Abstract
Mutation in TP53 is a common genetic alteration in human cancers. Certain tumor associated p53 missense mutants acquire gain-of-function (GOF) properties and confer oncogenic phenotypes including enhanced chemoresistance. The colorectal cancers (CRC) harboring mutant p53 are generally aggressive in nature and difficult to treat. To identify a potential gene expression signature of GOF mutant p53-driven acquired chemoresistance in CRC, we performed transcriptome profiling of floxuridine (FUdR) treated SW480 cells expressing mutant p53R273H (GEO#: GSE77533). We obtained several genes differentially regulated between FUdR treated and untreated cells. Further, functional characterization and pathway analysis revealed significant enrichment of crucial biological processes and pathways upon FUdR treatment in SW480 cells. Our data suggest that in response to chemotherapeutics treatment, cancer cells with GOF mutant p53 can modulate key cellular pathways to withstand the cytotoxic effect of the drugs. The genes and pathways identified in the present study can be further validated and targeted for better chemotherapy response in colorectal cancer patients harboring mutant p53.
Keywords: Mutant p53, Gain-of-function, Transcriptome, Colorectal cancer, Floxuridine, Chemoresistance
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human colorectal cancer cell line SW480 derived from primary adenocarcinoma of colon |
| Sex | Male |
| Sequencer or array type | Microarray: Illumina Human HT-12 V4.0 Expression BeadChip |
| Data format | Raw and processed data |
| Experimental factors | SW480 cells were treated (10 μg/ml) with floxuridine (FUdR) for 24 h and compared to control untreated (DMSO vehicle) cells |
| Experimental features | Total RNA was isolated from FUdR treated and untreated cells and subjected to microarray experiments to identify the genes differentially expressed in response to drug treatment. Three biological replicates were used for each of untreated and treated conditions. |
| Consent | NA |
| Sample source location | NA |
1. Direct link to deposited data
Deposited data can be found in the Gene Expression Omnibus (GEO) database:
2. Experimental design, materials and methods
2.1. Experimental design
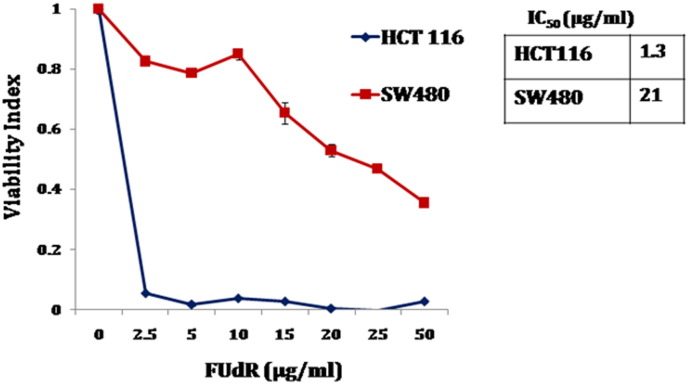
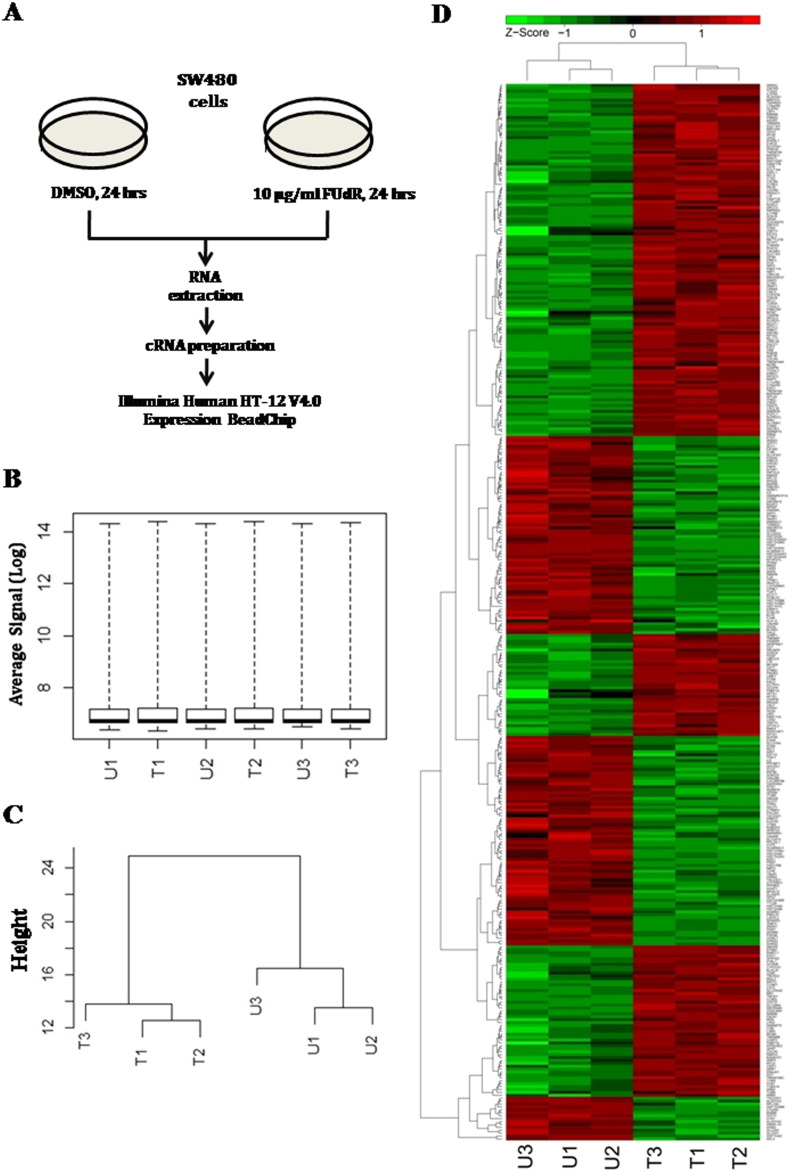
