Abstract
This paper describes the complete sequence of a giant lytic marine myophage, Vibrio phage ValKK3 that is specific to Vibrio alginolyticus ATCC® 17749™. Vibrio phage ValKK3 was subjected to whole genome sequencing on MiSeq sequencing platform and annotated using Blast2Go. The complete sequence of ValKK3 genome was deposited in DBBJ/EMBL/GenBank under accession number KP671755.
Keywords: Vibrio alginolyticus, Whole genome sequencing, Giant marine myophage
| Specifications | |
|---|---|
| Organism | Myoviridae |
| Genus | T4-Like |
| Sequencer | Illumina (MiSeq) |
| Data format | Analyzed |
| Experimental factors | Bacteriophage strain |
| Experimental features | Whole genome analysis and gene annotation of Vibrio phage ValKK3 |
| Consent | N/A |
| Sample source location | Marine sediment, Kota Kinabalu, N 06° 02.270′, E 116° 06.710′ |
1. Direct link to deposited data
2. Materials and methods
Vibrio phage ValKK3 was isolated from marine sediment of Kota Kinabalu, Malaysia that was capable to lyse fish and human pathogen, Vibrio alginolyticus ATCC® 17749™ [1]. The phage morphology was determined by a transmission electron microscope. The genome was extracted and purified using DNeasy Blood and Tissue Kit (Qiagen). The genome sequencing and assembly was performed by AITBiotech Pte. Ltd. (Singapore). The DNA library was prepared by Illumina Nextera XT kit. The samples were dual barcoded for multiplexing on the MiSeq instrument. The paired of 250 bp reads (2 × 250,000 samples) were generated and demultiplexed corresponding to the barcodes. The FASQ files were generated on board of MiSeq Instrument. The sequence assembly was performed using de novo assembly using Velvet assembly software version 1.1 (Zerbino, European Bioinformatics Institute, UK). The assembly producing the lowest number of contigs with the largest n50 value was chosen for further assembly with a > 30-fold sequence coverage into single contig. The complete contig sequence was subjected to BLASTn [2] and the sequence was annotated using Blast2GO [3].
3. Data description
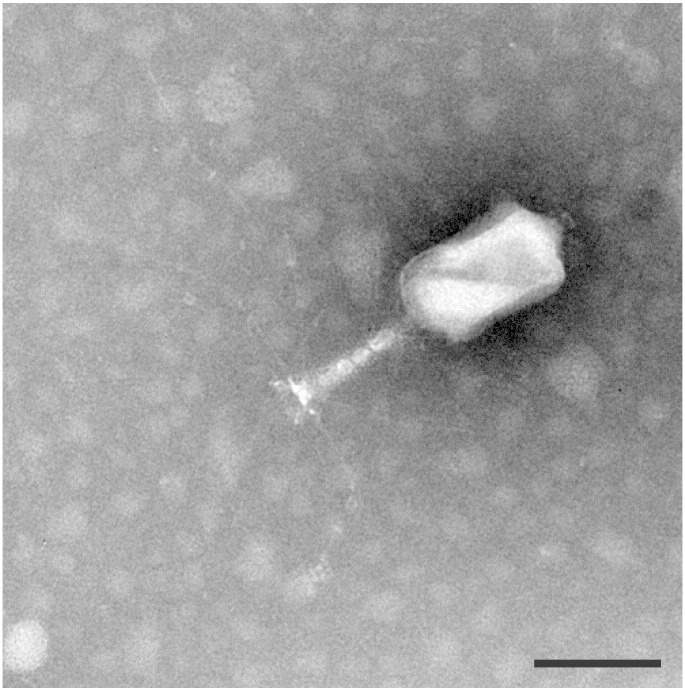
The morphological characteristics of ValKK3 (Fig. 1) showed that it belongs to the family of Myoviridae. To the best of our knowledge, among the giant T4-like phages reported so far (Table 1), the Vibrio phage ValKK3 is the only strain that can infect V. alginolyticus. The genome annotation analysis predicted 390 open reading frames which are numbered consecutively from ORF1 to ORF390 which represented 92.4% of the total genome. The putative genes varied from 111 bp (ORF268) to 4200 bp (ORF349). The protein function analysis showed that some CDSs involve in repression of host gene synthesis (ORF197, ORF202), defense system of own gene synthesis (ORF170, ORF176, ORF207) and quality control (ORF255), DNA synthesis (ORF61, ORF199, ORF367, ORF370, ORF241, ORF256, ORF381), DNA replication and recombination (ORF371, ORF372, ORF320, ORF178, ORF204, ORF106, ORF242, ORF257), structural protein: the head (ORF281–286), tail (ORF287–319) and tail fiber (ORF346–349), DNA packaging protein (ORF289, ORF290) and host lysis protein (ORF304). In this study, the putative functions of ValKK3 genome only represent 15.6% of the genes in the ValKK3. The other 84.4% in the ValKK3 are consisted of the genes which encoded proteins with unknown function or hypothetical proteins. The ValKK3 is similar to the Vibrio phage VH7D, Bacteriophage KVP40, Vibrio phage phi-pp2 and Vibrio phage nt-1 (Table 1). This assigns the ValKK3 to the T4-like phage genus of the Myoviridae family which belongs to the giant vibriophage group [7], [8] with even larger genome size. The blast search revealed that ValKK3 was homologous to vibriophage that is known to infect some members of Harveyi clade bacteria. It is also revealed that T4-like phage is widely spread in the marine environment infecting different Vibrio species. Finally, an additional genome information of ValKK3 is hoped to provide positive contributions to the study of Vibrio bacteriophage.
