Protein arginine methyltransferase 5 interacts with and methylates apoptosis signal–regulating kinase 1 at arginine residue 89, thereby negatively regulating its activity by promoting the interaction between ASK1 and Akt and thus phosphorylating ASK1 at serine residue 83.
Abstract
We describe a novel functional interaction between ASK1 and PRMT5. We show that PRMT5 interacts with and methylates ASK1 at arginine residue 89 and thereby negatively regulates its activity by promoting the interaction between ASK1 and Akt and thus phosphorylating ASK1 at serine residue 83. Furthermore, the association between ASK1 and Akt is enhanced by VEGF stimulation, and PRMT5 is required for this association. Moreover, PRMT5-mediated ASK1 methylation impaired the H2O2-induced activity of ASK1, and this inhibitory effect of PRMT5 was abolished by replacement of arginine 89 with Trp or depletion of PRMT5 expression by RNA interference. Together the results demonstrate cross-talk between arginine methylation and serine phosphorylation in ASK1.
INTRODUCTION
Apoptosis signal–regulating kinase 1 (ASK1), a 155-kDa protein, is a member of the mitogen-activated protein kinase (MAPK) kinase kinase family, which are activated in response to proinflammatory stimuli and cellular stress, leading to activation of MAPK c-Jun N-terminal kinase (JNK)/p38 signaling cascades (Ichijo et al., 1997; Hayakawa et al., 2006; Bunkoczi et al., 2007). For example, ASK1 is strongly activated in cells exposed to various oxidants, such as H2O2 and diamide (Matsuzawa et al., 2005; Soga et al., 2012). Oxidative stress–induced activation of ASK1 leads to apoptosis in various types of cells (Kadowaki et al., 2005; Hyun et al., 2010). Indeed, ASK1 is a central target of many cellular survival factors that bind to different domains of ASK1 to keep ASK1 in an inactive state (Du et al., 2004; Kim et al., 2012; Kosek et al., 2014). ASK1 can be phosphorylated at several sites, and these phosphorylation sites regulate ASK1 activity in both positive and negative manners (Kim et al., 2001; Tobiume et al., 2002; Fujii et al., 2004; Gu et al., 2009; Seong et al., 2010). In resting cells, ASK1 constantly forms an inactive complex with thioredoxin (Trx), but upon treatment of cells with tumor necrosis factor α or oxidants such as H2O2, ASK1 is dissociated from Trx and activated by subsequent modifications, including phosphorylation at the sites of Thr-845 (Fujino et al., 2007). Conversely, several serine/threonine protein kinases, such as Akt, directly phosphorylate ASK1 at Ser-83 to inhibit ASK1-induced apoptosis (Kim et al., 2001). Consistently, inhibition of Akt by phosphatidylinositol 3-kinase inhibitor significantly induces activation of p38 and JNK (Xie et al., 2009; Lu et al., 2010). The mechanism for ASK1 activation in response to apoptotic stimuli has not been fully elucidated.
Recent work shows that ASK1 is closely linked to cardiac diseases, such as cardiac hypertrophy, remodeling, and injury (Harding et al., 2010; Nako et al., 2012; Huang et al., 2014). In the left ventricle, ASK1 is activated by generation of angiotensin II–induced reactive oxygen species (ROS) through the angiotensin II type 1 receptor, resulting in cardiac hypertrophy and remodeling (Nako et al., 2012). Yokoi et al. (2006) reported that ASK1 mediates cellular senescence induced by high glucose in endothelial cells. They found that high glucose induces up-regulation of the ASK1 signaling in endothelial cells. However, transfection with an adenoviral construct including a dominant-negative form of the ASK1 gene significantly inhibited SA-β-gal activity induced by high glucose (Yokoi et al., 2006). Machino et al. (2003) reported that H2O2 induces ASK1 phosphorylation and concomitantly p38 MAPK and JNK phosphorylation, as well as activation of caspase-3, in pulmonary vascular endothelial cells. However, the dominant-negative form of ASK1 significantly inhibits the apoptosis response induced by H2O2 in endothelial cells (Machino et al., 2003). Vascular endothelial growth factor (VEGF) has been shown to increase endothelial resistance to H2O2 (Liu et al., 2002; Tao et al., 2005). Intriguingly, VEGF was induced by interleukin-6 through ASK1 in human osteosarcoma (Tzeng et al., 2013). Moreover, Nako et al. (2012) reported that oxidative stress–induced ASK1 activation led to endothelial apoptosis. However, VEGF treatment prevented oxidative stress–induced endothelial apoptosis by inhibiting ASK1 activation (Nako et al., 2012).
Protein arginine methyltransferases (PRMTs) are a group of proteins that catalyze the methyl transferal to the nitrogen of the terminal guanidine of arginines (Fisk and Read, 2011; Chen et al., 2014). PRMTs are divided into four types according to the terminal arginine modification. Types I–III all catalyze this first methyl transfer reaction (monomethylation), but, only type II PRMTs can generate symmetric dimethylarginine in target proteins (Fisk and Read, 2011). In this study, we sought to identify ASK1-interacting proteins to clarify new mechanisms of ASK1-mediated signaling. We identify PRMT5 (type II) as a novel ASK1-binding protein and show that it methylates Arg-89 in ASK1, and this methylation positively modulates Akt-mediated ASK1 phosphorylation at Ser-83, which attenuates H2O2-induced ASK1 activation.