We compared the sensitivity of two colorectal cancer cell lines with different p53 background to floxuridine (FUdR), a fluropyrimidine commonly used to treat CRC patients [1], [2], [3]. In agreement with the previous report, we found that HCT116 cells with wild type p53 were sensitive, whereas SW480 cells harboring mutant p53 were resistance to the drug [4] (Fig. 1). We hypothesized that there would be a discrete set of genes and pathways that renders mutant p53 expressing cancer cells resistance to the commonly used chemotherapeutic drugs. To address this question, we performed global gene expression profiling in SW480 cells upon FUdR treatment. We treated the cells either with FUdR (10 μg/ml) or with DMSO (0.1%) as vehicle control. Twenty-four hours post-treatment, total RNA was isolated from the cells and subsequently processed for the microarray experiments (Fig. 2A). The experiment was performed in triplicate. Differential gene expression analysis between FUdR treated and untreated cells was carried out from the normalized average signal intensities. Genes that showed significant fold changes (Adjusted p-value < 0.05 and fold change ≥ 1.5) were further analyzed in GeneCodis3 for functional characterization.
Fig. 1.
SW480 cells are resistant to FUdR treatment. Line chart showing the relative viability of HCT116 and SW480 cells in response to FUdR treatment in increasing doses. IC50 values are indicated.
Fig. 2.
Gene expression profiling in SW480 cells treated with FUdR (A) Schematic showing the overall design of the study. SW480 cells were treated with either 10 μg/ml FUdR or DMSO for 24 h before harvesting in biological triplicate for each group. Total RNA was isolated and subsequently converted to cRNAs followed by hybridization to Illumina Human HT-12 V4.0 Expression BeadChip array. (B) Quality control of the microarray data. Box plots showing normalized average signal intensities across the samples in log scale. (C) Hierarchical clustering (HC) of samples based on the normalized signal intensity data. (D) Heat map showing the normalized expression of differentially regulated genes (FC ≥ 1.5, FDR-adjusted p-value < 0.05) in FUdR treated and untreated SW480 cells. Color bar represents the Z-scores of normalized expression. Red color indicates high expression, green color indicates low expression. U1, U2, U3 represents three biological replicates of untreated (DMSO vehicle) and T1, T2, T3 represents three biological replicates of FUdR treated SW480 cells respectively.
