Supplemental Figure S2). Quantification of immunoblots showed that, when normalized to the total cell protein concentration, the senescent HFF-1 cells contained ∼25% RCC1 compared with early-passage cells (Figure 2B). However, because of their nearly three-times larger volume (1.27 ± 0.07 pl, passage 8 vs. 3.57 ± 0.30 pl, passage 30 HFF-1 fibroblasts), the late-passage cells contained <10% RCC1 concentration compared with the early-passage cells, confirming the strong reduction in the RCC1-dependent Ran⋅GTP generation in nondividing cells.
FIGURE 2:
RCC1 expression declines during cell cycle arrest. (A) Immunoblotting of total cell lysates of HFF-1 cells harvested from early (p 8) or late (p 30) passages of in vitro culture and of early-passage HFF-1 cells recovering from the treatment with doxorubicin. (B) Relative tubulin-normalized RCC1 protein expression in HFF-1 cells treated as in A (N = 3; means ± SD). (C) Micrographs of HFF-1 cells treated as in A and B and stained for the SABG. Scale bar: 100 μm.
RCC1 overexpression inhibits cell senescence
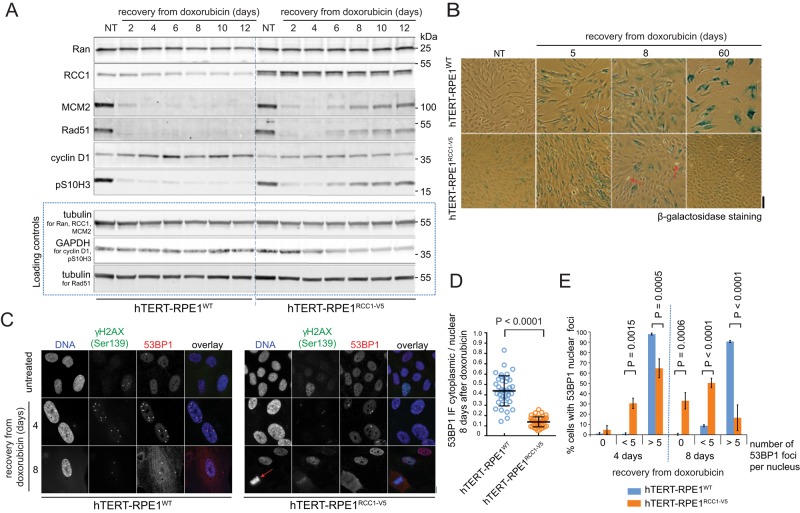
The cell cycle–promoting activity of RCC1 (Figure 1) indicated that increased RCC1 expression could attenuate DNA damage–induced cell cycle arrest. To test this idea, we compared the responses of hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells to doxorubicin treatment. Within the first 2–4 d after doxorubicin washout, most cells stopped dividing in both cultures, as indicated by the disappearance of interphase and mitotic markers (MCM2, Rad51, p-histone H3 [Ser-10] [pS10H3]). At the same time, the increase in cyclin D1 indicated cell cycle arrest (Figure 3A), and the appearance of the SABG signal marked the onset of senescence (Figure 3B). The cyclin D1 levels remained stable, and SABG positivity increased over time in the hTERT-RPE1WT cells (Figure 3B). In contrast, SABG-negative and proliferating cells gradually prevailed in the hTERT-RPE1RCC1-V5 cultures (Figure 3B, arrows), concomitant with increased interphase and mitotic markers and the decline in cyclin D1 expression (Figure 3A). The quantitative capillary immunoblotting (Simple Western) analysis confirmed that, 8 d after doxorubicin treatment, hTERT-RPE1WT cells accumulated cyclin D1, while hTERT-RPE1RCC1-V5 cells resumed expression of cyclin B1 (Supplemental Figure S3). As in the senescing fibroblasts (Supplemental Figure S2), the expression of Ran decreased to ∼65% in doxorubicin-treated hTERT-RPE1WT cells (Figure 3A). In contrast, Ran levels slightly increased in the hTERT-RPE1RCC1-V5 cells exposed to doxorubicin, indicating that RCC1 expression supported Ran stability in cells exposed to DNA damage (Figure 3A). Two months after the doxorubicin treatment, the hTERT-RPE1RCC1-V5 cells regained normal proliferation, while virtually no dividing cells were detectable in the hTERT-RPE1WT cell cultures (Figure 3B). To monitor the progress of DNA damage repair, we used immunofluorescence (IF) to quantify the 53BP1 nuclear foci that assemble at the sites of DNA double-strand break repair (Ciccia and Elledge, 2010). Most of the hTERT-RPE1RCC1-V5 cells had <5 nuclear foci after 8 d of recovery, and mitotic cells were already detectable (Figure 3C). In contrast, the nuclear 53BP1 foci persisted in nearly all doxorubicin-treated hTERT-RPE1WT cells (Figure 3C). At the same time, 53BP1 strongly accumulated in the cytoplasm of the hTERT-RPE1WT cells (Figure 3D), indicating delays in the 53BP1 nuclear import, which is Ran⋅GTP- and importin β–dependent (Moudry et al., 2012). It is possible that delays in nuclear import of cargoes other than 53BP1 limited the rate of DNA repair in the WT cells. However, these results strongly suggest that RCC1-dependent activation of NCT contributed to the efficient DNA damage repair and cell cycle reentry in the hTERT-RPE1RCC1-V5 cells. We observed similar differences in the response of hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells to γ-irradiation–induced DNA damage (Supplemental Figure S4), showing that the effects of RCC1 overexpression in doxorubicin-treated cells did not depend on accelerated drug efflux.
FIGURE 3:
RCC1 overexpression inhibits DNA damage–induced cell senescence in normal epithelial cells. (A) Immunoblotting of total lysates from the hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells recovering from doxorubicin treatment, showing the resumed expression of the cell cycle markers in the hTERT-RPE1RCC1-V5 cells. (B) Micrographs of cells treated as in A and stained for SABG. Scale bar: 100 μm. Arrows indicate dividing cells. (C) IF micrographs of γH2AX and 53BP1 staining in hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells recovering from doxorubicin. The arrow indicates a dividing cell. Scale bar: 10 μm. (D) Scatter plot of the cytoplasmic/nuclear ratios of the 53BP1 IF signal at 8 d of recovery from the doxorubicin. Individual cell data; means ± SD; t test representative of two experiments. (E) Column graph showing fractions of hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells that contained the indicated numbers of 53BP1 foci per nucleus during the recovery from doxorubicin treatment. Means ± SD from two independent experiments, adjusted p values from two-way analysis of variance (ANOVA) with Sidak’s multiple comparison tests.