RESULTS
Identification of PRMT5 as an ASK1-binding protein
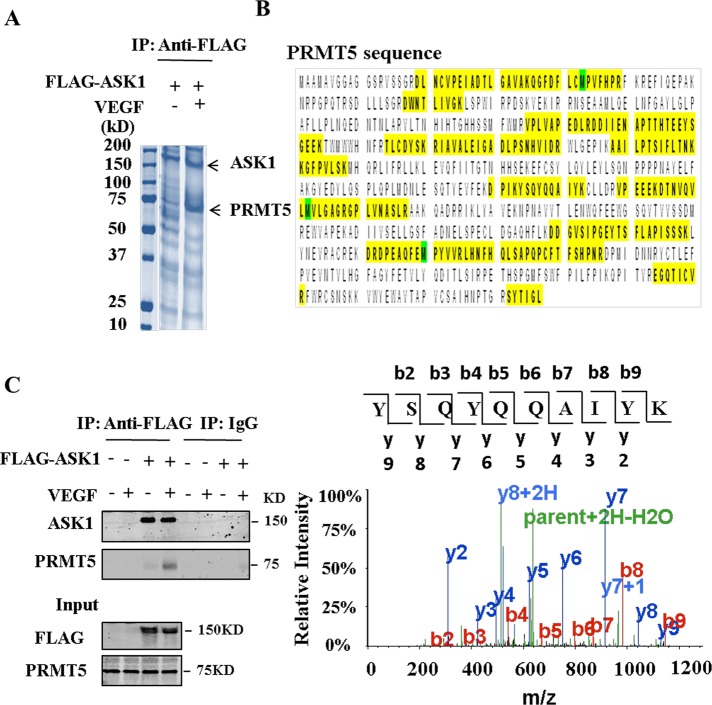
To identify binding partners that might interact with ASK1, we overexpressed FLAG-tagged ASK1 in EAhy926 cells under VEGF stimuli and undertook coimmunoprecipitation. We found that a band at ∼75 kDa was significantly enriched by VEGF stimulation (Figure 1A). We cut the slice and then tried to identify the ASK1-binding protein by liquid chromatography–tandem mass spectrometry (LC-MS/MS) after in-gel digestion. Surprisingly, the sequence identity of the peptides of this protein revealed the presence of PRMT5 (Figure 1B). Although it has been reported that when purifying a FLAG-tagged protein using FLAG antibody a major contaminant is the PRMT5/MEP50 protein complex (Chen and Gingras, 2007), we found that VEGF stimuli induced an abundance of PRMT5 in the immunoprecipitation complex (Figure 1, A and C), which indicates that the interaction is specific and functional.
FIGURE 1:
Identification of PRMT5 as an ASK1-binding protein. (A) EAhy926 cells were transfected with pFLAG-CMV-ASK1 and then treated with 100 ng/ml VEGF for 30 min. FLAG-tagged ASK1 protein was purified by anti-FLAG agarose. Purified proteins were electrophoresed and stained with Coomassie brilliant blue. PRMT5 (72 kDa) is indicated in the gel. (B) The 72-kDa band was cut and in-gel digested with trypsin. Peptides were subject to LC-MS/MS assay. Identified peptides that matched with the sequence of PRMT5 are shown in yellow. MS/MS spectrum of the tryptic peptide YSQYQQAIYK. Fragment ions are indicated as b and y ions. (C) Cell lysates from A were immunoprecipitated with anti-FLAG antibody or control immunoglobulin G (IgG) and then separated by 8% SDS–PAGE. Transferred membrane was immunoblotted with either anti-FLAG or PRMT5 antibody.
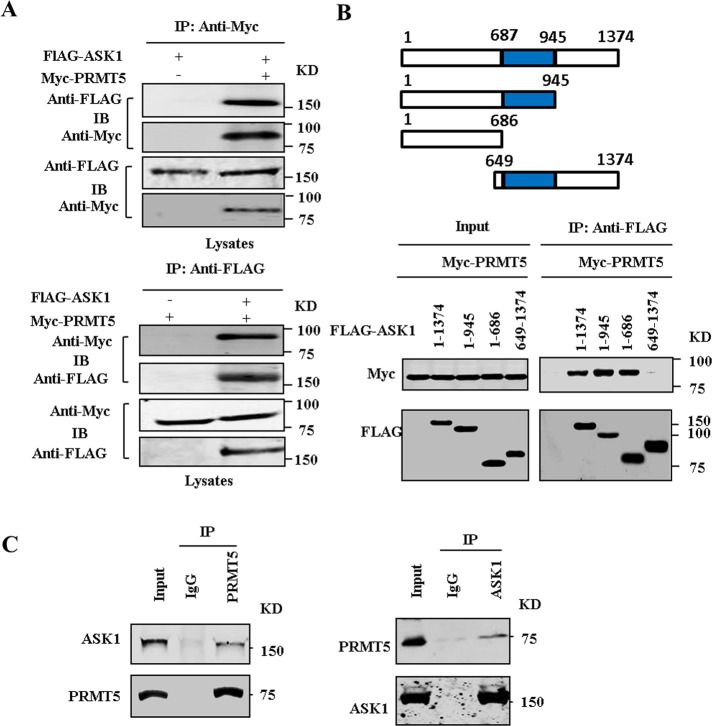
To confirm the interaction between ASK1 and PRMT5, we first examined the physical association between exogenous ASK1 and PRMT5. To this end, we performed coimmunoprecipitation experiments in HEK293T cells transfected with FLAG-ASK1 and Myc-PRMT5 expression vectors. As shown in Figure 2A, immunoprecipitation of Myc-PRMT5 led to coimmunoprecipitation of FLAG-tagged ASK1 when both proteins were cotransfected. As a control, the anti-Myc antibody did not immunoprecipitate FLAG-tagged ASK1 in the absence of Myc-PRMT5. Similarly, immunoprecipitation of FLAG-tagged ASK1 resulted in coimmunoprecipitation of Myc-PRMT5, whereas the anti-FLAG antibody did not immunoprecipitate Myc-PRMT5 in the absence of FLAG-tagged ASK1. To determine a critical region in ASK1 for PRMT5 association, we constructed several deletion mutants of ASK1. As shown in Figure 2B, deletion of the C-terminal region of ASK1 from residues 687 and 946 did not affect its ability to bind to PRMT5. However, deletion of the N-terminal region of ASK1 residues up to 648 abrogated the interaction with PRMT5. These results indicate that the N-terminal domain in ASK1 is crucial for its interaction with PRMT5. To further confirm an in vivo interaction between ASK1 and PRMT5, we carried out coimmunoprecipitation with human umbilical vein endothelial cells (HUVECs). We found that endogenous ASK1 formed a physical complex with endogenous PRMT5 in cells (Figure 2C).