2.2. Drug sensitivity assay
Cells were seeded at a density of ~ 50,000 cells/well in a 96 well plate and allowed to grow in complete Dulbecco's modified Eagle medium (DMEM, Thermo Fisher Scientific Inc., MA USA) for 16 h before drug treatment. Cells were treated either with DMSO (0.1%) or with increasing concentration of FUdR (# F0503, Sigma Aldrich, St. Louis, USA). After 72 h, cells were washed with 1 × PBS and incubated in complete medium containing WST-1 cell proliferation reagent (Roche, Basel, Switzerland) for 30 min in 37 °C CO2 incubator. The absorbance of the samples was measured at 450 nm in an ELISA plate reader and the viability index was calculated in MS-Excel from the average absorbance values obtained from three biological replicates.
2.3. Drug treatment, RNA isolation and quality control of the RNA samples for microarray
SW480 cells were seeded (~ 1.5 × 10 [6]) in 6 cm dishes in triplicate and were grown in complete DMEM medium for 16 h before drug treatment. Cells were treated either with 10 μg/ml of FUdR or with DMSO (0.1%) and allowed to grow for another 24 h before harvesting. Total RNA was isolated using RNeasy Plus Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Briefly, cells were washed with 1 × ice cold PBS and lysed by adding 600 μl of RLT lysis buffer directly to the dishes. The lysates were mixed well by pipetting and transferred to gDNA eliminator spin column followed by centrifugation at 13,000 rpm for 1 min. The flow-through was mixed with 70% ethanol and centrifuged in RNeasy spin column at 13,000 rpm for 30 s. The columns were washed subsequently with buffer RW1 and buffer RPE. Finally, the column bound RNA was eluted in 40 μl of RNase-free water by centrifugation at 13,000 rpm for 2 min. The concentration and purity of the RNA samples were measured in UV–Vis spectrophotometer. Quality of the RNA samples was further assessed by checking the RNA Integrity Number (RIN) in Agilent® 2100 Bioanalyzer using Agilent RNA 6000 Nano kit (Agilent Technologies, Santa Clara, California, USA). Samples with RIN > 7.5 were further considered for microarray profiling.
2.4. Preparation of biotinylated cRNA from total RNA
Quality checked RNA was amplified using Illumina® TotalPrep RNA Amplification Kit (Life Technologies, Thermo Fisher Scientific Inc., MA USA) according to the manufacturer's protocol. For first strand cDNA synthesis, 500 ng of total RNA in a volume of 11 μl was mixed with 9 μl reverse transcription master mix (1 μl of T7 oligo (dT) primer, 2 μl of 10 × first strand buffer, 1 μl of ribonuclease inhibitor, 4 μl of dNTP mix and 1 μl ArrayScript reverse transcriptase enzyme) followed by incubation at 42 °C for 2 h in a thermal cycler with 500 C lid temperature condition. Subsequently, second strand cDNA synthesis was carried out by adding 80 μl of second strand master mix (63 μl of nuclease free water, 10 μl of 10 × second strand buffer, 4 μl of dNTP mix, 2 μl of DNA Polymerase and 1 μl of RNase H) to the samples followed by incubation at 16 °C for 2 h in a thermal cycler. The double stranded (ds) cDNAs were purified using cDNA filter cartridges and finally eluted in 20 μl of nuclease free water of 55 °C. To prepare biotinylated cRNAs, 7.5 μl of in vitro transcription (IVT) master mix (2.5 μl of 10 × reaction buffer, 2.5 μl T7 enzyme mix, 2.5 μl biotin-dNTP mix) was added to 17.5 μl of purified ds-cDNA and incubated at 37 °C for 14 h. cRNAs were diluted with nuclease-free water and mixed with cRNA binding buffer and 100% ethanol. The samples were passed through cRNA filter cartridges by centrifugation at 10,000 × g for 1 min. The cartridges were washed with wash buffer and column bound cRNAs were finally eluted in 200 μl of 55 °C nucleasefree water. The concentration of the cRNAs was determined in Qubit® 2.0 fluorometer using Qubit RNA BR assay kit (Life Technologies, Thermo Fisher Scientific Inc., MA USA).
