Abstract
Objectives
Converging evidence suggests that the brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism affects brain structure. Yet the majority of studies have shown no effect of this polymorphism on hippocampal volumes, perhaps due to small effect size.
Methods
We performed a meta-analysis of studies investigating the association between Val66Met BDNF polymorphism and hippocampal volumes in healthy subjects by combining standardized differences between means (SDM) from individual studies using random effect models.
Results
Data from 399 healthy subjects (255 Val-BDNF homozygotes and 144 carriers of at least one Met-BDNF allele) in seven studies were meta-analysed. Both the left and right hippocampi were significantly larger in Val-BDNF homozygotes than in carriers of at least one Met-BDNF allele (SDM = 0.41, 95% Confidence Interval = 0.20; 0.62, z = 3.86, P = 0.0001; SDM = 0.41; 95% Confidence Interval = 0.20; 0.61, z = 3.81, P = 0.0001, respectively), with no evidence of publication bias.
Conclusions
Healthy carriers of BDNF gene Val66Met polymorphism show bilateral hippocampal volume reduction. The effect size was small, but the same direction of effect was seen in all meta-analyzed studies. The association with the BDNF gene Val66Met polymorphism makes hippocampal volume a potential candidate for an endophenotype of disorders presenting with reduced hippocampal volumes.
Keywords: Brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism, hippocampal volume, magnetic resonance imaging, meta-analysis, endophenotype
Introduction
Brain-derived neurotrophic factor (BDNF) belongs to the family of neurotrophins. Early in life it plays an important role in neurogenesis and neurodevelopment (Balu and Lucki 2009; Cohen-Cory et al. 2010). Later in life it facilitates neuronal plasticity and exerts neuroprotective effects (Yoshii and Constantine-Paton 2010). The BDNF gene is situated on chromosome 11. A common functional missense single nucleotide polymorphism located at nucleotide 196 produces a valine to methionine amino acid substitution in the region controlling dendritic trafficking and synaptic localization of BDNF. This leads to decreased interaction of BDNF with a sorting protein (sortilin), which attenuates activity related release of BDNF and results in decreased hippocampal volumes in animal models (Egan et al. 2003; Chen et al. 2006; Martinowich et al. 2007). In keeping with this, human studies have shown the effects of this polymorphism on memory (Egan et al. 2003; Hariri et al. 2003), hippocampal activation (Egan et al. 2003; Hariri et al. 2003), as well as hippocampal volumes in some studies (Bueller et al. 2006; Frodl et al. 2007).
The majority of studies investigating the effects of BDNF gene Val66Met polymorphism on hippocampal volume in human subjects have, however, been negative. This may be due to type II errors (false negative findings) caused by the modest to small effects of individual genetic polymorphisms on brain structure. Performing larger studies is one, albeit difficult solution to the problem. In the absence of such large studies, meta-analyzing data from multiple smaller investigations is another method of addressing problems with small sample and effect sizes.
Hippocampal volume decrease is a replicated feature of a number of neuropsychiatric disorders, including mood disorders, schizophrenia, Alzheimer’s dementia, post traumatic stress disorder, personality disorders (Hoschl and Hajek 2001; Geuze et al. 2005). Identifying a genetic basis for hippocampal volume changes would thus be of marked interest to neuropsychiatry. The effects of genetic polymorphisms on brain structure are however initially best studied among healthy subjects. Studying such effects in patients with major psychiatric disorders is confounded by the effects of medications, comorbid conditions and illness burden on brain structure (Hoschl and Hajek 2001). Furthermore, other genetic factors such as epistasis (Takahashi et al. 2008a), gene–environment interactions (Uher 2008) or effects of medications on gene expression (Duman et al. 1997) may further complicate genetic neuroimaging studies of clinical populations. Establishing an association between a particular genetic polymorhism and brain structure in healthy subjects and obtaining effect size estimates would help to design studies testing this association in psychiatric disorders, where additional confounding factors need to be controlled for.
We thus performed a meta-analysis of hippocampal volumes in healthy subjects genotyped for the BDNF Val66Met polymorphism, in order to achieve a greater statistical power and a more precise estimate of effect size relative to individual studies. We expected to find smaller hippocampal volumes among Met carriers relative to Val homozygotes.
Methods
Study ascertainment
Studies were considered for inclusion if they (1) were indexed in Medline as published or in press in peer-reviewed journals by October 31, 2010 (2) investigated brain structure in healthy subjects genotyped for BDNF gene Val66Met polymorphism, and (3) reported manual volumetric measurements of the left or the right hippocampus. We analyzed the left and the right hippocampus separately, as most of the available studies did not provide mean and variance estimates for the whole hippocampal volume. When a study reported means and standard deviations of a structure adjusted for confounds, these were used in the meta-analysis in place of the raw means.