RCC1 promotes doxorubicin resistance in colorectal carcinoma cells
Because the overexpression of RCC1 prevented the onset of DNA damage–induced cell senescence in normal cells (Figure 3 and Supplemental Figure S4), RCC1 could play a role in cancer cell resistance to DNA damage. Consistent with this idea, the expression of RCC1 was found to be activated by a superenhancer element in colorectal carcinoma HCT116 cells (Hnisz et al., 2013), which is a well-studied model of resistance to DNA damage–inducing chemotherapy (Chang et al., 2002; Sliwinska et al., 2009). We used several approaches to test the role of RCC1 in HCT116 cells.
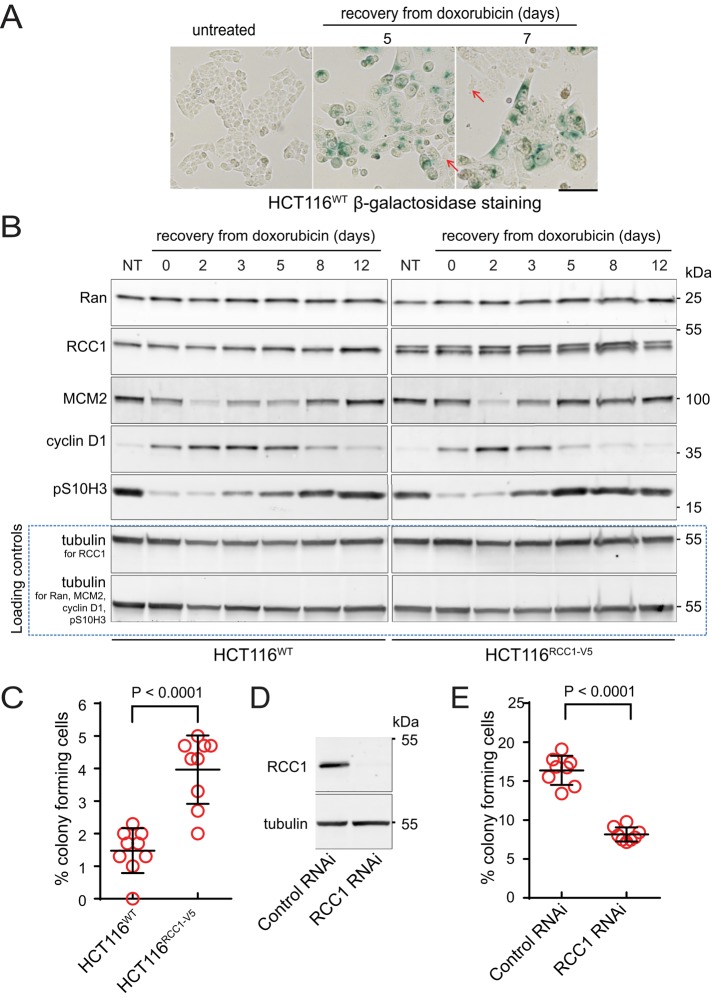
First, we compared the effects of doxorubicin on HCT116WT and an HCT116RCC1-V5 cell line that stably expressed V5-tagged RCC1 (Figure 4). As previously reported (Chang et al., 2002; Sliwinska et al., 2009) the doxorubicin-treated HCT116WT cells initially stopped dividing, increased in size, and became SABG-positive (Figure 4A). Small, rapidly dividing, and SABG-negative cells prevailed in the cultures 5–7 d later (Figure 4A, arrows). A transient peak in cyclin D1 expression and a decline in S and G2/M markers (MCM2, pS10H3; Figure 4B, left) indicated a temporary cell cycle arrest. The phenotypic changes induced by doxorubicin in the HCT116RCC1-V5 cells were similar to those seen in the HCT116WT cells. However, the shorter duration of cyclin D1 expression and earlier recovery of the S and G2/M markers indicated that the overexpressed RCC1 slightly accelerated cell cycle reentry after DNA damage in the HCT116RCC1-V5 cell line (Figure 4B).
FIGURE 4:
Endogenous RCC1 expression inhibits DNA damage–induced cell senescence in HCT116 colorectal carcinoma cells. (A) Micrographs of HCT116WT cells recovering from doxorubicin and stained for the SABG. Scale bar: 100 μm. Arrows indicate SABG activity–free cells. (B) Immunoblotting of total lysates from HCT116WT and HCT116RCC1-V5 cells recovering from doxorubicin treatment. (C) Scatter plot of the fractions of colony-forming HCT116WT and HCT116RCC-V5 cells recovering from doxorubicin. Single-culture data; means ± SD from three experiments performed in triplicate; t test. (D) Immunoblotting of total lysates of HCT116WT cells treated with control scramble or RCC1-directed RNAi. (E) Scatter plot of the fractions of colony-forming HCT116WT cells treated with doxorubicin, followed by control or Ran-directed RNAi. Single-culture data; means ± SD from two experiments with four replicates each; t-test.
We used clonogenic assays to validate the role of RCC1 expression in DNA damage recovery. After doxorubicin treatment, the fraction of colony-forming cells was nearly twice as high in HCT116RCC1-V5 compared with HCT116WT cells (Figure 4C), demonstrating that RCC1 overexpression strongly increased cell survival following DNA damage. By analyzing cultures derived from single doxorubicin-treated HCT116WT cells, we found that rapidly proliferating clones contained RCC1 concentrations similar to the untreated HCT116 cells, in contrast to a slow-proliferating clone (Supplemental Figure S5), indicating that the recovery from DNA damage selects for cells with high RCC1 expression. The requirement for endogenous RCC1 expression in recovery from DNA damage was further confirmed by strongly reduced colony formation in the doxorubicin-treated HCT116WT cells upon RCC1 knockdown by RNAi (Figure 4E). Based on these results, RCC1 functions as the DNA damage resistance–promoting factor in HCT116 cells, indicating a potential role of RCC1 in tumor cell proliferation in vivo. Indeed, previously reported RNA microarray data sets contained evidence for statistically significantly higher RCC1 expression in ovarian (Scotto et al., 2008), colorectal (Alhopuro et al., 2012), and carboplatin-resistant cervical tumors (Peters et al., 2005), compared with normal tissues (Supplemental Figure S6). Because moderate RCC1 overexpression was sufficient to accelerate cell cycle and DNA damage repair (Figures 1 and 3), relatively small increases in tumor RCC1 could correspond to the activation of Ran pathways restricted to the proliferative fraction of tumor cells.