2.5. Hybridization of labeled cRNAs to BeadChip and microarray
cRNA samples were hybridized to Illumina Human HT-12 v 4.0 Expression BeadChip whole genome array following manufacturer's protocol (Illumina Inc., San Diego, USA). Briefly, hybridization buffer (HYB) was added to 750 ng of each cRNA sample and the samples were loaded in the HT-12 v 4.0 BeadChip placed in hybridization chamber. The hybridization reaction was carried out in Illumina hybridization oven at 58 °C for 14 h. The BeadChips were subsequently washed, blocked and conjugated with Cy3-Streptavidin. The bioarrays were scanned in iScan system (Illumina Inc., San Diego, USA) and extracted raw intensity values were saved as intensity data (*.idat) files.
2.6. Data analysis
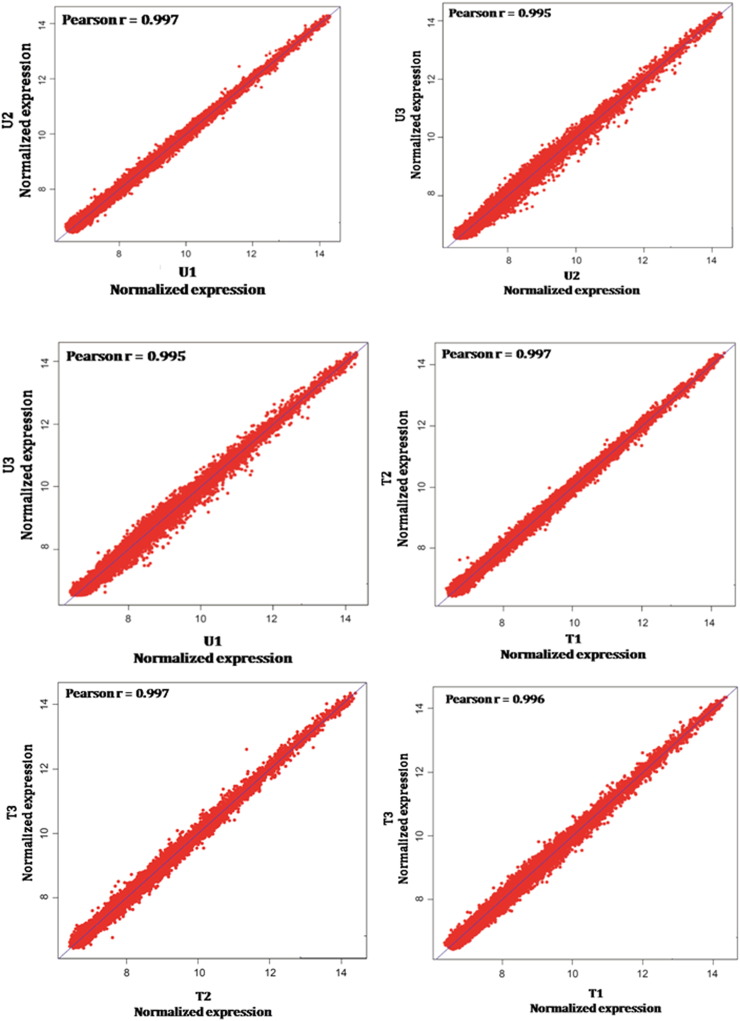
Background subtracted data were extracted using Genome Studio V2011.1 software tool (Illumina Inc., San Diego, USA) and the quality control (QC) of the data was performed using its in-built plotting features. The data were further processed in R statistical environment (http://www.r-project.org) using Lumi package to generate Box Plots of normalized signal intensities across the samples (Fig. 2B). Variance-stabilizing transformation (VST) algorithm was used for all 6 microarray samples to achieve identical distribution of signal intensities for comparison [5]. Next, the data was normalized using robust spline normalization (RSN) method of lumi package [6]. Probes showing detection p-value < 0.01 in all samples were considered for further analysis. Correlation analysis of normalized intensities between biological replicates showed good correlation with average Pearson correlation coefficient > 0.9 (Supplementary Fig. S1). Hierarchical clustering based on the pattern of gene expression showed clear separation of the untreated and treated cells (Fig. 2C). Differential expression analysis was carried out in R Bioconductor Limma package and the p-values were corrected for multiple testing using Benjamini and Hochberg false discovery rate (FDR) algorithm [7]. A linear model was fitted for each gene given a series of arrays using lmFit function. We found 208 genes were up-regulated and 155 genes were down-regulated by at least 1.5 fold with FDR-adjusted p-value < 0.05 (Fig. 2D) upon FUdR treatment in SW480 cells. Unsupervised hierarchical clustering was performed using average linkage and Euclidean distance.