FIGURE 2:
PRMT5 interacts with ASK1. (A) FLAG-tagged ASK1 and Myc-tagged PRMT5 expression vectors were cotransfected into HEK293T cells. Extracted proteins were precipitated by anti-FLAG and anti-Myc, respectively, and then separated by 8% SDS–PAGE. The transferred membrane was immunoblotted with either anti-FLAG or anti-Myc antibody. (B) Schematic depiction of ASK1-domain mutants. HEK293T cells were transfected with Myc-tagged PRMT5 vectors together with the indicated FLAG-tagged ASK1 deletion mutants. Proteins were immunoprecipitated with anti-FLAG antibody. (C) Cell lysates obtained from HUVECs were immunoprecipitated with anti-ASK1, anti-PRMT5 antibody, or control IgG and then separated by 8% SDS–PAGE. Transferred membrane was immunoblotted with either anti-ASK1 or PRMT5 antibody.
PRMT5 mediates arginine methylation of ASK1
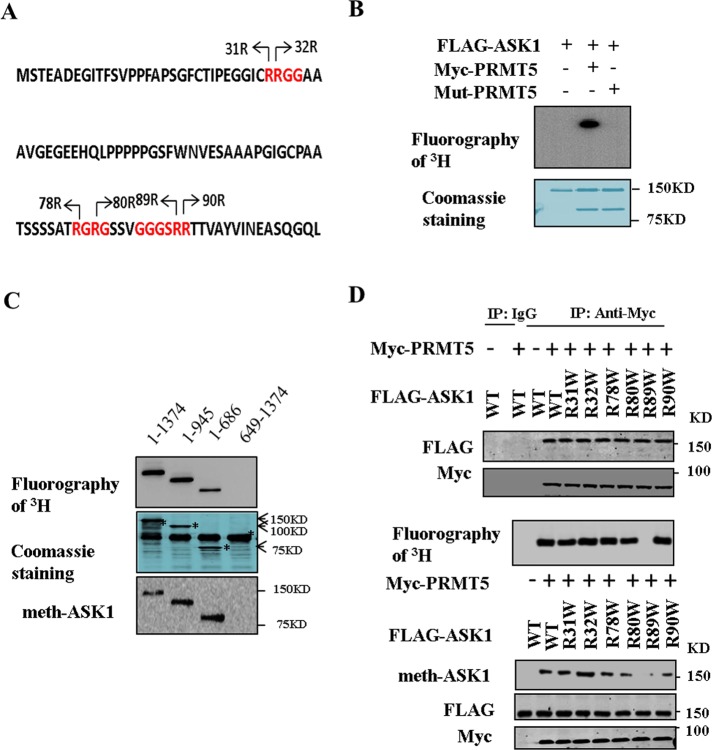
ASK1 is methylated and modulated by PRMTs. Cho et al. (2012), for example, reported the PRMT1-mediated methylation of ASK1 at Arg-78 and Arg-80. Analysis of the amino acid sequence of human ASK1 revealed that six arginine residues reside in RG-enriched motifs in the N-terminal region of ASK1, which are potential methylation sites for PRMT5 (Figure 3A). We hypothesized that ASK1 proteins might also be methylated by PRMT5. We examined whether PRMT5 was able to mediate the arginine methylation of ASK1 in an in vitro methylation assay in which FLAG-tagged ASK1 was incubated with Myc-tagged PRMT5 or mutant PRMT5 in the presence of S-adenosyl-l-[methyl-3H]methionine ([3H]SAM) as the methyl donor. As shown in Figure 3B, Myc-PRMT5 methylated FLAG-ASK1. In contrast, methylation in ASK1 could not be detected with the mutant PRMT5 or the absence of PRMT5. Subsequently we found that deletion of the N-terminal region of ASK1 from residues 1–648 extremely attenuates the methylation of ASK1 (Figure 3C). These data indicated that PRMT5 mediates arginine methylation of ASK1 at the N-terminal region.
FIGURE 3:
PRMT5 methylates ASK1 at Arg-89. (A) The N-terminal sequence of human ASK1. RG-rich residues are in red, and potential methylation sites are indicated by arrows. (B) HEK293T cells were transfected for 48 h with expression vectors encoding FLAG-ASK1 or wild-type or mutant PRMT5. Purified FLAG-ASK1 and wild-type or mutant PRMT5 incubated in the presence of [3H]SAM. Reaction products were analyzed by fluorography. (C) Wild-type or mutant fragment ASK1 (5 μg) and Myc-PRMT5 (10 μg) incubated in the presence of [3H]SAM. Reaction products were analyzed by fluorography. The gel was also stained with Coomassie brilliant blue. (D) HEK293T cells were transfected for 48 h with expression vectors encoding FLAG-ASK1, mutant of ASK1, and Myc-PRMT5 as indicated. Cell lysates were then subjected to immunoprecipitation with anti-Myc or control IgG and then separated by 8% SDS–PAGE (top). Wild-type or mutant ASK1 and Myc-PRMT5 incubated in the presence of [3H]SAM. Reaction products were analyzed by fluorography (middle). Cell lysates were subjected to Western blot and examined by immunoblot with anti–dimethylation symmetric, anti-FLAG, and anti-Myc antibodies (bottom).
PRMT5 methylates ASK1 preferentially at Arg-89
The N-terminal of ASK1 contains a series of arginine residues, six of which (Arg-31, Arg-32, Arg-78, Arg-80, Arg-89, and Arg-90) reside in an RG-enriched motif that serves as a methylation motif for PRMT5 (Figure 3A). We next examined which arginine residue of ASK1 is the methylation site. We replaced these six arginine residues with Trp by site-directed mutagenesis and examined whether the mutant proteins were methylated by PRMT5 in vitro. As shown in Figure 3D, the Arg mutant into Trp has no effect on the association of ASK1 and PRMT5; however, replacement of Arg-89 with Trp markedly reduced the extent of PRMT5-mediated methylation of ASK1 (Figure 3D and Supplemental Figure S1). In contrast, replacement of Arg-31, Arg-32, Arg-78, Arg-80, and Arg-90 with Trp did not affect the level of PRMT5-mediated methylation of ASK1. These data suggested that PRMT5 methylates ASK1 preferentially at Arg-89.