Ran promotes DNA damage signaling and repair
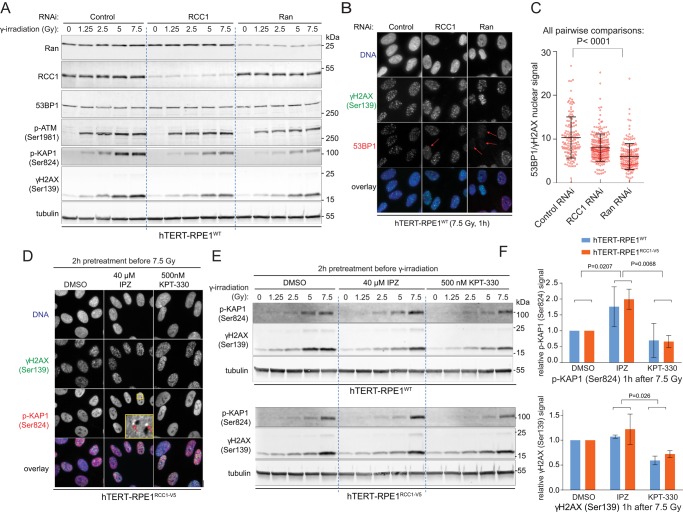
Apart from accelerating the cell cycle, RCC1 and Ran⋅GTP could support the survival from DNA damage by activating DNA repair. Consistent with the requirement for Ran activity in DNA damage signaling, immunoblotting showed that the knockdown of Ran or RCC1 by RNAi reduced the ATM-dependent phosphorylation of histone H2AX (γH2AX, Ser-139) and KAP1 (Iyengar and Farnham, 2011; p-KAP1, Ser-824) in γ-irradiated hTERT-RPE1WT cells (Figure 5A). We applied IF in cells that were fixed with paraformaldehyde and copermeabilized with Triton X-100 to detect changes in the recruitment of 53BP1 to the chromatin, following DNA damage (Jackson and Bartek, 2009). Although the 53BP1 expression was not affected (Figure 5A), both RCC1 and Ran knockdowns significantly reduced the ratio of nuclear 53BP1 and γH2AX signals in the irradiated cells (Figure 5C), indicating defects in 53BP1 association with the DNA damage sites marked by γH2AX signal. The knockdown of Ran had a stronger effect (Figure 5C) and caused the virtual disappearance of 53BP1 foci in a fraction of cells (Figure 5B, arrows). These results indicate that Ran activity is required for the recruitment of 53BP1 to DNA double-strand breaks, which is an essential step for their subsequent repair (Schultz et al., 2000; Callen et al., 2013).
FIGURE 5:
Ran-regulated NCT promotes the cellular response to DNA damage. (A) Immunoblotting of total cell lysates of hTERT-RPE1WT cells treated with control, RCC1-, or Ran-directed RNAi oligos and harvested 1 h after increasing doses of γ-irradiation. Notice the decreased intensity of the p-KAP1 and γH2AX signals in the RCC1 or Ran RNAi-treated cells. (B) IF micrographs of the γ-irradiated (5 Gy) hTERT-RPE1WT cells showing the reduced 53BP1 recruitment to the chromatin (arrows) upon Ran or RCC1-directed RNAi. (C) Scatter plot of the 53BP1/γH2AX IF signal ratios in cells treated as in B. Individual cell data; means ± SD; ANOVA with Tukey’s test. (D) IF micrographs of hTERT-RPE1RCC1-V5 cells pretreated with 40 μM IPZ or 500 nM KPT-330 and fixed 1 h after γ-irradiation (7.5 Gy). The inset shows the p-KAP1 heterochromatin-associated foci (arrow). (E) Immunoblotting of total cell lysates of hTERT-RPE1WT (top panel) and hTERT-RPE1RCC1-V5 (bottom) cells treated with 40 μM IPZ or 500 nM KPT-330 and harvested 1 h after increasing doses of γ-irradiation. (F) Quantification of the tubulin-normalized p-KAP1 and γH2AX signals detected on immunoblots of total lysates of cells treated as described and harvested 1 h after the irradiation with 7.5 Gy. Means ± SD from two experiments; ANOVA with Tukey’s posttest.
Next we used inhibitors of importin β and exportin 1 to test the role of two major NTRs in DNA damage signaling. To that end, we pretreated cells for 2 h with the exportin 1 inhibitor KPT-330 (Muqbil et al., 2013) or the importin β inhibitor importazole (IPZ; Soderholm et al., 2011) and collected samples for IF and immunoblots 1 h after γ-irradiation. IF indicated an increased number of p-KAP1 nuclear foci associated with the heterochromatin in IPZ-treated cells, while KPT-330 treatment had an opposite effect (Figure 5D). Although this trend was reproducible, the frequency of the IPZ-induced p-KAP1 foci varied between experiments. However, consistent with the IF data, immunoblotting showed that treatment with IPZ induced increased p-KAP1 signal upon irradiation, particularly in hTERT-RPE1RCC1-V5 cells (Figure 5, E and F). Also in agreement with the IF data, KPT-330 caused a strong reduction in p-KAP1 signal and a more moderate decrease in γH2AX signal on immunoblots (Figure 5, E and F). Reciprocal effects of the inhibitors indicate that the interplay between exportin 1– and importin β–regulated NCT modulates the ATM-dependent phosphorylation of KAP1, which has a role in the heterochromatin DNA repair (White et al., 2012).
Because the activation of Ran promotes cell cycle progression (Figure 1), the RNAi knockdowns or inhibitor treatments described above could have induced cell cycle changes that contribute to the observed effects on DNA damage signaling and repair processes. However, the above results demonstrate that, whether directly or also through modulating the cell cycle, the activity of Ran⋅GTP-regulated NCT mechanisms strongly affects essential steps of DDR in cell populations.
RCC1-induced activation of Ran-regulated NCT terminates DDR
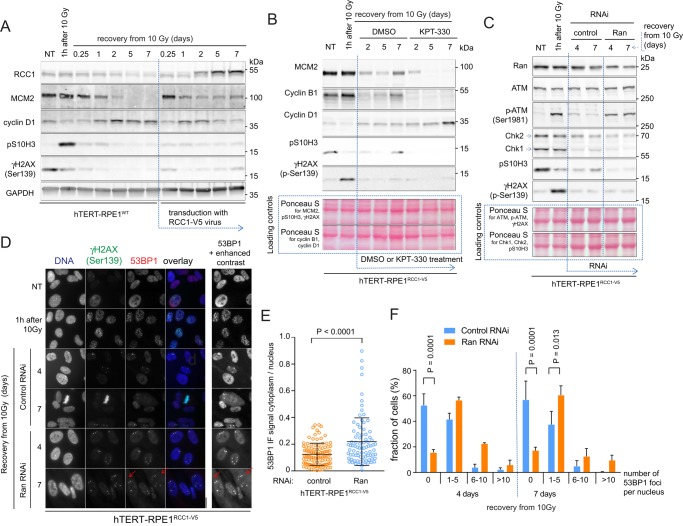
The perturbations of DNA damage signaling induced by Ran or RCC1 knockdowns (Figure 5) indicate that the mechanism of RCC1-induced recovery from DNA damage could require high Ran activity during the initial response to the damage. We examined this possibility by manipulating Ran function in cells that already sustained γ-irradiation–induced (10 Gy) DNA damage. First, we followed the irradiation of the hTERT-RPE1WT cells by transduction with control or RCC1-V5–expressing lentiviruses (Figure 6A). As shown by the strongly reduced expression of S-phase and G2/M markers (MCM2, pS10H3), the majority of irradiated control cells stopped proliferating, as expected. The rise of transiently expressed RCC1-V5 was paralleled by a drop in cyclin D1 levels, and the renewed expression of interphase and mitotic markers (MCM2, pS10H3; Figure 6A), indicating cell cycle reentry. This result showed that the activation of Ran in cells recovering from DNA damage is sufficient to terminate the DDR and enable the continuation of cell proliferation (Figure 5).