Studies were excluded, if (1) only overall hippocampal volumes rather than separate results for the left and the right side were provided, (2) gray matter density (voxel based morphometry), rather than actual volumes were reported, (3) noncontiguous slices were used for the measurements, (4) the effect size for hippocampal volume changes could not be extracted from the manuscript, (5) the study investigated a mixture of healthy subjects and patients with psychiatric/neurological conditions without providing separate results for the healthy controls. Two reviewers (TH, MK) assessed each study to ensure that all inclusion and exclusion criteria were met and all data were transcribed correctly.
We carried out a systematic search of MEDLINE, EMBASE, SCOPUS databases for articles published between 1999 (the first report about the effects of BDNF on brain structure (Wassink et al. 1999)) and October 31, 2010 in any language using the following Medical Subjects Heading categories: magnetic resonance imaging and BDNF. This is an over inclusive search strategy which, however, ensures identification of all available studies. Review articles relating to genetic neuroimaging and reference lists of included studies were also searched for published articles.
Statistical analyses
Meta-analyses were performed using Comprehensive Meta Analysis software, Version 2. As a measure of effect size, we used standardized difference between means (SDM), which is identical to Cohen’s d measure of effect size. Since we cannot expect constant population effect size across studies (fixed effects), we decided to use the random effects model, with study as the random effect. This assumes that the “population” of studies has variable true effects that are normally distributed.
No studies have reported hippocampal volumes separately for Met-BDNF homozygotes, as these are rare. Therefore similar to individual studies, we grouped subjects as either Val-homozygotes, or Met-carriers (a combined group of Met-BDNF homozygotes and Met-BDNF heterozygotes).
We calculated I2, to provide an easily interpretable measure of consistency between studies. The I2 is an estimate of the percentage of the total variation across studies due to true heterogeneity rather than chance. I2 is placed between 0 and 100%. A value of 0% indicates no observed heterogeneity, and larger values show increasing heterogeneity (Higgins et al. 2003). I2 values of 25, 50 and 75% denote low, moderate and large heterogeneity.
We performed a jack-knife sensitivity analysis, omitting one study at a time, to assess whether the results would change after exclusion of any study and whether they remain significant in replication studies, e.g., after exclusion of the first published positive study.
We used the Egger’s regression test of funnel plot asymmetry (Egger et al. 1997), as well as the classic fail-safe N to examine the risk of publication bias. To test for association between age, proportion of females, publication year and SDM of hippocampal volumes, we performed metaregression, using non-parametric rank correlation (Kendall Tau). We adopted a significance level of p = 0.05, two tailed for all of the above-mentioned analyses.
Departures from Hardy–Weinberg equilibrium (HWE) in studies which did not provide this information directly were tested using chi-square test.
Results
Results of the systematic search
Out of 126 studies initially found by the systematic search, 14 studies measured hippocampal volumes in healthy subjects genotyped for BDNF gene Val66Met polymorphism (Szeszko et al. 2005; Agartz et al. 2006; Bueller et al. 2006; Frodl et al. 2007; Takahashi et al. 2008b; Chepenik et al. 2009; Dutt et al. 2009; Jessen et al. 2009; Joffe et al. 2009; Koolschijn et al. 2010; Benjamin et al. 2010; Gonul et al. 2011; Haukvik et al. 2010; Karnik et al. 2010). The study by (Haukvik et al. 2010) overlapped with the (Agartz et al. 2006) study, thus yielding 13 non-overlaping datasets. The studies by Joffe et al. (2009) and Karnik et al. (2010) did not use manual tracing, but rather automated segmentation and registration based procedures for hippocampal volumetry, thus failing inclusion criterion 3. Manual tracing continues to be the gold standard for hippocampal volumetry. Directly comparing results obtained with manual volumetry and automated segmentation and registration-based procedures, such as automated anatomical labelling is not appropriate. The automated methods typically yield means and variance estimates for hippocampal volumes which significantly differ from those obtained by manual volumetry as well as show low intra-class correlations with manual volumetry (Tae et al. 2008). These studies also provided total hippocampal volumes only (exclusion criterion number 1). There was no association between hippocampal volume and Val-66Met BDNF polymorphism in either of these studies. Two other volumetric studies measured the whole hippocampal volume only, without providing separate results for the left and the right hippocampus (Agartz et al. 2006; Chepenik et al. 2009). Only one of these studies provided