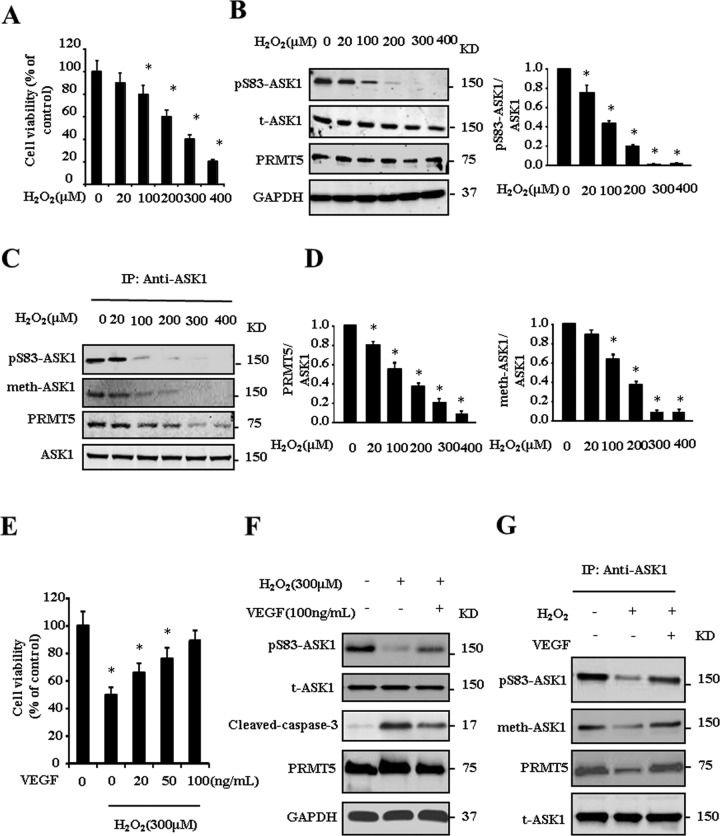
VEGF stimulation attenuates H2O2-induced ASK1 activation
Given that PRMT5 mediates the arginine methylation of ASK1, we next examined whether PRMT5 modulates ASK1 activity. VEGF has been reported to increase endothelial resistance to H2O2 (Liu et al., 2002), and thus we used the cell model of HUVECs stimulated with H2O2 and VEGF. As shown in Figure 4A, H2O2 stimulation induces HUVECs apoptosis. As expected, activation of ASK1 induced by H2O2 was observed by determining the phosphorylation of Ser-83, which inhibits the activity of ASK1 (Figure 4B). Intriguingly, H2O2 treatment decreases the interaction between ASK1 and PRMT5 in a coimmunoprecipitation analysis, which results in attenuation of the methylation of ASK1 (Figure 4, C and D). The H2O2-induced endothelial cell apoptosis and dissociation of ASK1 and PRMT5 were reversed by VEGF pretreatment (Figure 4, E–G). These results suggest that VEGF-induced methylation of ASK1 positively regulates phosphorylation of Ser-83 and thus negatively regulates H2O2-induced activation of ASK1.
FIGURE 4:
VEGF attenuates H2O2-induced ASK1 activation. HUVECs were treated with different concentrations of H2O2 for 24 h. Cell viability (A), phosphorylation of ASK1 at Ser-83 (B), and methylation of ASK1 (C, D) were determined by the indicated antibodies. HUVECs were pretreated with VEGF for 30 min and then treated with 300 μm H2O2 for 24 h. Cell viability (E) and phosphorylation of ASK1at Ser-83, cleaved caspase-3 (F) and methylation of ASK1 (G) were determined by the indicated antibodies. *p < 0.05 compared with control group.
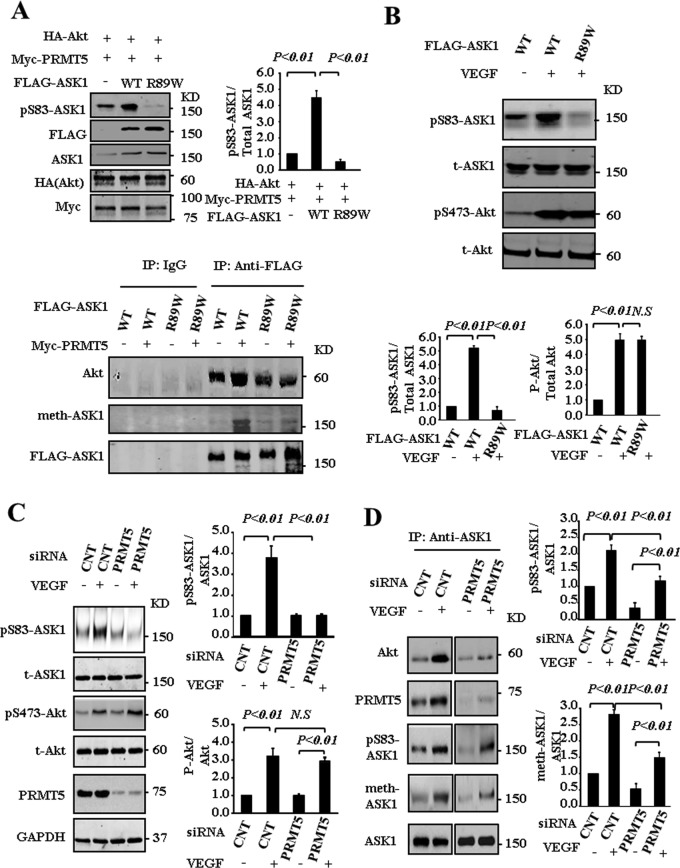
PRMT5-mediated methylation of ASK1 at Arg-89 promotes the association between ASK1 and Akt
On the basis of our finding that methylation of ASK1 negatively regulates H2O2-induced activation of ASK1, we next asked whether methylation of ASK1 at Arg-89 affects Ser-83 in vitro and in vivo. To address this issue, we used a HEK293T cell line cotransfected with Akt, PRMT5, and ASK1 or mutant ASK1 (R89W). As shown in Figure 5A (top and left), Akt phosphorylates wild-type ASK1 at Ser-83 but does not phosphorylate mutant ASK1 (R89W). In addition, phosphorylation of ASK1 at Ser-83 was inhibited at a basal level. Immunoprecipitation analysis revealed that both ASK1 and mutant ASK1(R89W) interact with endogenous Akt, indicating the possibility that mutant ASK1(R89W) competed with endogenous ASK1 for interaction with Akt. PRMT5 enhances the association between ASK1 and Akt; however, this association is significantly inhibited by mutant ASK1 (R89W), further indicating the dominant-negative effect of mutant ASK1(R89W) on the phosphorylation level of endogenous ASK1 at Ser-83 (Figure 5A, bottom) by competing with endogenous ASK1 for the interaction with Akt. This effect was also examined in EAhy926 cells transfected ASK1 and mutant ASK1. As shown in Figure 5B, VEGF stimulation resulted in activation of Akt and increased the phosphorylation of ASK1 at Ser-83. However, overexpression of mutant ASK1(R89W) significantly inhibited the phosphorylation of Ser-83 but had no effect on the activity of Akt(pS473-Akt), which was induced by VEGF stimulus. These data imply that posttranslational modification (methylation) of ASK1 at Arg-89 contributes to the phosphorylation of ASK1 at Ser-83 mediated by Akt. We next examined the effects of PRMT5 knockdown on the phosphorylation of ASK1 Ser-83 in endothelial cells stimulated with VEGF. As shown in Figure 5C, VEGF stimulus increases the activity of Akt and phosphorylation