FIGURE 6:
RCC1-induced activation of NCT promotes the completion of the DNA repair and cell cycle reentry. (A–C) Immunoblots showing the changes of the cell cycle and DDR markers in cells exposed to different treatments after the γ-irradiation (10 Gy). (A) The γ-irradiated hTERT-RPE1WT cells were transduced with control or RCC1-V5 expressing lentiviruses. (B) The γ-irradiated hTERT-RPE1RCC1-V5 cells were treated with dimethyl sulfoxide (control) or 500 nM KPT-330. (C) Control or Ran-directed RNAi was applied after the γ-irradiation of the hTERT-RPE1RCC1-V5 cells. (D) Micrographs of 53BP1 and γH2AX IF staining in the hTERT-RPE1RCC1-V5 treated as in C. Inset with the enhanced contrast shows the accumulation of the cytoplasmic 53BP1 signal in the Ran RNAi-treated cells (arrows). (E) Scatter plot of the cytoplasmic/nuclear ratios of the 53BP1 IF signal in the control or Ran RNAi-treated cells at 7 d of recovery from the γ-irradiation. Individual cell data; means ± SD; t test, representative of two experiments. (F) Fractions of hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells recovering from γ-irradiation that contained the indicated numbers of 53BP1 foci per nucleus. Means ± SD from two independent experiments; two-way ANOVA with Bonferroni’s posttest.
To determine whether RCC1-induced DDR termination requires the function of exportin 1, we treated γ-irradiated hTERT-RPE1RCC1-V5 cells with KPT-330. The untreated hTERT-RPE1RCC1-V5 cells returned to cell cycle after irradiation, as shown by the expression of MCM2, pS10H3, and cyclin B1 (Figure 6B). KPT-330 treatment robustly reversed these trends and induced the accumulation of cyclin D1, indicating that DDR termination requires exportin 1– dependent nuclear export.
To verify that Ran mediates RCC1-induced DDR termination, we treated irradiated hTERT-RPE1RCC1-V5 cells with control or Ran-directed RNAi. As expected, RNAi controls resumed the cell cycle, as indicated by increased pS10H3 signal and reexpression of the Chk1 kinase, whose activity is essential for S-phase progression (Toledo et al., 2013; Figure 6C). The treatment with Ran RNAi in hTERT-RPE1RCC1-V5 cells reduced Ran levels by ∼50% (Figure 6C). However, even the partial reduction in Ran expression induced severe delays in DNA repair, as shown by the persistence of phosphorylated ATM on immunoblots (Figure 6C) and 53BP1 nuclear foci in cells stained for IF (Figure 6D). Cell cycle arrest was also confirmed by continuously low levels of Chk1 and pS10H3 (Figure 6C). Similar to hTERT-RPE1WT cells treated with doxorubicin (Figure 3), the irradiated and Ran RNAi-treated hTERT-RPE1RCC1-V5 cells accumulated 53BP1 in the cytoplasm (Figure 6, D and E). Therefore the reduced nuclear import of large molecular cargoes, such as 53BP1, appears to be a hallmark of cells failing to complete DNA repair as a consequence of insufficient Ran activity. In summary, these results demonstrate that RCC1-dependent activation of Ran-regulated nuclear import and export pathways facilitated the termination of DDR and cell cycle reentry.
DISCUSSION
The key finding of our study is that increased RCC1 expression accelerates cell cycle progression and DNA damage repair in a mechanism that involves raised cellular Ran⋅GTP levels and the function of importin β and exportin 1. Our results indicate that the overall activity of Ran⋅GTP-regulated nuclear export and import pathways modulates the cell cycle pace and cellular responses to DNA damages. Many of the underlying cellular and molecular mechanisms have yet to be clarified. However, our results indicate that, through the role of Ran⋅GTP in NCT of the cell cycle and DDR regulators, the interphase cell cycle checkpoints are sensing the RCC1-dependent Ran⋅GTP levels. If the cellular Ran⋅GTP concentration reaches sufficient threshold, Ran⋅GTP-regulated NCT reactions approach rates that support normal DDR function and cell cycle transitions. Consequently, DNA damage in such cells is successfully repaired, followed by termination of DDR and reentry into the cell cycle. If Ran⋅GTP levels fall below the required threshold, or a required NTR pathway is defective, the mechanisms of DNA repair or cell cycle reentry are delayed, triggering cell cycle arrest, as we documented, or possibly cell death.
The identity of the cell cycle transition(s) that was enabled by the RCC1 overexpression is an outstanding question raised by our results. Upon the induction of DNA damage, cell cycle reentry was induced by RCC1 in cells that exited mitosis (Figures 3 and 4), indicating that, in the majority of cells, RCC1 induced DNA damage repair and cell cycle reentry through its interphase role in NCT. The transient accumulation of cyclin D1 and absence of mitotic markers indicated that RCC1 overexpression enabled the exit from G1/S arrest. However, cells can enter senescence also from G2/M cell cycle arrest associated with high cyclin D1 expression (Gire and Dulic, 2015). Moreover, single-cell imaging studies showed that the timing of the individual cell’s commitment to S-phase reentry from the G0/G1 boundary depends on the mitosis-promoting signals in the preceding cell cycle (Spencer et al., 2013). Therefore it remains possible that, in RCC1-overexpressing cells exposed to DNA damage, activated mitotic function of Ran (Kalab and Heald, 2008; Forbes et al., 2015) enables G2/M transition and accelerates the S-phase commitment in the next cell cycle. The involvement of Ran in facilitating NCT of DNA replication factors (Liku et al., 2005; Ghosh et al., 2011) suggests that the intra S-phase cell cycle checkpoint also could be sensitive to the disruptions or activation of NCT as well. Because the WT and RCC1-overexpressing hTERT-RPE1 cells contain similar fractions of cells in G1, S, and G2/M stages (Supplemental Figure S1), it is possible that RCC1 accelerates several cell cycle phases. Future research involving synchronized cells in single-cell studies with cell cycle markers is needed to identify the specific cell cycle transitions targeted by Ran activation.