sufficient details for calculation of effect sizes (Chepenik et al. 2009), thus not allowing for a separate meta-analysis. This study showed increased total hippocampal volume in Val-BDNF homozygotes relative to Met-BDNF carriers. The remaining study showed no association between Val66Met BDNF polymorphism and hippocampal volume but did not provide sufficient details to extract the direction and extent of the effect sizes (Agartz et al. 2006). Two additional volumetric studies provided data only on a mixture of healthy controls and patients without separating the results (Szeszko et al. 2005; Benjamin et al. 2010). Seven of the studies thus met inclusion criteria for this meta-analysis (Bueller et al. 2006; Frodl et al. 2007; Takahashi et al. 2008b; Dutt et al. 2009; Jessen et al. 2009; Koolschijn et al. 2010; Gonul et al. 2011). The study by (Dutt et al. 2009) included four subjects with personal history of depression. Since this represented a minority among the 61 recruited control subjects, we included this study in the meta-analysis. All of the other meta-analyzed studies excluded subjects with a personal history of Axis I psychiatric disorders. Four of the meta-analyzed studies excluded subjects with family history of psychiatric Axis I disorders. All meta-analyzed studies included both men and women. The average age of healthy controls ranged from 24 ± 6 to 44 ± 9 years. Due to small number of Met-BDNF homozygotes, no study separated Met-BDNF homozygotes from Met-BDNF heterozygotes. In all studies, results for carriers of at least one Met-BDNF allele – a combined group of Met-BDNF homozygotes and heterozygotes – were thus provided. In six out of seven studies included in the meta-analysis the BDNF Val66Met genotype distributions were in Hardy–Weinberg equilibrium (results directly provided: Frodl et al. 2007; Takahashi et al. 2008b; Dutt et al. 2009; Koolschijn et al. 2010), results calculated from genotype frequencies (Bueller et al. 2006; Gonul et al. 2011). A single study did not provide information on HWE or sufficient details to calculate it (Jessen et al. 2009). Slice thickness in the meta-analyzed studies ranged from 1 to 2 mm, with magnet field strength of 1.5 T in all studies with exception of 3 T magnet in Jessen et al. (2009), please see Table I for details.
Table 1.
Summary of reviewed studies.
| Study | N Val/Val; Met/Val; Met/Met |
Magnet strength (Tesla) |
Slice thickness (mm) |
Age (mean ± SD) |
Females/ Males |
Interrater reliability for tracings |
Blinding | Results for hippocampus (positive study - at least one statistically significant difference) |
Included in meta-analysis; reason why not |
Exclusion of family history of Axis I psychiatric disorders |
|---|---|---|---|---|---|---|---|---|---|---|
| Takahashi et al. 2008b | 13; 11; 5 | 1.5 | 1.0 | 24.2 ± 6.1 | 12/17 | 0.92 | Yes | Negative | Yes | Yes |
| Agartz et al. 2006 | 73; 27; 4 | 1.5 | 1.5 | 41.6 ± 8.9 | 35/69 | Not mentioned | Yes | Negative | No; volumes of both hippocampi only | No |
| Chepenik et al. 2009 | 12; 6; 0 | 1.5 | 1.2 | range 18–58 | 12/6 | 0.90–0.92 | Yes | Positive; Val/Val > Met + | No; volumes of both hippocampi only | No |
| Frodl et al. 2007 | 40; 19; 1 | 1.5 | 1.5 resampled to 1.0 | 41.6 ± 12.3 | 29/31 | 0.97 | Yes | Positive; Val/Val > Met + | Yes | Yes |
| Bueller et al. 2006 | 21; 15; 0 | 1.5 | 1.5 | 27 ± 19 | 22/14 | no | Yes | Positive; Val/Val > Met + | Yes | Yes |
| Koolschijn et al. 2010 | 59; 26; 5 | 1.5 | 1.2 | 38.2 ± 13.6 | 34/56 | 0.87–0.96 | Not mentioned | Negative | Yes | Yes |
| Szeszko et al. 2005 | 15; 10; 0 | 1.5 | 1.5 | 27.1 ± 6.7 | 15/10 | 0.87–0.94 | Yes | Positive; Val/Val > Met + when controls combined with FE schizophrenia, negative in controls | No; mix of patients with schizophrenia and healthy volunteers | No |
| Benjamin et al. 2010 | 67; 21 Met carriers | 1.5 | 3.0 | 70.13 ± 5.6 | 62/26 | 0.7–0.8 | Not mentioned | Negative | No; mix of patients with geriatric depression and healthy volunteers | No |
| Dutt et al. 2009 | 44 (hippocampal volumes provided for 43 subjects); 17 Met carriers | 1.5 | 1.5 | 40.8 ± 15.1 | 33/28 | 0.86–0.87 | Yes | Negative | Yes | No |
| Joffe et al. 2009 | 68; 43; 2 | 1.5 | 1.0 | not mentioned for MRI subgroup | not mentioned for MRI subgroup | Automated Anatomical Labeling | Not mentioned | Negative | No; volumes of both hippocampi only, not manual volumetry | Yes |
| Jessen et al. 2009 | 55; 29 Met carriers | 1.5 for 30 subjects, 3.0 for 54 subjects | 1.0 | 43.9 ± 8.7 | 40/44 | 0.97 | Not mentioned | Negative | Yes | No |
| Karnik et al. 2010 | 87; 41; 1 | 1.5 | 1.0 | 51.3 ± 25.8 (Val/Val); 45.2 ± 23.9 (Met + group) | 70/59 | semi-automated procedure | Not mentioned | Negative | No; volumes of both hippocampi only, not manual volumetry | Excluded first-degre relative with psychosis in 88 subjects) |
| Gonul et al. 2011 | 24; 16; 0 | 1.5 | 2.0 | 30.8 ± 7.2 (Val/Val group) 28.2 ± 4.5 (Met + group) | 23/17 | 0.95–0.96 | Yes | Negative | Yes | Yes |
FE, first episode; Met+, carriers of at least one Met allele of the BDNF gene Val66Met polymorphism; MRI, magnetic resonance imaging.