of ASK1 at Ser-83; knockdown of PRMT5 substantially suppresses the Ser-83 phosphorylation induced by VEGF but has no effect on the VEGF-induced Akt activity. These results indicate that PRMR5 plays an important role in VEGF-induced phosphorylation of ASK1 at Ser-83. Furthermore, immunoprecipitation analysis shows that VEGF stimulation enhances the interaction of ASK1 with both Akt and PRMT5, resulting in increased arginine methylation and phosphorylation (at Ser-83) of ASK1. However, PRMT5 knockdown attenuates VEGF-induced methylation and phosphorylation (at Ser-83) of ASK1, as well as decreases the interaction of ASK1 and Akt (Figure 5D). Taken together, these data indicate that PRMT5 is required for VEGF-induced interaction of ASK1 and Akt in endothelial cells and PRMT5-mediated methylation of ASK1 at Arg-89 positively regulates its phosphorylation at Ser-83 by Akt.
FIGURE 5:
PRMT5-mediated methylation of ASK1 at Arg-89 promotes the phosphorylation of ASK1 at Ser-83. (A) HEK293T cells were transfected for 48 h with the indicated combination of expression vectors for HA-Akt, FLAG-ASK1, FLAG-ASK1(R89W), and Myc-PRMT5. Cell lysates were examined directly by immunoblot analysis (top and left) or immunoprecipitation (bottom) with the indicated antibodies. (B) EAhy926 cells were transfected for 48 h with the indicated expression vectors for FLAG-ASK1 and FLAG-ASK1(R89W). Cells were then left untreated or treated with 100 ng/ml VEGF for 30 min. Cell lysates were examined directly by immunoblot analysis with the indicated antibodies. (C–D) HUVECs were transfected for 72 h with green fluorescent protein siRNA (CNT) or PRMT5 siRNA. The cells were then left untreated or treated with 100 ng/ml VEGF for 30 min. Cell lysates were examined directly by immunoblot analysis with the indicated antibodies (C) or subjected to immunoprecipitation with antibody to ASK1 (D).
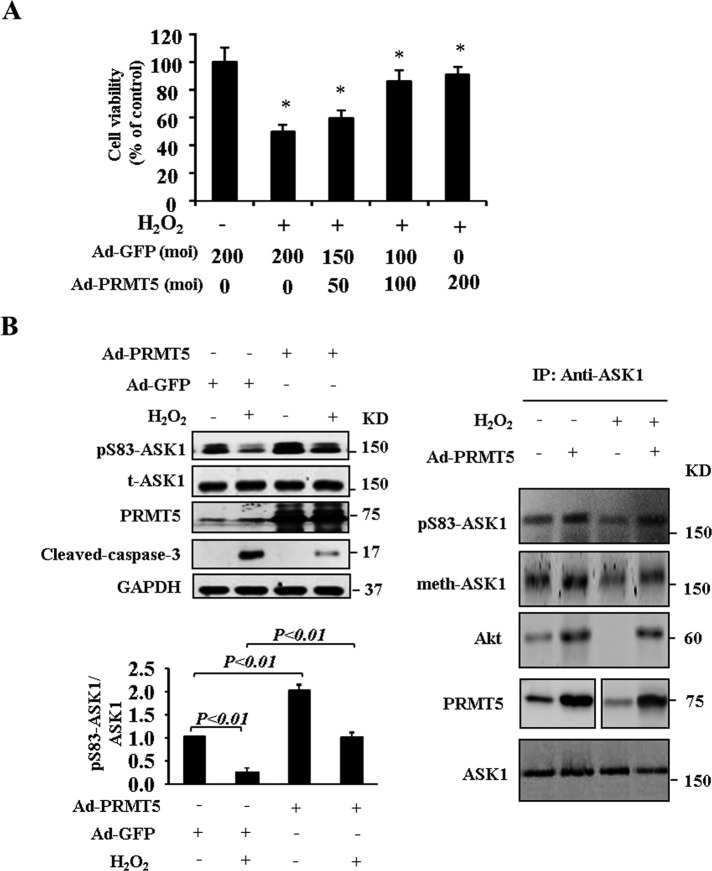
PRMT5 suppresses H2O2-induced activation of ASK1
The ASK1 signaling pathway mediates the induction of apoptosis by oxidants such as H2O2. Deletion of PRMT5 by small interfering RNA (siRNA) increased the activation of ASK1 by inhibiting Ser-83 phosphorylation (Figure 5C). We next examined whether overexpression of PRMT5 attenuates H2O2-induced ASK1 activation in endothelial cells. As shown in Figure 6A, H2O2 treatment resulted in increased HUVEC apoptosis. Moreover, ectopic expression of PRMT5 inhibited H2O2-induced apoptosis in HUVECs. We determine the phosphorylation of ASK1 at Ser-83; the results show that H2O2 treatment inhibits phosphorylation of Ser-83 in ASK1 and activates caspase-3. Overexpression of PRMT5 significantly promotes Ser-83 phosphorylation in both basal and H2O2 treatment conditions, but H2O2-induced ASK1 kinase activity was inhibited. The H2O2-induced activation of caspase-3 was reduced by overexpression of PRMT5 (Figure 6B). We conducted further immunoprecipitation experiments in endothelial cells to study the mechanism(s) of how PRMT5 regulates ASK1 phosphorylation and methylation. We verified that H2O2 treatment decreased the interaction between ASK1 and Akt in coimmunoprecipitation analysis in HUEVCs, and H2O2-induced dissociation was reversed by overexpression of PRMT5 (Figure 6B). The inhibition of arginine methylation (at Arg-89) and phosphorylation (at Ser-83) of Ask1 due to H2O2 stimulus was reversed by overexpression of PRMT5 (Figure 6B), suggesting that arginine methylation of ASK1 and interaction with Akt may be involved in the inhibitory effect of PRMT5 on H2O2-induced apoptotic cell death.