Fig. S1.
Scatter plots showing the correlation of normalized signal intensities between biological replicates of FUdR treated and untreated SW480 cells. Pearson correlation coefficients (r) are indicated.
2.7. Functional classification of differentially regulated genes in GeneCodis
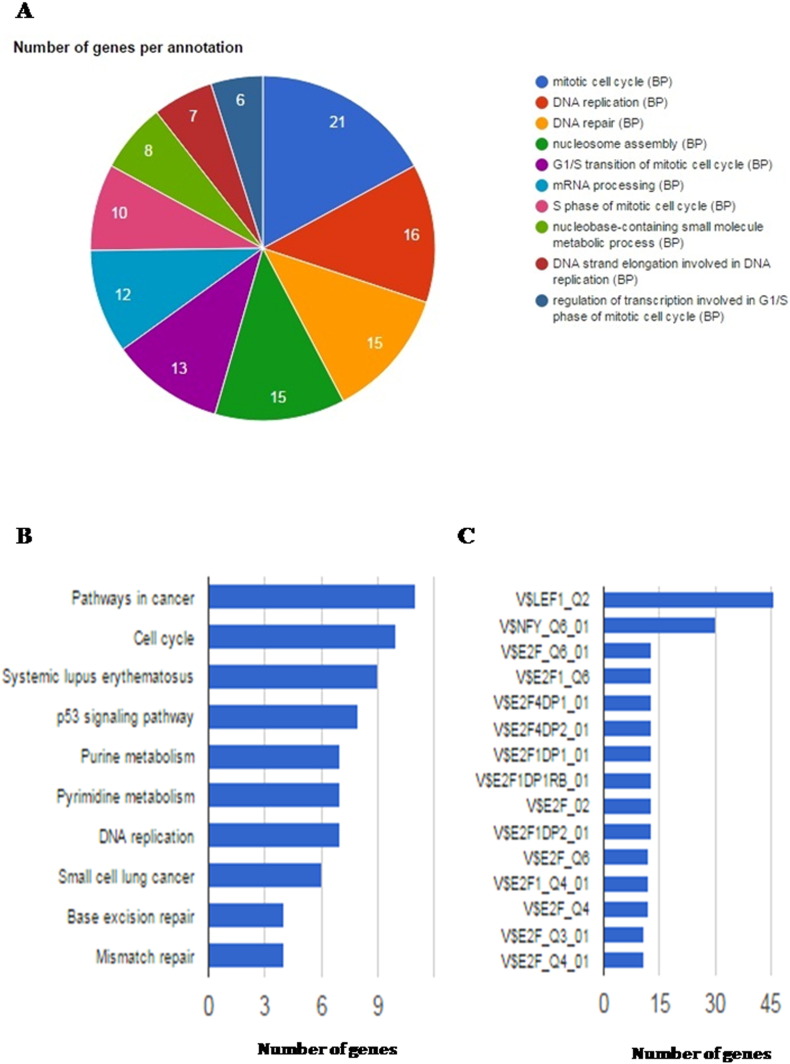
The differentially regulated genes were analyzed in GeneCodis3 Gene Ontology software (http://genecodis.cnb.csic.es/) with the default settings [8], [9], [10]. Crucial biological processes (GO terms) including mitotic cell cycle, DNA replication and repair, nucleosome assembly, mRNA processing and transcriptional regulation in G1/S phase of cell cycle were found to be significantly enriched (FDR corrected Hypergeometric p-value < 0.05, Fig. 3A and Supplementary Table 1). Further, KEGG pathway enrichment analysis showed that pathways involved in cancer and cell cycle were significantly overrepresented (FDR corrected Hypergeometric p-value < 0.05) by the deregulated genes upon drug treatment in SW480 cells (Fig. 3B and Supplementary Table 2). Using TF enrichment tool, we also identified the transcription factors (TF) potentially involved in regulation of these drug induced genes (Fig. 3C and Supplementary Table 3).
Fig. 3.
Functional classification of differentially regulated genes upon FUdR treatment in SW480 cells. (A) Pie chart represents GO biological processes significantly enriched (FDR corrected Hypergeometric p-value < 0.05) by the differentially regulated genes between FUdR treated and untreated SW480 cells. (B) Bar plot showing the enrichment of KEGG pathways by the differentially expressed genes. The pathways are shown in Y-axis and the number of genes in each pathway is represented in X-axis. (C) Bar plot showing the transcription factors enriched for the deregulated genes upon FUdR treatment. Y-axis and X-axis represent the transcription factors and the number of their target genes respectively.