Results of the meta-analysis
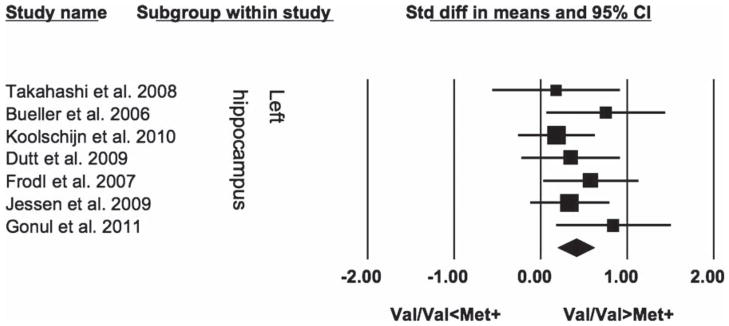
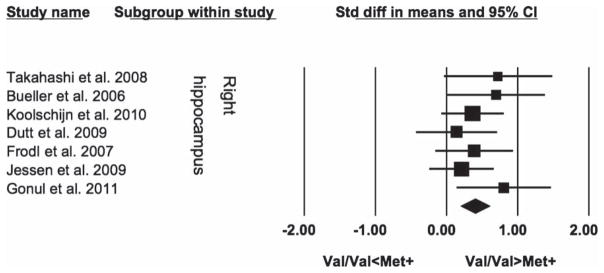
Overall we meta-analyzed data from 399 healthy subjects (255 Val-BDNF homozygotes and 144 carriers of at least one Met-BDNF allele). Both the left and right hippocampi were significantly larger in Val-BDNF homozygotes than in carriers of at least one Met-BDNF allele (SDM = 0.41, SE = 0.11, 95% CI = 0.20; 0.62, z = 3.86, P = 0.0001; SDM = 0.41; SE = 0.11, 95% CI = 0.20; 0.61, z = 3.81, P = 0.0001 respectively), for details see Figures 1 and 2.
Figure 1.
Comparison of the left hippocampal volumes between healthy Val homozygotes and carriers of BDNF gene Val66Met polymorphism.
Figure 2.
Comparison of the right hippocampal volumes between healthy Val homozygotes and carriers of BDNF gene Val66Met polymorphism.
Interestingly all studies, even the negative ones, for both the left and right hippocampi showed the same direction of changes (i.e. Val-BDNF homozygotes having larger hippocampi than Met-BDNF carriers). There was no statistical heterogeneity among the studies for the left (I2 = 0), or the right hippocampus (I2 = 0%).
Sensitivity analysis
The results for either the left or the right hippocampus remained significant after exclusion of any individual study, including the first study (Bueller et al. 2006), the study with the largest effect (Bueller et al. 2006) or sample size (Koolschijn et al. 2010) or the study of Japanese subjects (Takahashi et al. 2008b). Exclusion of individual studies yielded effect sizes ranging from 0.36 to 0.48 for the left and 0.36 to 0.45 for the right hippocampus.
Meta-regression
There was no statistically significant association between age, proportion of females, publication year (although the first study showed the largest effect size) and SDM for either the left or the right hippocampus. Only five studies included in the meta-analysis separately listed the proportion of Met-BDNF homozygotes and three of these contained only Met heterozygotes. We thus did not perform meta-regression and only visually inspected the results. The smallest effect size was found in the study with the largest proportion of Met-BDNF homozygotes (Takahashi et al. 2008b) and the largest effects size was seen in the study with the smallest proportion of Met-BDNF homozygotes (Gonul et al. 2011).
Publication bias
There was no evidence for publication bias as tested by the visual inspection of funnel plots and by Egger’s regression intercept. The lack of publication bias was also evidenced by the fact that the majority of included studies were negative. The number of additional studies needed to bring the P value above 0.05 was 23 for both the left and right hippocampi.
Sample size calculations
A study would need to recruit 95 subjects per group to detect an effect size of 0.41 as seen in this meta-analysis for the differences in hippocampal volumes between Val-BDNF homozygotes and carriers of at least one Met-BDNF allele as statistically significant (P = 0.05, power = 80%, two-tailed test).