FIGURE 6:
Overexpression of PRMT5 suppresses H2O2-induced activation of ASK1. (A) HUVECs were infected with either Ad-PRMT5 or Ad-GFP for 48 h and then treated with 300 μm H2O2 for 24 h. Cell viability assay (*p < 0.05 compared with control group). (B) Cell lysates obtained from A were subjected to Western blot. Phosphorylation of ASK1 at Ser-83 and cleaved-caspase-3 was determined. Methylation of ASK1 was determined by immunoprecipitation with antibody to ASK1.
DISCUSSION
In the present study, using a proteomics strategy, we successfully identified PRMT5 as an ASK1-binding protein in endothelial cells. The association of PRMT5 and ASK1 was further confirmed in vitro and in vivo. Further investigation indicated that the N-terminal of ASK1 was critical for association with PRMT5. PRMT5 catalyzes the methylation of ASK1 at Arg-89, and this PRMT5-mediated arginine methylation promotes the association between ASK1 and Akt and leads to increased ASK1 phosphorylation at Ser-83.
ASK1 mediated H2O2-induced apoptosis in endothelial cells (Machino et al., 2003; Zhang et al., 2005). ASK1 is activated during the cellular response to ROS and then activates MAPK kinase 4 (MKK4)/MKK7 and MKK3/ MKK6, which in turn activate JNK and p38 kinase, respectively (Wang et al., 1998; Takeda et al., 2003). ASK1 can be regulated by phosphorylation at several sites, and at least seven phosphorylation sites have been identified. For example, ASK1 is regulated by phosphorylation or dephosphorylation at Ser-83, Ser-967, Ser-1034, Tyr-718, and Thr-845 (Kim et al., 2001; Tobiume et al., 2002; Fujii et al., 2004; Gu et al., 2009; Seong et al., 2010). Phosphorylation of Thr-845 is required for ASK1 kinase activity. Of interest, the N-terminal domain of ASK1 surrounding the Ser-83 residue contains the Akt phosphorylation motif RxRxxS/T (Kim et al., 2001). More recently, the proto-oncogene serine/threonine kinase, PIM1, was also shown to interact with and to phosphorylate ASK1 at Ser83 (Gu et al., 2009). PIM1 phosphorylation of ASK1 decreased ASK1 activity and attenuated H2O2-induced ASK1-mediated activation of JNK/p38 and caspase-3. Thus phosphorylation of ASK1 at Ser-83 by Akt or PIM1 maintains ASK1 in an inactive state and suppresses ASK1-mediated p38/JNK downstream signaling.
Arginine methylation occurred in the consensus sequences containing an RxRxxS/T motif has been reported in several Akt target proteins, such as FOXO and BAD (Yamagata et al., 2008; Sakamaki et al., 2011), indicating possible cross-talk between arginine methylation and phosphorylation to regulate Akt target protein’s activity. PRMTs have been reported to methylate ASK1 in cancer cells (Cho et al., 2012). Cho et al. (2012) reported that PRMT1 mediated the methylation of ASK1 at Arg-78 and Arg-80, which potentiated the interaction between ASK1 and thioredoxin. However, PRMT1-mediated arginine methylation had no effect on Akt-mediated phosphorylation of ASK1 at Ser-83. By mass spectrometry, we identified PRMT5, a type II protein arginine methyltransferase that catalyzes the symmetric dimethylation of arginine residues within target proteins, an ASK1-binding protein (Figure 1, A–C). Unlike PRMT1, we found that PRMT5-mediated methylation of ASK1 at Arg-89 potentiated the interaction between ASK1 and Akt (Figures 4 and 5). Indeed, our results show that arginine methylation of ASK1 promoted the binding of ASK1 to Akt in the presence of VEGF (Figure 5D). It is well known that VEGF increases endothelial resistance to H2O2; however, the mechanism is obscure (Liu et al., 2002). VEGF prevented endothelial cell from apoptosis induced by ROS, in which activation of p42/p44 Ca2+-calmodulin–dependent protein kinase, together with partial inhibition of JNK activity, was considered to account for the antiapoptotic effect of VEGF. In the present study, we showed that PRMT5-mediated arginine methylation occurs beyond the RxRxxS/T motif. Of interest, this methylation of arginine is induced by VEGF, and thus positively regulated Akt-mediated ASK1 phosphorylation at Ser-83, indicating cross-talk between arginine methylation and phosphorylation restricted to H2O2-induced ASK1 activity. PRMT5 has been reported to translocate from nucleus to cytoplasm (Chen et al., 2014). The mechanism of how VEGF regulates the interaction of PRMT5 and ASK1, however, is unknown. Thus it would be interesting to investigate whether VEGF regulates the translocation of PRMT5 in endothelial cells.
In summary, H2O2-induced ASK1 activity can be inhibited by PRMT5 methylation of ASK1 at Arg-89, which in turn promotes interaction between ASK1 and Akt and enhances phosphorylation of ASK1 at Ser-83, which inhibits endothelial cells apoptosis. Our data clearly demonstrate cross-talk between arginine methylation and serine phosphorylation in ASK1 and suggest that PRMT5 may be a potential molecular target for developing new antiapoptotic agents.
MATERIALS AND METHODS
Cells and antibody
HEK293T, HUVECs, and EAhy926 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Anti-dimethylation symmetric antibody was purchased from Millipore (Billerica, MA; 07-412-200KL). Anti-ASK1 antibody was purchased from Abcam (Cambridge, MA; ab131506). Other antibodies were purchased from Cell Signaling Technology (Danvers, MA).