3. Discussion
Mutations in TP53 result in both loss of tumor suppressor functions and gain of new oncogenic properties [11], [12]. Certain “hotspot” missense mutations enable p53 proteins to gain oncogenic functions (GOFs) that actively drive tumorigenesis [12], [13], [14], [15], [16]. Tumors bearing p53 mutations are generally aggressive in nature and confer increased resistance to commonly used chemotherapeutics [17], [18], [19]. About half of the colorectal cancers harbor p53 mutation (43.2%, IARC TP53 database) which is associated with the poor survival of patients [14], [18], [20], [21]. In this study, using a genome wide approach, we identified potential gene expression signatures and cellular pathways that may be involved in conferring drug resistance to colorectal cancer cells expressing GOF mutant p53. In response to drug induced DNA damage, oncogenic p53 mutants can transcriptionally regulate key cellular genes by cooperating with other transcription factors and confer chemo-resistance [4], [12], [22]. Our analysis showed that, the differentially regulated genes upon drug treatment in SW480 cells are mostly targeted by the transcription factors (such as NFY, E2F1) known to interact with mutant p53 [12], [23]. This observation indicates that some of these genes may be transactivated by GOF mutant p53 in response to drug treatment. Collectively, this study provides a comprehensive picture of the cellular genes and pathways deregulated upon chemotherapy treatment in colon cancer cells harboring mutant p53. Our data can be a valuable resource for future studies investigating the molecular mechanisms underlying mutant p53 driven chemoresistance in human cancers.
The following are the supplementary data related to this article.
GO biological processes enriched by the differentially expressed genes upon FUdR treatment in SW480 cells.
Kegg pathways enriched by the differentially expressed genes upon FUdR treatment in SW480 cells.