Discussion
In this meta-analysis, we found significantly larger bilateral hippocampal volumes in healthy Val homozygotes relative to carriers of at least one Met allele of the BDNF gene Val66Met polymorphism. Contrary to the results of this meta-analysis 11 of the 13 individual studies reported no association between hippocampal volumes and BDNF Val66Met polymorphism in healthy subjects. Biological, as well as statistical issues could underlie this discrepancy.
Regarding the biological heterogeneity, the prevalence of BDNF polymorphisms differs between Asian and Caucasian samples which, however, does not mean that the effects of these alleles differ as well. Excluding the single study investigating Japanese subjects (Takahashi et al. 2008b) did not change the results. Furthermore, the direction of the effect in this study was the same as in all of the other meta-analyzed studies, i.e. larger hippocampal volumes in Val homozygotes relative to Met carriers. Gene–environment interactions, as well as epistasis are likely to play a role in determining regional brain morphometry. At this time it is impossible to address all of these effects as sufficient data are missing.
A more parsimonious explanation for the discrepancy between the meta-analysis and the individual studies seems to be the lack of statistical power due to a small sample size (false negative finding, type II error). The overall effect size for differences between Val-BDNF homozygotes and Met-BDNF allele carriers was 0.41, thus requiring 95 subjects per group to detect. The individual studies, the largest of which recruited a total of 90 healthy subjects (Koolschijn et al. 2010), were thus underpowered to detect the association as statistically significant. Further in support of this, all studies included in the meta-analysis reported numerically larger left as well as right hippocampi in Val-BDNF homozygotes relative to carriers of the Met-BDNF allele, thus yielding no statistical heterogeneity. Furthermore, overall statistical significance of the meta-analysis was retained after exclusion of each individual study.
The consistency of findings is noteworthy. It has previously been shown that individual association studies are liable to type I errors and that these can be propagated by use of loose definitions of replication (Sullivan 2007). In this respect, the meta-analyzed studies approach criteria for the strict definition of replication, i.e. looked at the same SNP, the same phenotype, found association with the same allele and reported the same direction of findings, albeit non-significant due to a small sample size.
The smaller hippocampal volumes in the Val66Met BDNF polymorphism detected in this meta-analysis have been also found in some (Pezawas et al. 2004; Matsuo et al. 2009; Schofield et al. 2009), but not other (Ho et al. 2006; Nemoto et al. 2006; Joffe et al. 2009; Montag et al. 2009) studies using voxel-based morphometry. Converging lines of evidence from basic science as well as clinical studies further corroborate the effects of Val66Met BDNF polymorphism on hippocampal volumes. The dentate gyrus of hippocampus is one of the regions of the brain where neuronal proliferation is evident even in adults (Lledo et al. 2006). Stress paradigms which decrease the levels of BDNF also lead to hippocampal volume decreases in animal models. Conversely increasing the levels of BDNF, by for example antidepressants, results in hippocampal volume increases (Groves 2007). Last but not least, studies in human subjects have shown the effects of BDNF gene Val66Met polymorphism on decreased memory performance, which is a hippocampal mediated task (Egan et al. 2003; Hariri et al. 2003).
The effect of the BDNF gene Val66Met polymorphism on hippocampal volume has implications for psychiatric disorders associated with decreased hippocampal volumes, including mood disorders, schizophrenia, Alzheimer’s dementia, post traumatic stress disorder, personality disorders (Hoschl and Hajek 2001; Geuze et al. 2005). A complex scenario emerges in which the hippocampus may be a target for genetic effects (Pezawas et al. 2008), disrupting the development of this critical structure, as well as for environmental insults which can be further aggravated by the presence of the Val66Met BDNF polymorphism (Ho et al. 2007; McIntosh et al. 2007). A single gene may thus be implicated in both hippocampal volume changes presenting as a risk factor for psychosis (Prasad and Keshavan 2008), but also in hippocampal volume decline in response to illness burden (Hoschl and Hajek 2001; Hajek et al. 2005; Macqueen et al. 2005).
The cumulative effect size from this meta-analysis has practical implications for future genetic neuroimaging studies. For a sufficiently powered study of association between the BDNF gene Val66Met polymorphism and hippocampal volume, 95 subjects per group would be needed. This is in keeping with the concept of endophenotype (Gottesman and Gould 2003). Rather than requiring thousands or tens of thousands of subjects to test for associations between genetic polymorphisms and complex behavioral syndromes, 1 to 2 orders of magnitude smaller sample sizes may be required to test for the association between a particular allele, such as the Val66Met BDNF polymorphism and an intermediate phenotype, such as hippocampal volume.
This estimate applies to healthy subjects and may vary in clinical samples. Chepenik et al. (2009) have shown, that the effects of the BDNF gene Val66Met polymorphism on brain structure may be enhanced in patients with bipolar disorders. On the other hand, recruiting psychiatric patients might introduce additional heterogeneity and thus require a larger sample size.
It is also of note that a non-quantitative review of the effects of the BDNF gene Val66Met polymorphism on brain structure would have to conclude that the majority of studies showed no association. The quantitative meta-analytical approach thus, in this case, provides information not readily evident from the individual studies or from non-quantitative reviews.