Fig. 1.
Electron micrograph of negative-stained ValKK3. Bars = 100 nm.
Table 1.
Genome comparison among giant vibriophages.
| Bacteriophage strain | Vibrio phage ValKK3 | Vibrio phage VH7D | Bacteriophage KVP40 | Vibrio phage phi-pp2 | Vibrio phage nt-1 |
|---|---|---|---|---|---|
| Host species | V. alginolyticus | Vibrio strain 7D | V. parahaemolyticus | V. parahaemolyticus | V. natriegens |
| GenBank accession no. | KP671755 | KC131129 | AY283928 | JN849462 | HQ317393 |
| Head length (nm) | 140 | 155 | 140 | 150 | 120 |
| Head diameter (nm) | 79 | 75 | 70 | 90 | 70 |
| Tail length (nm) | 120 | Not reported | 130 | 120 | 110 |
| Genome size (bp) | 248,088 | 246,964 | 244,834 | 246,421 | 247,511 |
| Blastn identity (%) | 100 | 99 | 81 | 81 | 85 |
| ORF number | 390 | 378 | 386 | 383 | 379 |
| G + C level (%) | 41.2 | 41.3 | 42.6 | 42.7 | 41.3 |
| Reference | This study | Luo et al. [4] | Miller et al. [5], Matsuzaki et al. [6] | Lin and Lin [7] | Comeau et al. [8], Zachary [9] |
4. Nucleotide accession number
This complete genome sequence has been deposited at DDBJ/EMBL/GenBank under accession no. KP671755.
Conflict of interest
The authors clarified that this work and writing has no conflict of interest.
Acknowledgments
This study was financially supported by the Universiti Malaysia Sabah's Research Priority Area Scheme (SBK0110-STWN-2013) and Malaysia Ministry of Education's Fundamental Research Grant Scheme (FRG0338-STWN-1/2013).
References
- 1.Liu X.F., Cao Y., Zhang H.L., Chen Y.J., Hu C.J. Complete genome sequence of Vibrio alginolyticus ATCC 17749T. Genome Announc. 2015;3(1):e01500–e01514. doi: 10.1128/genomeA.01500-14. (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 4.Luo Z.H., Yu Y.P., Jost G., Xu W., Huang X.L. Complete genome sequence of a giant Vibrio bacteriophage VH7D. Mar. Genomics. 2015;3:293–295. doi: 10.1016/j.margen.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Miller E.S., Heidelberg J.F., Eisen J.A., Nelson W.C., Scott Durkin A., Ciecko A., Feldblyum T.V., White O., Paulsen I.T., Nierman W.C., Lee J., Szczypinski B., Fraser C.M. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 2003;185(17):5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuzaki S., Tanaka S., Koga T., Kawata T. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol. Immunol. 1992;36:93–97. doi: 10.1111/j.1348-0421.1992.tb01645.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y.R., Lin C.S. Genome-wide characterization of vibrio phage ϕpp2 with unique arrangements of the mob-like genes. BMC Genomics. 2012;13:224. doi: 10.1186/1471-2164-13-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comeau A.M., Arbiol C., Krish H.M. Composite conserved promoter-terminator motifs (PeSLs) that mediate modular shuffling in the diverse T4-like myovirus. Genome Biol. Evol. 2014;6(7):1611–1619. doi: 10.1093/gbe/evu129. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zachary A. Physiology and ecology of bacteriophages of marine bacterium Beneckea natriegens: salinity. Appl. Environ. Microbiol. 1976;31(3):415–422. doi: 10.1128/aem.31.3.415-422.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]