Transcription factors associated with the differentially expressed genes by FUdR treatment in SW480 cells.
Conflict of interest
All authors read the manuscript and declare no conflict of interests.
Acknowledgements
We acknowledge the Director, National Institute of Biomedical Genomics, Kalyani, India for kindly providing the Illumina iScan facility. This work was supported by CSIR—Mayo Clinic Collaboration for Innovation and Translational Research Grant CMPP-08 and CSIR-NMITLI project TLP 0007 awarded to S. Roychoudhury. AD, PD and SKA are supported by predoctoral fellowship from the Council of Scientific and Industrial Research, India. SD is supported by postdoctoral fellowship from Department of Biotechnology (DBT), India.
References
- 1.Geng L., Huehls A.M., Wagner J.M., Huntoon C.J., Karnitz L.M. Checkpoint signaling, base excision repair, and PARP promote survival of colon cancer cells treated with 5-fluorodeoxyuridine but not 5-fluorouracil. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028862. (Epub 2011 Dec 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemeny N.E. Regional chemotherapy of colorectal cancer. Eur. J. Cancer. 1995 Jul–Aug;31A(7–8):1271–1276. doi: 10.1016/0959-8049(95)00162-c. [DOI] [PubMed] [Google Scholar]
- 3.Power D.G., Kemeny N.E. The role of floxuridine in metastatic liver disease. Mol. Cancer Ther. 2009 May;8(5):1015–1025. doi: 10.1158/1535-7163.MCT-08-0709. (Epub 2009 Apr 21) [DOI] [PubMed] [Google Scholar]
- 4.Alam S.K. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2015 Oct 23 doi: 10.1038/cdd.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008 Feb;36(2) doi: 10.1093/nar/gkm1075. (Epub 2008 Jan 4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing illumina microarray. Bioinformatics. 2008 Jul 1;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. (Epub 2008 May 8) [DOI] [PubMed] [Google Scholar]
- 7.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to Multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 8.Carmona-Saez P., Chagoyen M., Tirado F., Carazo J.M., Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1):R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogales-Cadenas R. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W317–W322. doi: 10.1093/nar/gkp416. (Epub 2009 May 22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. (Epub 2012 May 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 12.Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013 Jan;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 13.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010 Jan;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabapathy K. The contrived mutant p53 oncogene — beyond loss of functions. Front Oncol. 2015 Dec 10;5:276. doi: 10.3389/fonc.2015.00276. (eCollection 2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walerych D., Lisek K., Del Sal G. Mutant p53: one, no one, and one hundred thousand. Front Oncol. 2015;5:289. doi: 10.3389/fonc.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leroy B. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res. 2013 Jan;41(Database issue):D962–D969. doi: 10.1093/nar/gks1033. (Epub 2012 Nov 17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buttitta F. p53 alterations are predictive of chemoresistance and aggressiveness in ovarian carcinomas: a molecular and immunohistochemical study. Br. J. Cancer. 1997;75(2):230–235. doi: 10.1038/bjc.1997.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X.L., Zhou J., Chen Z.R., Chng W.J. P53 mutations in colorectal cancer — molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015 Jan 7;21(1):84–93. doi: 10.3748/wjg.v21.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller P.A., Vousden K.H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014 Mar 17;25(3):304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh H.S., Yao J., Smith D.R. p53 point mutation and survival in colorectal cancer patients. Cancer Res. 1995 Nov 15;55(22):5217–5221. [PubMed] [Google Scholar]
- 21.Iacopetta B. TP53 mutation in colorectal cancer. Hum. Mutat. 2003 Mar;21(3):271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 22.Di Agostino S. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006 Sep;10(3):191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012 Jun 15;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO biological processes enriched by the differentially expressed genes upon FUdR treatment in SW480 cells.
Kegg pathways enriched by the differentially expressed genes upon FUdR treatment in SW480 cells.
Transcription factors associated with the differentially expressed genes by FUdR treatment in SW480 cells.