Plasmid and site-directed mutagenesis
Site-directed mutagenesis was performed by PCR with the use of a QuikChange Kit (Stratagene, La Jolla, CA). The arginine mutants of ASK1 were constructed with the following mutagenic primers: R31W, 5′-GGCATCTGCUGGAGGGGAGGAGCGGCG-3′; R32W, 5′-GGCATCTGCAGGUGGGGAGGAGCGGCG-3′; R78W, 5′-GAGCAGTGCCACCUGGGGCCGGGGCAGC-3′; R80W, 5′-GCCACCCGAGGCUGGGGCAGCTCTGTTG-3′; R89W, 5′-GGGGGCAGCUGGCGGACCACGGTGGCA-3′; R89K, 5′-GGGGGCAGCAAACGGACCACGGTGGCA-3′; and R90W, 5′-GGGGGCAGCCGAUGGACCACGGTGGCA-3′. Mutant G367A/R368A pCS2-PRMT5 was constructed with the primer 5′-GTGCTGGGAGCAGCAGCGGGACCCCTGGTG-3′ (mismatches with the template are underlined). All mutations were verified by automatic DNA sequencing.
Coimmunoprecipitation assay and Western blotting
The coimmunoprecipitation assay was carried out as described previously (Daitoku et al., 2004). Protein extract (20 μg) was fractionated on SDS–PAGE gels and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membrane was blocked with 1× PBS/0.3% Tween 20 containing 5% dry milk and incubated with primary antibody overnight at 4°C. The immune complexes were detected by chemiluminescence (ECL; Amersham International).
Proteomic analysis
FLAG-tagged ASK1 were overexpressed in EAhy926 cells, after which the cells were treated with 100 ng/ml VEGF for 30 min. Cell lysates were first precleared and then incubated with anti-FLAG antibody. Next protein A/G-agarose beads were added to the cell lysates and incubated for 4 h. The bound proteins were analyzed by SDS–PAGE and Coomassie brilliant blue staining. Gel slices were excised and subjected to in-gel digestion with trypsin. Peptides were extracted from the gel pieces. Sample was resuspended in 23 μl of 50 mM (NH4)2CO3 and analyzed by LC-MS/MS (Finnigan LCQ Deca XP Plus ion trap mass spectrometer). All MS/MS spectra were searched against the NCBInr_20130221 database. Database searching was performed allowing for differential modification on cysteine residues and methionine residues and full tryptic cleavage, with peptide mass tolerance of 1.5 Da and fragment tolerance 0.8 Da.
siRNA transfection
The siRNAs used were, for human PRMT5, 5′-AUCAUCUUUUAGGAAGUGCUGGGCUCC-3′ and 5′-AAUGGUAUAUGAGCGGCCUGUGGGGUU-3′, and for green fluorescent protein, 5′-CUACAACAGCCACAACGUC-3′. Cells were transfected by using Lipofectamine RNAiMAX reagent according to the manufacturer’s instructions (Invitrogen).
In vitro methylation assay
FLAG-ASK1 was incubated with Myc-PRMT5 in the presence of SAM (55 Ci/μmol) at 37°C for 6 h. After washing of the beads, the reaction products were analyzed by fluorography and Coomassie brilliant blue staining.
Adenoviral constructs
Recombinant adenoviral vectors Ad-GFP and Ad-PRMT5 were amplified in 293 cells and purified by cesium chloride gradient. After vector purification, adenoviral vectors were kept at −80°C in 10 mM Tris containing 20% glycerol.
Cell viability assay
Cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide assay as described previously (Cho et al., 2012).
Statistical analyses
Data are expressed as mean ± SE. The statistical significance of differences was assessed by Student’s t test or analysis of variance, as appropriate; p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the Chinese Natural Science Foundation (81260479 to W.S. and 81302415 to Z.Z.), the Guangxi Natural Science Foundation (2014GXNSFAA118259 to W.S.), and the Tianjin Natural Science Foundation (12JCYBJC15800 to Z.Z.).
Abbreviations used:
- ASK1
apoptosis signal–regulating kinase 1
- HUVEC
human umbilical vein endothelial cell
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- PRMT
protein arginine methyltransferase
- ROS
reactive oxygen species
- VEGF
vascular endothelial growth factor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0738) on February 24, 2016.
REFERENCES
- Bunkoczi G, Salah E, Filippakopoulos P, Fedorov O, Muller S, Sobott F, Parker SA, Zhang H, Min W, Turk BE, et al. Structural and functional characterization of the human protein kinase ASK1. Structure. 2007;15:1215–1226. doi: 10.1016/j.str.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Chen M, Yi B, Sun J. Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J Biol Chem. 2014;289:24325–24335. doi: 10.1074/jbc.M114.577494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Lee MK, Yoon KW, Lee J, Cho SG, Choi EJ. Arginine methylation-dependent regulation of ASK1 signaling by PRMT1. Cell Death Differ. 2012;19:859–870. doi: 10.1038/cdd.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Cai SH, Shi Z, Nagase F. Binding activity of H-Ras is necessary for in vivo inhibition of ASK1 activity. Cell Res. 2004;14:148–154. doi: 10.1038/sj.cr.7290214. [DOI] [PubMed] [Google Scholar]
- Fisk JC, Read LK. Protein arginine methylation in parasitic protozoa. Eukaryotic Cell. 2011;10:1013–1022. doi: 10.1128/EC.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Goldman EH, Park HR, Zhang L, Chen J, Fu H. Negative control of apoptosis signal-regulating kinase 1 through phosphorylation of Ser-1034. Oncogene. 2004;23:5099–5104. doi: 10.1038/sj.onc.1207668. [DOI] [PubMed] [Google Scholar]
- Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27:8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009;28:4261–4271. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SJ, Browne GJ, Miller BW, Prigent SA, Dickens M. Activation of ASK1, downstream MAPKK and MAPK isoforms during cardiac ischaemia. Biochim Biophys Acta. 2010;1802:733–740. doi: 10.1016/j.bbadis.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Matsuzawa A, Noguchi T, Takeda K, Ichijo H. The ASK1-MAP kinase pathways in immune and stress responses. Microbes Infect. 2006;8:1098–1107. doi: 10.1016/j.micinf.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wu L, Wu J, Li Y, Hou L. Cellular FLICE-like inhibitory protein protects against cardiac hypertrophy by blocking ASK1/p38 signaling in mice. Mol Cell Biochem. 2014;397:87–95. doi: 10.1007/s11010-014-2175-3. [DOI] [PubMed] [Google Scholar]