Because Ran⋅GTP regulates ∼20 different NTRs, multiple NTRs acting in parallel could contribute to the RCC1-induced acceleration of the cell cycle. However, it remains possible that the rate of cell cycle transitions could be particularly sensitive to NCT of only a subset of essential cargoes. The identity of such cell cycle rate-limiting NTR cargoes is not known, although published literature offers a list of many candidates. During the G1/S transition, such cargoes could include the mRNAs for many cyclins, including cyclin D1, whose eIF4e adaptor-dependent nuclear export requires exportin 1 (Culjkovic et al., 2006; Osborne and Borden, 2015). The S-phase entry and transition also involve Ran in the nuclear–cytoplasmic shuttling of proteins of the cyclin D, A, and E families (Malumbres and Barbacid, 2001; Jackman et al., 2002). Exportin 1 is required for the nuclear–cytoplasmic shuttling of cyclin B1 (Toyoshima et al., 1998; Gavet and Pines, 2010) and the Gwl/Mastl kinase that supports the function of the CDK1/cyclin B1 kinase (Alvarez-Fernandez et al., 2013; Wang et al., 2013). Potentially, Ran activity could modulate the timing of the activation of the cyclin B/CDK1 and Gwl/Mastl kinases during entry into mitosis. The loss of RCC1 function in temperature-sensitive tsBN2 cells induced premature entry to mitosis via the nuclear accumulation of the CDC25B phosphatase (Ohtsubo et al., 1987; Nishijima et al., 1997; Karlsson et al., 1999). Therefore the CDC25 phosphatase family could have a role in the Ran-regulated cell cycle rate. Because the phosphatases Wip1 and PP4 promote the termination of DDR signaling (Kleiblova et al., 2013; Shaltiel et al., 2014), the potential involvement of NCT in their activation is also of interest.
As indicated by the studies of Ran function in aging cells (Snow et al., 2013), large size could be one of the common features of cargoes whose NCT is particularly sensitive to Ran activity. Consistent with this idea, we observed that the knockdown of Ran induced cytoplasmic accumulation of 53BP1 (240–250 kDa; Figures 3, 5, and 6). In addition to 53BP1, many other prominent DNA repair regulators are large, multidomain proteins whose nuclear import, and potentially also export, could depend on sufficient Ran⋅GTP levels. However, the NCT of only a few of these regulators was studied in detail (53BP1; Moudry et al., 2012), TopBP1 (179 kDa; Bai et al., 2014), and BRCA1 (220 kDa; Thompson, 2010).
In addition to its role in NCT, the activation of Ran could promote changes in cell cycle or DDR indirectly, through gene expression changes induced by the NCT of transcriptional and epigenetic factors (Nemergut et al., 2001; Turner et al., 2012; Muqbil et al., 2014). Such transcriptionally mediated effects of Ran possibly contribute to the activation of the cell cycle and DDR in RCC1-overexpressing cell lines. However, our results indicate that, even within the potentially diverse cellular context of cells with different RCC1 expression (Figures 5 and 6), the activity of the Ran-regulated NCT pathways was still required for the cellular response to DNA damage.
The crucial role of cellular RCC1 concentrations is a salient feature of the Ran system, as shown by computational modeling (Figure 1 and Supplemental Figure S1) and our results. Because RCC1-mediated guanine nucleotide exchange reaction on Ran is the slowest step in the GTP/GDP cycle on Ran (Supplemental Figure S1), excessive RCC1 concentration sequesters Ran, explaining the saturable effect of RCC1 expression on Ran⋅GTP (Supplemental Figure S1). Interestingly, RCC1 overexpression in normal cells (Figure 3) or high endogenous RCC1 expression in carcinoma cells (Figure 4) stabilized endogenous Ran levels in cells recovering from DNA damage. The underlying mechanism remains to be determined. However, the computational models of the Ran system predict that changes in RCC1 expression are amplified through such coregulation, expanding the amplitude of cellular Ran⋅GTP levels and their cellular effects. Consequently, DNA damage–induced decline of RCC1 in normal cells could lead to a severe impairment of NCT function as documented by the stalled nuclear import of 53BP1 (Figures 3 and 6). Because cell cycle reentry requires Ran⋅GTP-dependent NCT activity, the depletion of Ran⋅GTP levels could function as a universal mechanism that enforces stable cell cycle arrest in senescent cells.
MATERIALS AND METHODS
Cell culture
Human foreskin fibroblasts (HFF-1) from the American Type Culture Collection (ATCC, Manassas, VA), hTERT-immortalized retinal pigment epithelial cells (hTERT-RPE1; ATCC), and human embryonic kidney cells, 293FT (HEK-293FT; Invitrogen) were cultured as previously described (Kalab et al., 2006; Hasegawa et al., 2013). Human colorectal carcinoma HCT116 cells were cultured in DMEM with 10% fetal bovine serum (FBS). Normal human fibroblasts CRL1474 (ATCC) were cultured in MEM with 15% FBS and normal lung fibroblasts Wi38 (ATCC) in MEM with 10% FBS. Media of cells expressing RCC1-V5 was supplemented with 10 μg/ml blasticidin (Invitrogen). All cells were grown in a 37°C humidified incubator with 5% CO2 and in media supplemented with 100 U/ml penicillin and 10 μg/ml streptomycin. Cells were treated with Importazole (IPZ; Sigma-Aldrich, St. Louis, MO; final concentration 40 μM) and KPT-330 (Karyopharm, Newton, MA; final concentration 500 nM) by supplementing their complete cell culture with aliquots of 10 mM solutions of the drugs in dimethyl sulfoxide.
Cell count and cell volume measurement
For cell count measurement, aliquots of 50,000 cells/well were seeded in six-well plates. At different time points, cells were collected by trypsinization, washed in Dulbecco’s phosphate-buffered saline (DPBS), and fixed in Trypan Blue (15250; Invitrogen) before the cell count and average diameter were determined with Cellometer Auto T4 (Nexcelom, Lawrence, MA). The PDT was calculated from the growth curves using the equation PDT = t × log(2)/[(log(Ct) − log(C0))], where t is the time of culture, Ct is the cell count at time t, and C0 is the cell count at the initial t = 0. t = 0 was selected as the first time point after the initial decline in cell number after plating.