Our study has several limitations. Every meta-analysis depends on the quality of the primary data, comparability of methods and control for known confounding variables. The methodological comparability with regards to MRI methods was good. All studies used at least 1.5-T scanners, maximum 2 mm slice thickness and 3D acquisitions. These methods are optimal and sufficient for volumetry of the hippocampus. Exclusion of the only study which used a 3-T scanner (Jessen et al. 2009) or 2 mm slice thickness (Gonul et al. 2011) in the sensitivity analysis did not change the results. Furthermore it has been shown that slice thickness does not affect hippocampal volumes (Campbell et al. 2004). In keeping with this we, as well as others have previously combined data obtained with a range of slice thicknesses for meta-analytical purposes (Hajek et al. 2008, 2009; Kempton et al. 2008). Five out of seven volumetric analyses were blinded to the genotype of the participants. When reported, the inter-rater reliability estimates were sufficiently high. With seven studies in 399 subjects we were above the cut-off of three studies in at least 50 subjects set up in previous meta-analysis (McDonald et al. 2004). Meta-analytical techniques could be misled by preferential publications of positive findings. There was no evidence for publication bias in the reviewed studies and, in fact, most of the meta-analyzed studies reported nominally non-significant results. Six of the identified studies did not meet inclusion criteria for this meta-analysis. Only one of these studies reported a statistically significant difference between Val-BDNF homozygotes and Met-BDNF carriers. In keeping with the overall results of our meta-analysis, the Val-homozygotes had larger hippocampal volumes than Met-BDNF carriers (Chepenik et al. 2009). None of the other studies showed statistically significant association between Val66Met BDNF polymorphism and total hippocampal volume. One of these studies showed numerically larger (Szeszko et al. 2005), whereas two studies reported non-significantly smaller hippocampal volumes (Joffe et al. 2009; Benjamin et al. 2010) in Val homozygotes relative to Met-BDNF carriers. Two studies did not provide details about the direction of the non-significant effect (Agartz et al. 2006; Karnik et al. 2010). Based on our analyses, the number of additional studies needed to bring the p value above 0.05 was 23 for both the left and right hippocampi. Therefore, the four studies with either opposite or unknown statistically non-significant effect direction would be unlikely to change the results. We included only healthy subjects, as the complex nature of psychiatric syndromes with oligo or polygenic underpinnings and frequent gene environment and epistatic effects, as well as other factors affecting brain structure (burden of illness, medication, comorbid conditions) would introduce noise into the meta-analysis, which would be impossible to account for. Clarifying the association between regional brain volumes and individual genes is thus a necessary first step before more complex interactions can be studied. We were unable to formally test whether Met-BDNF homozygotes differed from Met-BDNF heterozygotes (dose–response effect). Met allele is rare in western populations which were studied in most of the included studies. Only five studies provided proportion of Met-BDNF homozygotes, two of which contained no met homozygotes. Furthermore, no studies reported hippocampal volumes separately for Met homozygotes. Visual inspection of the data, however, did not provide support for the dose response effect as in fact the largest effects size was seen in the study with the smallest proportion of Met-BDNF homozygotes. The studies did not provide results separately for males and females. We thus at least looked at association between proportion of females and effect sizes using a meta-regression. There was no statistically significant association between proportion of females and SDM for either the left or the right hippocampus.
Overall, the available evidence suggests that there are bilateral hippocampal volume reductions in healthy carriers of the BDNF gene Val66Met polymorphism. The effect size of this difference is small (ES = 0.41), but the same direction of findings was seen in all studies included in the meta-analysis, regardless of statistical significance. Association with the BDNF gene Val66Met polymorphism makes hippocampal volume a potential candidate for an endophenotype of disorders presenting with hippocampal volume decreases. Because of the small effect size, a sample size of at least 95 subjects per group would ensure sufficient power to further investigate the effects of BDNF gene on brain structure.
Acknowledgments
This study was supported by funding from Canadian Institutes of Health Research, Nova Scotia Health Research Foundation, Ministry of Health of the Czech Republic (grant MZ0PCP2005) and Ministry of Education, Youths and Sports of the Czech Republic (grant MSM0021620816).
Footnotes
Statement of Interest
None of the authors has any conflict of interest to declare.