- Hyun MS, Hur JM, Mun YJ, Kim D, Woo WH. BBR induces apoptosis in HepG2 cell through an Akt-ASK1-ROS-p38MAPKs-linked cascade. J Cell Biochem. 2010;109:329–338. doi: 10.1002/jcb.22384. [DOI] [PubMed] [Google Scholar]
- Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- Kadowaki H, Nishitoh H, Urano F, Sadamitsu C, Matsuzawa A, Takeda K, Masutani H, Yodoi J, Urano Y, Nagano T, et al. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 2005;12:19–24. doi: 10.1038/sj.cdd.4401528. [DOI] [PubMed] [Google Scholar]
- Kim AH, Khursigara G, Sun X, Franke TF, Chao MV. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol Cell Biol. 2001;21:893–901. doi: 10.1128/MCB.21.3.893-901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Shim JH, Chun E, Lee KY. Reciprocal inhibition between the transforming growth factor-beta-activated kinase 1 (TAK1) and apoptosis signal-regulating kinase 1 (ASK1) mitogen-activated protein kinase kinase kinases and its suppression by TAK1-binding protein 2 (TAB2), an adapter protein for TAK1. J Biol Chem. 2012;287:3381–3391. doi: 10.1074/jbc.M111.317875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek D, Kylarova S, Psenakova K, Rezabkova L, Herman P, Vecer J, Obsilova V, Obsil T. Biophysical and structural characterization of the thioredoxin-binding domain of protein kinase ASK1 and its interaction with reduced thioredoxin. J Biol Chem. 2014;289:24463–24474. doi: 10.1074/jbc.M114.583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WL, Guo X, Chen QQ, Guo ZG. VEGF protects bovine aortic endothelial cells from TNF-alpha- and H2O2-induced apoptosis via co-modulatory effects on p38-and p42/p44-CCDPK signaling. Acta Pharmacol Sin. 2002;23:45–49. [PubMed] [Google Scholar]
- Lu X, Masic A, Li Y, Shin Y, Liu Q, Zhou Y. The PI3K/Akt pathway inhibits influenza A virus-induced Bax-mediated apoptosis by negatively regulating the JNK pathway via ASK1. J Gen Virol. 2010;91:1439–1449. doi: 10.1099/vir.0.018465-0. [DOI] [PubMed] [Google Scholar]
- Machino T, Hashimoto S, Maruoka S, Gon Y, Hayashi S, Mizumura K, Nishitoh H, Ichijo H, Horie T. Apoptosis signal-regulating kinase 1-mediated signaling pathway regulates hydrogen peroxide-induced apoptosis in human pulmonary vascular endothelial cells. Crit Care Med. 2003;31:2776–2781. doi: 10.1097/01.CCM.0000098027.49562.29. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Saegusa K, Noguchi T, Sadamitsu C, Nishitoh H, Nagai S, Koyasu S, Matsumoto K, Takeda K, Ichijo H. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6:587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- Nako H, Kataoka K, Koibuchi N, Dong YF, Toyama K, Yamamoto E, Yasuda O, Ichijo H, Ogawa H, Kim-Mitsuyama S. Novel mechanism of angiotensin II-induced cardiac injury in hypertensive rats: the critical role of ASK1 and VEGF. Hypertens Res. 2012;35:194–200. doi: 10.1038/hr.2011.175. [DOI] [PubMed] [Google Scholar]
- Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci USA. 2011;108:6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong HA, Jung H, Ichijo H, Ha H. Reciprocal negative regulation of PDK1 and ASK1 signaling by direct interaction and phosphorylation. J Biol Chem. 2010;285:2397–2414. doi: 10.1074/jbc.M109.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga M, Matsuzawa A, Ichijo H. Oxidative stress-induced diseases via the ASK1 signaling pathway. Int J Cell Biol. 2012;2012:439587. doi: 10.1155/2012/439587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Matsuzawa A, Nishitoh H, Ichijo H. Roles of MAPKKK ASK1 in stress-induced cell death. Cell Struct Funct. 2003;28:23–29. doi: 10.1247/csf.28.23. [DOI] [PubMed] [Google Scholar]
- Tao Q, Spring SC, Terman BI. Comparison of the signaling mechanisms by which VEGF, H2O2, and phosphatase inhibitors activate endothelial cell ERK1/2 MAP-kinase. Microvasc Res. 2005;69:36–44. doi: 10.1016/j.mvr.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J Cell Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- Tzeng HE, Tsai CH, Chang ZL, Su CM, Wang SW, Hwang WL, Tang CH. Interleukin-6 induces vascular endothelial growth factor expression and promotes angiogenesis through apoptosis signal-regulating kinase 1 in human osteosarcoma. Biochem Pharmacol. 2013;85:531–540. doi: 10.1016/j.bcp.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Wang XS, Diener K, Tan TH, Yao Z. MAPKKK6, a novel mitogen-activated protein kinase kinase kinase, that associates with MAPKKK5. Biochem Biophys Res Commun. 1998;253:33–37. doi: 10.1006/bbrc.1998.9749. [DOI] [PubMed] [Google Scholar]
- Xie D, Gore C, Zhou J, Pong RC, Zhang H, Yu L, Vessella RL, Min W, Hsieh JT. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc Natl Acad Sci USA. 2009;106:19878–19883. doi: 10.1073/pnas.0908458106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Yokoi T, Fukuo K, Yasuda O, Hotta M, Miyazaki J, Takemura Y, Kawamoto H, Ichijo H, Ogihara T. Apoptosis signal-regulating kinase 1 mediates cellular senescence induced by high glucose in endothelial cells. Diabetes. 2006;55:1660–1665. doi: 10.2337/db05-1607. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zheng S, Storz P, Min W. Protein kinase D specifically mediates apoptosis signal-regulating kinase 1-JNK signaling induced by H2O2 but not tumor necrosis factor. J Biol Chem. 2005;280:19036–19044. doi: 10.1074/jbc.M414674200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.