Recombinant constructs
The pLenti-PGK-RCC1-V5 construct (pK439) was prepared by the modification of the PGK-histone H2B-mCherry (21217; Addgene, Cambridge, MA; Kita-Matsuo et al., 2009). Using PCR cloning and restriction digests, the histone H2B was replaced with an RCC1 open reading frame. The resulting pLenti-PGK-RCC1-mCherry was digested with ScaI and SacII and ligated with the ScaI and SacII fragment from pLenti X2 Blast (17391; Addgene; Campeau et al., 2009), introducing blasticidin resistance. mCherry was excised with Age1 and BsRG1 and replaced with a V5 sequence-coding oligo. The construct was validated by sequencing.
Lentiviral expression of RCC1-V5
Lentiviruses for the expression from pLenti-PGK-RCC1-V5 were produced as previously described (Kalab et al., 2006; Hasegawa et al., 2013). For transduction, normal cell media was replaced with media supplemented with lentiviral suspension (50–100 μl/ml) and 0.6/μl Polybrene/ml (Sigma-Aldrich). Following overnight treatment, cells were washed with warm DPBS (Life Technologies) and returned to normal growth medium. Stably expressing cells were selected by culture in media containing 10 μg/ml blasticidin. For transient expression, cells were transduced with pLenti-PGK-RCC1-V5 lentivirus 6 h after irradiation.
Induction of senescence by replicative exhaustion or DNA damage
For induction of senescence by replicative exhaustion, HFF-1 cells were kept in culture until passage 30. For induction of DNA damage by doxorubicin, normal cell media was supplemented with doxorubicin; this was followed by washes with warm DPBS and return to normal media. HFF-1, HCT116WT, HCT116RCC1-V5, CRL1474, and Wi38 cells were treated with 100 nM doxorubicin for 24 h. The hTERT-RPE1WT and hTERT-RPE1RCC1-V5 cells were treated for 16 h with 500 nM doxorubicin. For γ-irradiation–induced DNA damage, we used a J. L. Shepherd & Associates (San Fernando, CA) Mark-1-Model 68 Irradiator with a concealed 137Cs source and calibrated lead shields to modulate the dose.
SABG staining
We used a previously described protocol (Debacq-Chainiaux et al., 2009) to detect β-galactosidase activity. Bright-field micrographs were acquired using a Nikon Digital Sight color camera on a Nikon Eclipse T5100 microscope.
Electrophoresis and immunoblotting
For Supplemental Figures S1 and S2, cells were harvested and cell lysates were prepared for electrophoresis as described previously (Kalab et al., 2006; Hasegawa et al., 2013). For all other experiments, cells were trypsinized, washed in 37°C FBS-containing growth media, resuspended in 30–100 μl Clear Sample Buffer (CSB; 120 mM Tris, pH 6.8, 4% SDS, 20% glycerol), and immediately heated at 100°C for 5 min. Protein concentration was measured using a Micro BCA Protein Assay Kit (Pierce Thermo-Fisher, Carlsbad, CA) and adjusted to 4 mg/ml with CSB. Before electrophoresis, samples were mixed 1:1 with SDS-free CSB, 10% β-mercaptoethanol, and 0.5% bromophenol blue and heated at 100°C for 2 min. For Supplemental Figures S1 and S2, proteins were separated by homemade SDS–PAGE gels (Hasegawa et al., 2013). In all other experiments, we used 4–20% Criterion gels (Bio-Rad, Hercules, CA). Gels were blotted to polyvinylidene difluoride transfer membrane (Immobilon-FL; MIllipore, Bedford, MA) or LI-COR Odyssey (LI-COR Biotechnology, Lincoln, NE) nitrocellulose. Immunoblots were blocked and incubated with primary antibodies and IRDye 800– and/or IRDye 680–conjugated donkey secondary antibodies using Li-COR Odyssey buffers and protocols. The fluorescence emission was acquired using the Odyssey Imaging System (LI-COR). For each primary antibody, tubulin, actin, GAPDH, or Ponceau S–stained images of the blots were used as a loading control. The primary antibodies used for immunoblotting were from BD Biosciences (San Jose, CA; Ran, 610340; importin β, 610559), Bethyl (Montgomery, TX; Ran, A304-298A-M; p-KAP1(Ser-824), A300-767A; ATM, A300-136A), Genetex (Irvine, CA; RCC1,GTX63356; MCM2, GTX6238; cyclin B1, GTX6139; p-ATM (Ser-1981), GTX61739), Novus Biologicals (Littleton, CO; 53BP1, NB100-304), Santa Cruz Biotechnology (Dallas, TX; RanGAP1, sc-1862; cyclin D1, sc-753; GAPDH, sc-32233; Rad51, sc-8349; RanBP2, sc-74518; 53BP1, sc-22760; Chk1, sc-8408), Abcam (Cambridge, MA; RCC1, ab109379; TPX2 ab32785), Cell Signaling Technology (Danvers, MA; RanBP1, 8780; Chk2, 3440), Epitomics (Burlingame, CA; pS10H3, 1173-1), Developmental Studies Hybridoma Bank (Iowa City, IA; tubulin E7, E7), Millipore (γH2AX(Ser-193), 05-636), and GenScript (Piscataway, NJ; β-actin, A00702).
IF
Cells were grown on cleaned 12-mm round coverslips or on 32-mm ibidi chambers for RNAi-treated cells. For most experiments, cells were prepared for IF as previously described (Hasegawa et al., 2013), using antibodies to γH2AX (05-636; Millipore), 53BP1 (sc-22760; Santa Cruz Biotechnology), and p-KAP1 (A300-767A; Bethyl). For the detection of 53BP1 recruitment to γH2AX (Figure 5B), cells were fixed in 4% paraformaldehyde and 1% Triton X-100 in PBS for 10 min at room temperature.
53BP1 nuclear foci counting
The cells stained for 53BP1 and γH2AX were photographed on a wide-field Olympus IX81 microscope (Olympus USA, Waltham, MA) equipped with a UplanSApo 60×/1.35 oil-immersion lens and a Hamamatsu C4742 charge-coupled device (CCD) camera. For each sample, we acquired images of the DNA stain (Hoechst-33342), 53BP1 and γH2AX in at least 85 cells in randomly selected fields. Using the γH2AX foci as a control for a DNA damage–specific signal, the number of 53BP1 foci detected in each nucleus was manually counted and recorded in Excel spreadsheets by at least two independent observers.