References
- Agartz I, Sedvall GC, Terenius L, Kulle B, Frigessi A, Hall H, et al. BDNF gene variants and brain morphology in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:513–523. doi: 10.1002/ajmg.b.30338. [DOI] [PubMed] [Google Scholar]
- Balu DT, Lucki I. Adult hippocampal neurogenesis: regulation, functional implications, and contribution to disease pathology. Neurosci Biobehav Rev. 2009;33:232–252. doi: 10.1016/j.neubiorev.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin S, McQuoid DR, Potter GG, Payne ME, MacFall JR, Steffens DC, et al. The brain-derived neurotrophic factor Val66Met polymorphism, hippocampal volume, and cognitive function in geriatric depression. Am J Geriatr Psychiatry. 2010;18:323–331. doi: 10.1097/JGP.0b013e3181cabd2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Campbell S, Marriott M, Nahmias C, MacQueen GM. Lower hippocampal volume in patients suffering from depression: a meta-analysis. Am J Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepenik LG, Fredericks C, Papademetris X, Spencer L, Lacadie C, Wang F, et al. Effects of the brain-derived neurotrophic growth factor val66met variation on hippocampus morphology in bipolar disorder. Neuropsychopharmacology. 2009;34:944–951. doi: 10.1038/npp.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Cory S, Kidane AH, Shirkey NJ, Marshak S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Dev Neurobiol. 2010;70:271–288. doi: 10.1002/dneu.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Heninger GR, Nestler EJ. A molecular and cellular theory of depression. Arch Gen Psychiatry. 1997;54:597–606. doi: 10.1001/archpsyc.1997.01830190015002. [DOI] [PubMed] [Google Scholar]
- Dutt A, McDonald C, Dempster E, Prata D, Shaikh M, Williams I, et al. The effect of COMT, BDNF, 5-HTT, NRG1 and DTNBP1 genes on hippocampal and lateral ventricular volume in psychosis. Psychol Med. 2009;39:1783–1797. doi: 10.1017/S0033291709990316. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Schule C, Schmitt G, Born C, Baghai T, Zill P, et al. Association of the brain-derived neurotrophic factor Val-66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–416. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 2. Findings in neuropsychiatric disorders. Mol Psychiatry. 2005;10:160–184. doi: 10.1038/sj.mp.4001579. [DOI] [PubMed] [Google Scholar]
- Gonul AS, Kitis O, Eker MC, Eker OD, Ozan E, Coburn K. Association of the brain-derived neurotrophic factor Val66Met polymorphism with hippocampus volumes in drug-free depressed patients. World J Biol Psychiatry. 2011;12:110–118. doi: 10.3109/15622975.2010.507786. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Groves JO. Is it time to reassess the BDNF hypothesis of depression? Mol Psychiatry. 2007;12:1079–1088. doi: 10.1038/sj.mp.4002075. [DOI] [PubMed] [Google Scholar]
- Hajek T, Carrey N, Alda M. Neuroanatomical abnormalities as risk factors for bipolar disorder. Bipolar Disord. 2005;7:393–403. doi: 10.1111/j.1399-5618.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- Hajek T, Kozeny J, Kopecek M, Alda M, Hoschl C. Reduced subgenual cingulate volumes in mood disorders: a meta-analysis. J Psychiatry Neurosci. 2008;33:91–99. [PMC free article] [PubMed] [Google Scholar]
- Hajek T, Kopecek M, Kozeny J, Gunde E, Alda M, Hoschl C. Amygdala volumes in mood disorders – Meta-analysis of magnetic resonance volumetry studies. J Affect Disord. 2009;115:395–410. doi: 10.1016/j.jad.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, et al. Brain-derived neurotrophic factor val-66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukvik UK, Saetre P, McNeil T, Bjerkan PS, Andreassen OA, Werge T, et al. An exploratory model for G x E interaction on hippocampal volume in schizophrenia; obstetric complications and hypoxia–related genes. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1259–1265. doi: 10.1016/j.pnpbp.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Milev P, O’Leary DS, Librant A, Andreasen NC, Wassink TH. Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Arch Gen Psychiatry. 2006;63:731–740. doi: 10.1001/archpsyc.63.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Dawson JD, Wassink TH. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am J Psychiatry. 2007;164:1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoschl C, Hajek T. Hippocampal damage mediated by corticosteroids – a neuropsychiatric research challenge. Eur Arch Psychiatry Clin Neurosci. 2001;251(Suppl 2):II81–88. doi: 10.1007/BF03035134. [DOI] [PubMed] [Google Scholar]
- Jessen F, Schuhmacher A, von Widdern O, Guttenthaler V, Hofels S, Suliman H, et al. No association of the Val66Met polymorphism of the brain-derived neurotrophic factor with hippocampal volume in major depression. Psychiatr Genet. 2009;19:99–101. doi: 10.1097/YPG.0b013e32832080ce. [DOI] [PubMed] [Google Scholar]
- Joffe RT, Gatt JM, Kemp AH, Grieve S, Dobson-Stone C, Kuan SA, et al. Brain derived neurotrophic factor Val66Met polymorphism, the five factor model of personality and hippocampal volume: Implications for depressive illness. Hum Brain Mapp. 2009;30:1246–1256. doi: 10.1002/hbm.20592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik MS, Wang L, Barch DM, Morris JC, Csernansky JG. BDNF polymorphism rs6265 and hippocampal structure and memory performance in healthy control subjects. Psychiatry Res. 2010;178:425–429. doi: 10.1016/j.psychres.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempton MJ, Geddes JR, Ettinger U, Williams SC, Grasby PM. Meta-analysis, database, and meta-regression of 98 structural imaging studies in bipolar disorder. Arch Gen Psychiatry. 2008;65:1017–1032. doi: 10.1001/archpsyc.65.9.1017. [DOI] [PubMed] [Google Scholar]
- Koolschijn PC, van Haren NE, Bakker SC, Hoogendoorn ML, Pol HE, Kahn RS. Effects of brain-derived neurotrophic factor Val66Met polymorphism on hippocampal volume change in schizophrenia. Hippocampus. 2010;20:1010–1017. doi: 10.1002/hipo.20699. [DOI] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Macqueen GM, Hajek T, Alda M. The phenotypes of bipolar disorder: relevance for genetic investigations. Mol Psychiatry. 2005;10:811–826. doi: 10.1038/sj.mp.4001701. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Walss-Bass C, Nery FG, Nicoletti MA, Hatch JP, Frey BN, et al. Neuronal correlates of brain-derived neurotrophic factor Val66Met polymorphism and morphometric abnormalities in bipolar disorder. Neuropsychopharmacology. 2009;34:1904–1913. doi: 10.1038/npp.2009.23. [DOI] [PubMed] [Google Scholar]
- McDonald C, Zanelli J, Rabe-Hesketh S, Ellison-Wright I, Sham P, Kalidindi S, et al. Meta-analysis of magnetic resonance imaging brain morphometry studies in bipolar disorder. Biol Psychiatry. 2004;56:411–417. doi: 10.1016/j.biopsych.2004.06.021. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Moorhead TW, McKirdy J, Sussmann JE, Hall J, Johnstone EC, et al. Temporal grey matter reductions in bipolar disorder are associated with the BDNF Val66Met polymorphism. Mol Psychiatry. 2007;12:902–903. doi: 10.1038/sj.mp.4002044. [DOI] [PubMed] [Google Scholar]
- Montag C, Weber B, Fliessbach K, Elger C, Reuter M. The BDNF Val66Met polymorphism impacts parahippocampal and amygdala volume in healthy humans: incremental support for a genetic risk factor for depression. Psychol Med. 2009;39:1831–1839. doi: 10.1017/S0033291709005509. [DOI] [PubMed] [Google Scholar]
- Nemoto K, Ohnishi T, Mori T, Moriguchi Y, Hashimoto R, Asada T, et al. The Val66Met polymorphism of the brain-derived neurotrophic factor gene affects age-related brain morphology. Neurosci Lett. 2006;397:25–29. doi: 10.1016/j.neulet.2005.11.067. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Goldman AL, Verchinski BA, Chen G, Kolachana BS, et al. Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry. 2008;13:709–716. doi: 10.1038/mp.2008.32. [DOI] [PubMed] [Google Scholar]
- Prasad KM, Keshavan MS. Structural cerebral variations as useful endophenotypes in schizophrenia: do they help construct “extended endophenotypes”? Schizophr Bull. 2008;34:774–790. doi: 10.1093/schbul/sbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, et al. Disturbances in selective information processing associated with the BDNF Val66Met polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biol Psychol. 2009;80:176–188. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. Spurious genetic associations. Biol Psychiatry. 2007;61:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, et al. Brain-derived neurotrophic factor val-66met polymorphism and volume of the hippocampal formation. Mol Psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- Tae WS, Kim SS, Lee KU, Nam EC, Kim KW. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology. 2008;50:569–581. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Suzuki M, Tsunoda M, Kawamura Y, Takahashi N, Maeno N, et al. The association of genotypic combination of the DRD3 and BDNF polymorphisms on the adhesio interthalamica and medial temporal lobe structures. Prog Neuropsychopharmacol Biol Psychiatry. 2008a;32: 1236–1242. doi: 10.1016/j.pnpbp.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Suzuki M, Tsunoda M, Kawamura Y, Takahashi N, Tsuneki H, et al. Association between the brain-derived neurotrophic factor Val66Met polymorphism and brain morphology in a Japanese sample of schizophrenia and healthy comparisons. Neurosci Lett. 2008b;435:34–39. doi: 10.1016/j.neulet.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Uher R. The implications of gene-environment interactions in depression: will cause inform cure? Mol Psychiatry. 2008;13:1070–1078. doi: 10.1038/mp.2008.92. [DOI] [PubMed] [Google Scholar]
- Wassink TH, Nelson JJ, Crowe RR, Andreasen NC. Heritability of BDNF alleles and their effect on brain morphology in schizophrenia. Am J Med Genet. 1999;88:724–728. doi: 10.1002/(sici)1096-8628(19991215)88:6<724::aid-ajmg25>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Yoshii A, Constantine-Paton M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol. 2010;70:304–322. doi: 10.1002/dneu.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]