IF quantification
MetaMorph 7.0.5 (Molecular Devices, Sunnyvale, CA) was used to quantify the IF signal intensity in images acquired using the UplanSApo 60×/1.35 oil-immersion lens and the Hamamatsu C4742 CCD camera. IF staining was performed as described above. Before quantification, the image-specific average out of cell fluorescence was subtracted from all images. For quantification of the cellular distribution of 53BP1, DNA images were thresholded for light objects to enable automatic creation of nuclear regions that were then transferred to 53BP1 images. After cytoplasmic 53BP1 regions for each cell were manually selected, region measurement data for each cell was logged into Excel, and the ratio of the average cytoplasmic and nuclear signal was calculated. Similarly, for Figure 5C, the DNA signal was used to create nuclear regions in the 53BP1 and γH2AX images, and Excel was used to calculate the ratios of average 53BP1 and γH2AX signal for each nucleus.
Immunoblotting quantification
To quantify protein expression on immunoblots, we used Odyssey, version 3 (LI-COR). The background-subtracted signal of individual protein bands was normalized to the GADPH or tubulin signal that was detected in the same lane.
Simple Western analysis
Simple Western (ProteinSimple, San Jose, CA) analyses were performed as previously described (Chen et al., 2013). Cell lysates (20 or 40 ng protein/capillary) were mixed with 1X SDS sample buffer, dithiothreitol, and fluorescent standards (ProteinSimple); heated at 70°C for 10 min before loading into the Peggy Sue instrument; separated by molecular weight; and immobilized using UV light. The capillaries were incubated with a blocking reagent (ProteinSimple), followed by primary and horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). A chemiluminescence signal was developed, captured by a CCD camera, and quantified as the peak area by the Compass software according to the ProteinSimple protocols. The primary antibodies used were Ran (610340; BD Biosciences), RCC1 (63356; GeneTex), cyclin D1 and cyclin B1 (sc-753 and sc-752; Santa Cruz Biotechnology), and vinculin (V9131; Sigma-Aldrich).
FACS analysis
Cells (500 μl of suspension in DPBS) were fixed with 4.5 ml ice-cold 70% ethanol, centrifuged (1200 rpm, 5 min, 4°C), washed with 5 ml ice-cold DPBS (1200 rpm, 5 min, 4°C), resuspended in 1 ml staining solution (20 μg/ml propidium iodide [PI; Sigma-Aldrich], 200 μg/ml RNaseA [Sigma-Aldrich], 0.1% Triton X-100/PBS), and incubated for 30 min at 37°C. After filtration with 70-μm nylon mesh, cells were sorted using FACSCalibur (Becton-Dickinson, Franklin Lakes, NJ). Cell debris and doublets were gated out, and G0/G1, S, and G2/M populations were quantified using the ModFit, version 3.0 (Becton-Dickinson). Images showing the total PI signal count versus count distribution were displayed using FlowJo-VX.
RCC1 and Ran RNAi
Control siRNA oligo (5′-UGGUUUACAUGUCGACUAA-3′; ON-TARGET plus nontargeting siRNA #1), RCC1 siRNA SMARTpools (5′-GGAGAACCGUGUGGUCUUA-3′, 5′-CAGCAGCCCUCACCGAUGA-3′, 5′-GCACAGAACCCGGCUUGGU-3′ and 5′-GGACAAUAACGGUGUGAUU-3′; Chen et al., 2007), and Ran siRNA (5′-GAAAUUCGGUGGACUGAGA-3′ and 5′-CCAACAGAGGACCUAUUAA-3′) were obtained from Thermo-Fisher/Dharmacon. HCT116WT or hTERT-RPE1RCC1-V5 cells were plated in six-well plates or 100-mm dishes in antibiotic-free medium and transfected with a 5 nM final siRNA concentration using Dharmafect (Thermo-Fisher) according to manufacturer’s instructions.
FLIM imaging, analysis, and display
Spatially resolved, time-correlated single-photon counting (TCSPC) data sets were acquired as described previously (Kalab et al., 2006; Hasegawa et al., 2013), using a Zeiss LSM710 NLO (Zeiss USA, Thornwood, NY) microscope equipped with an SPC-830 (Becker & Hickl GmbH, Berlin, Germany) TCSPC controller. Imaging conditions were chosen to limit the emission to (0.2–1) × 106 counts/s. Images of 128 × 128 pixels (1024 time bins/pixel) were acquired over 60–120 s and analyzed with SPCI software (Becker & Hickl), while assuming incomplete two-exponential decay and using trapezoid integration and no prefixed parameters. We used SPCI to prepare the RGB pseudocolor images of the median τdonor values of RBP-4 detected in cells. The pseudocolor τdonor images were displayed within the range of 300 ps that was centered at cell-specific average τdonor.
Statistical analysis and image processing
We analyzed data with Excel or GraphPad Prism, version 6.05 (GraphPad Software, La Jolla, CA). Images were prepared for publication with Adobe Photoshop and Illustrator CC (Adobe Systems, San Jose, CA).
Supplementary Material
Acknowledgments
We thank R. Heald and K. Weis for comments and suggestions on the manuscript; P. Scaffidi, B. Vogelstein, C. Harris, and A. Schetter for cell lines, Karyopharm (Newton, MA) for the donation of KPT-330; and J. Lee and S. J. Ryu for technical assistance. Imaging was performed at the Confocal Microscopy Core Facility of the Center for Cancer Research, National Cancer Institute (NCI), in Bethesda, Maryland. P.C., K.H., Y.P., J.-Q.C., M.A.H., S.K., and P.K. were supported by the Intramural Research Program of the Center for Cancer Research, NCI. D.O. and E.T. were supported by National Institutes of Health grant R01 GM071522.
Abbreviations used:
- ANOVA
analysis of variance
- ATCC
American Type Culture Collection
- ATM
ataxia telangiectasia–mutated
- ATR
ataxia telangiectasia and Rad3–related
- CSB
Clear Sample Buffer
- DDR
DNA damage response
- DPBS
Dulbecco’s phosphate-buffered saline
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- FLIM
fluorescence lifetime imaging microscopy
- FRET
Förster resonance energy transfer
- HEK-293FT
human embryonic kidney cells, 293FT
- HFF-1
human foreskin fibroblasts
- hTERT-RPE1
hTERT-immortalized retinal pigment epithelial cells
- IF
immunofluorescence
- IPZ
importazole
- NCT
nuclear–cytoplasmic transport
- NTR
nuclear transport receptor
- PDT
population doubling time
- PGK
phosphoglycerate kinase
- PI
propidium iodide
- pS10H3
p-histone H3 (Ser-10)
- RNAi
RNA interference
- SABG
senescence-associated β-galactosidase
- TCSPC
time-correlated single-photon counting.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0025) on February 10, 2016.
REFERENCES
- Alhopuro P, Sammalkorpi H, Niittymaki I, Bistrom M, Raitila A, Saharinen J, Nousiainen K, Lehtonen HJ, Heliovaara E, Puhakka J, et al. Candidate driver genes in microsatellite-unstable colorectal cancer. Int J Cancer. 2012;130:1558–1566. doi: 10.1002/ijc.26167. [DOI] [PubMed] [Google Scholar]
- Alvarez-Fernandez M, Sanchez-Martinez R, Sanz-Castillo B, Gan PP, Sanz-Flores M, Trakala M, Ruiz-Torres M, Lorca T, Castro A, Malumbres M. Greatwall is essential to prevent mitotic collapse after nuclear envelope breakdown in mammals. Proc Natl Acad Sci USA. 2013;110:17374–17379. doi: 10.1073/pnas.1310745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Michael WM, Yan S. Importin beta-dependent nuclear import of TopBP1 in ATR-Chk1 checkpoint in Xenopus egg extracts. Cell Signal. 2014;26:857–867. doi: 10.1016/j.cellsig.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc Natl Acad Sci USA. 2002;99:389–394. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Heldman MR, Herrmann MA, Kedei N, Woo W, Blumberg PM, Goldsmith PK. Absolute quantitation of endogenous proteins with precision and accuracy using a capillary Western system. Anal Biochem. 2013;442:97–103. doi: 10.1016/j.ab.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9:598–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciarello M, Roscioli E, Di Fiore B, Di Francesco L, Sobrero F, Bernard D, Mangiacasale R, Harel A, Schinina ME, Lavia P. Nuclear reformation after mitosis requires downregulation of the Ran GTPase effector RanBP1 in mammalian cells. Chromosoma. 2010;119:651–668. doi: 10.1007/s00412-010-0286-5. [DOI] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- Forbes DJ, Travesa A, Nord MS, Bernis C. Nuclear transport factors: global regulation of mitosis. Curr Opin Cell Biol. 2015;35:78–90. doi: 10.1016/j.ceb.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 2010;189:247–259. doi: 10.1083/jcb.200909144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. J Biol Chem. 2011;286:23831–23841. doi: 10.1074/jbc.M110.215988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire V, Dulic V. Senescence from G2 arrest, revisited. Cell Cycle. 2015;14:297–304. doi: 10.1080/15384101.2014.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Ryu SJ, Kalab P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J Cell Biol. 2013;200:151–161. doi: 10.1083/jcb.201206142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Kubota Y, den Elzen N, Hagting A, Pines J. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol Biol Cell. 2002;13:1030–1045. doi: 10.1091/mbc.01-07-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Karlsson C, Katich S, Hagting A, Hoffmann I, Pines J. Cdc25B and Cdc25C differ markedly in their properties as initiators of mitosis. J Cell Biol. 1999;146:573–584. doi: 10.1083/jcb.146.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiblova P, Shaltiel IA, Benada J, Sevcik J, Pechackova S, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013;201:511–521. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement K, Goodarzi AA. DNA double strand break responses and chromatin alterations within the aging cell. Exp Cell Res. 2014;329:42–52. doi: 10.1016/j.yexcr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Liku ME, Nguyen VQ, Rosales AW, Irie K, Li JJ. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol Biol Cell. 2005;16:5026–5039. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zak J, Chen S, Sanchez-Pulido L, Severson DT, Endicott J, Ponting CP, Schofield CJ, Lu X. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell. 2014;157:1130–1145. doi: 10.1016/j.cell.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Moudry P, Lukas C, Macurek L, Neumann B, Heriche JK, Pepperkok R, Ellenberg J, Hodny Z, Lukas J, Bartek J. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 2012;19:798–807. doi: 10.1038/cdd.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqbil I, Bao B, Abou-Samra AB, Mohammad RM, Azmi AS. Nuclear export mediated regulation of microRNAs: potential target for drug intervention. Curr Drug Targets. 2013;14:1094–1100. doi: 10.2174/1389450111314100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqbil I, Kauffman M, Shacham S, Mohammad RM, Azmi AS. Understanding XPO1 target networks using systems biology and mathematical modeling. Curr Pharm Des. 2014;20:56–65. doi: 10.2174/13816128113199990611. [DOI] [PubMed] [Google Scholar]
- Nagai M, Yoneda Y. Downregulation of the small GTPase Ras-related nuclear protein accelerates cellular ageing. Biochim Biophys Acta. 2012;1830:2813–2819. doi: 10.1016/j.bbagen.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Nishijima H, Nishitani H, Seki T, Nishimoto T. A dual-specificity phosphatase Cdc25B is an unstable protein and triggers p34(cdc2)/cyclin B activation in hamster BHK21 cells arrested with hydroxyurea. J Cell Biol. 1997;138:1105–1116. doi: 10.1083/jcb.138.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Sekiguchi M, Hayashida H, Kuma K, Miyata T, Fukushige S, Murotsu T, et al. Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- Osborne MJ, Borden KL. The eukaryotic translation initiation factor eIF4E in the nucleus: taking the road less traveled. Immunol Rev. 2015;263:210–223. doi: 10.1111/imr.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Peters D, Freund J, Ochs RL. Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells. Mol Cancer Ther. 2005;4:1605–1616. doi: 10.1158/1535-7163.MCT-04-0311. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- Shaltiel IA, Aprelia M, Saurin AT, Chowdhury D, Kops GJ, Voest EE, Medema RH. Distinct phosphatases antagonize the p53 response in different phases of the cell cycle. Proc Natl Acad Sci USA. 2014;111:7313–7318. doi: 10.1073/pnas.1322021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel IA, Krenning L, Bruinsma W, Medema RH. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J Cell Sci. 2015;128:607–620. doi: 10.1242/jcs.163766. [DOI] [PubMed] [Google Scholar]
- Sliwinska MA, Mosieniak G, Wolanin K, Babik A, Piwocka K, Magalska A, Szczepanowska J, Fronk J, Sikora E. Induction of senescence with doxorubicin leads to increased genomic instability of HCT116 cells. Mech Ageing Dev. 2009;130:24–32. doi: 10.1016/j.mad.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- Snow CJ, Dar A, Dutta A, Kehlenbach RH, Paschal BM. Defective nuclear import of Tpr in Progeria reflects the Ran sensitivity of large cargo transport. J Cell Biol. 2013;201:541–557. doi: 10.1083/jcb.201212117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME. BRCA1 16 years later: nuclear import and export processes. FEBS J. 2010;277:3072–3078. doi: 10.1111/j.1742-4658.2010.07733.x. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83:1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Galan JA, Normandin K, Bonneil E, Hickson GR, Roux PP, Thibault P, Archambault V. Cell cycle regulation of Greatwall kinase nuclear localization facilitates mitotic progression. J Cell Biol. 2013;202:277–293. doi: 10.1083/jcb.201211141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ., 3rd The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol Cancer Res. 2012;10:401–414. doi: 10.1158/1541-7786.MCR-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.