Abstract
p53 is a central mediator of cellular stress responses, and its precise regulation is essential for the normal progression of hematopoiesis. MYSM1 is an epigenetic regulator essential for the maintenance of hematopoietic stem cell (HSC) function, hematopoietic progenitor survival, and lymphocyte development. We recently demonstrated that all developmental and hematopoietic phenotypes of Mysm1 deficiency are p53-mediated and rescued in the Mysm1−/−p53−/− mouse model. However, the mechanisms triggering p53 activation in Mysm1−/− HSPCs, and the pathways downstream of p53 driving different aspects of the Mysm1−/− phenotype remain unknown. Here we show the transcriptional activation of p53 stress responses in Mysm1−/− HSPCs. Mechanistically, we find that the MYSM1 protein associates with p53 and colocalizes to promoters of classical p53-target genes Bbc3/PUMA (p53 upregulated modulator of apoptosis) and Cdkn1a/p21. Furthermore, it antagonizes their p53-driven expression by modulating local histone modifications (H3K27ac and H3K4me3) and p53 recruitment. Using double-knockout mouse models, we establish that PUMA, but not p21, is an important mediator of p53-driven Mysm1−/− hematopoietic dysfunction. Specifically, Mysm1−/−Puma−/− mice show full rescue of multipotent progenitor (MPP) viability, partial rescue of HSC quiescence and function, but persistent lymphopenia. Through transcriptome analysis of Mysm1−/−Puma−/− MPPs, we demonstrate strong upregulation of other p53-induced mediators of apoptosis and cell-cycle arrest. The full viability of Mysm1−/−Puma−/− MPPs, despite strong upregulation of many other pro-apoptotic mediators, establishes PUMA as the essential non-redundant effector of p53-induced MPP apoptosis. Furthermore, we identify potential mediators of p53-dependent but PUMA-independent Mysm1−/−hematopoietic deficiency phenotypes. Overall, our study provides novel insight into the cell-type-specific roles of p53 and its downstream effectors in hematopoiesis using unique models of p53 hyperactivity induced by endogenous stress. We conclude that MYSM1 is a critical negative regulator of p53 transcriptional programs in hematopoiesis, and that its repression of Bbc3/PUMA expression is essential for MPP survival, and partly contributes to maintaining HSC function.
p53 is a central regulator of cell fate and cellular responses to physiological and exogenous stresses.1 It is a DNA-binding protein that primarily acts as a transcriptional regulator.2 Acute p53 activation can trigger cell-cycle arrest, cellular senescence, or apoptosis. Different combinations of p53-induced effectors can execute these distinct cell fate decisions. For example, cyclin-dependent kinase inhibitor Cdkn1a/p21 is a significant p53-induced mediator of cell-cycle arrest,3, 4 whereas BH3-domain protein Bbc3/PUMA is an important mediator of p53-dependent apoptosis.5, 6
Knowledge of cell-type-specific features of the p53 pathway is important for the understanding of physiological stress responses and the development of clinical strategies for targeted p53 activation.7, 8 Many studies in this area have focused on hematopoietic stem and progenitor cells (HSPCs).9, 10 p53 was shown to have specialized roles in homeostatic regulation of hematopoietic stem cell (HSC) quiescence, self-renewal, and differentiation.11, 12, 13 p53-target gene Cdkn1a/p21 was originally implicated in enforcing homeostatic HSC quiescence,14 although this role was questioned in recent studies.15 Other novel p53-target genes were identified as regulators of HSC quiescence, including Gfi1 and Necdin.11, 16, 17
In hematopoietic responses to stress, Bbc3/PUMA was identified as a major mediator of p53-dependent apoptosis,18, 19, 20, 21 with some redundancy with other BH3-only family members such as Pmaip1/NOXA.22, 23, 24, 25 Other studies highlighted specific differences in the stress responses of HSCs and more mature hematopoietic progenitors,10 with HSCs in mouse bone marrow being less prone to induction of pro-apoptotic genes and cell death than progenitor cells.26, 27, 28 Despite a growing understanding of the specialized features of p53 responses in HSCs and hematopoiesis, and their obvious biological and clinical importance, the underlying molecular mechanisms remain poorly understood.
Myb-like SWIRM and MPN domains 1 (MYSM1) is a chromatin-binding protein29 previously implicated in transcriptional regulation through histone H2AK119 (histone H2A lysine 119) deubiquitination.30 Mysm1 loss in mice results in the arrest of lymphocyte development and severe defects in HSPC functions, including loss of HSC quiescence and impaired survival of multipotent progenitors (MPPs).31, 32, 33 Patients with rare MYSM1 mutations display lymphopenia and HSC exhaustion, confirming the essential role of MYSM1 in hematopoiesis and lymphocyte development in humans.34, 35
There are two perspectives on the role of MYSM1 in hematopoiesis. In a series of recent papers, Mysm1−/− hematopoietic defects were linked to impaired expression of various hematopoietic lineage differentiation genes, namely Ebf1, Id2, Gfi1, and Flt3.32, 33, 36, 37 In contrast, we and others demonstrated that Mysm1−/− phenotypes are rescued in the Mysm1−/−p53−/− mouse model,38, 39 indicating that p53 activation is the common mechanism mediating HSC dysfunction, MPP depletion, and lymphopenia in Mysm1 deficiency. However, the mechanisms leading to p53 activation and its effects in Mysm1-deficient HSPCs remain unclear. Although Gatzka M et al.39 indicated induction of Cdkn2a/p19ARF as the cause of p53 activation in the Mysm1−/− thymus, it is unknown whether these findings extend to other Mysm1−/− hematopoietic cells, particularly HSPCs. Furthermore, the specific p53-induced effectors that trigger dysfunction in Mysm1-deficient HSPCs remain unknown.
Here we show that MYSM1 is a novel regulator of p53 transcriptional programs that antagonizes the expression of p53 stress-response genes through direct effects on chromatin at their promoters. We further show that Bbc3/PUMA is the primary non-redundant mediator of MPP depletion in Mysm1 deficiency and contributes to HSC dysfunction, whereas depletion of lymphoid-lineage cells involves PUMA-independent p53 activities.
Results
p53 stress-response activation in Mysm1-deficient HSPCs
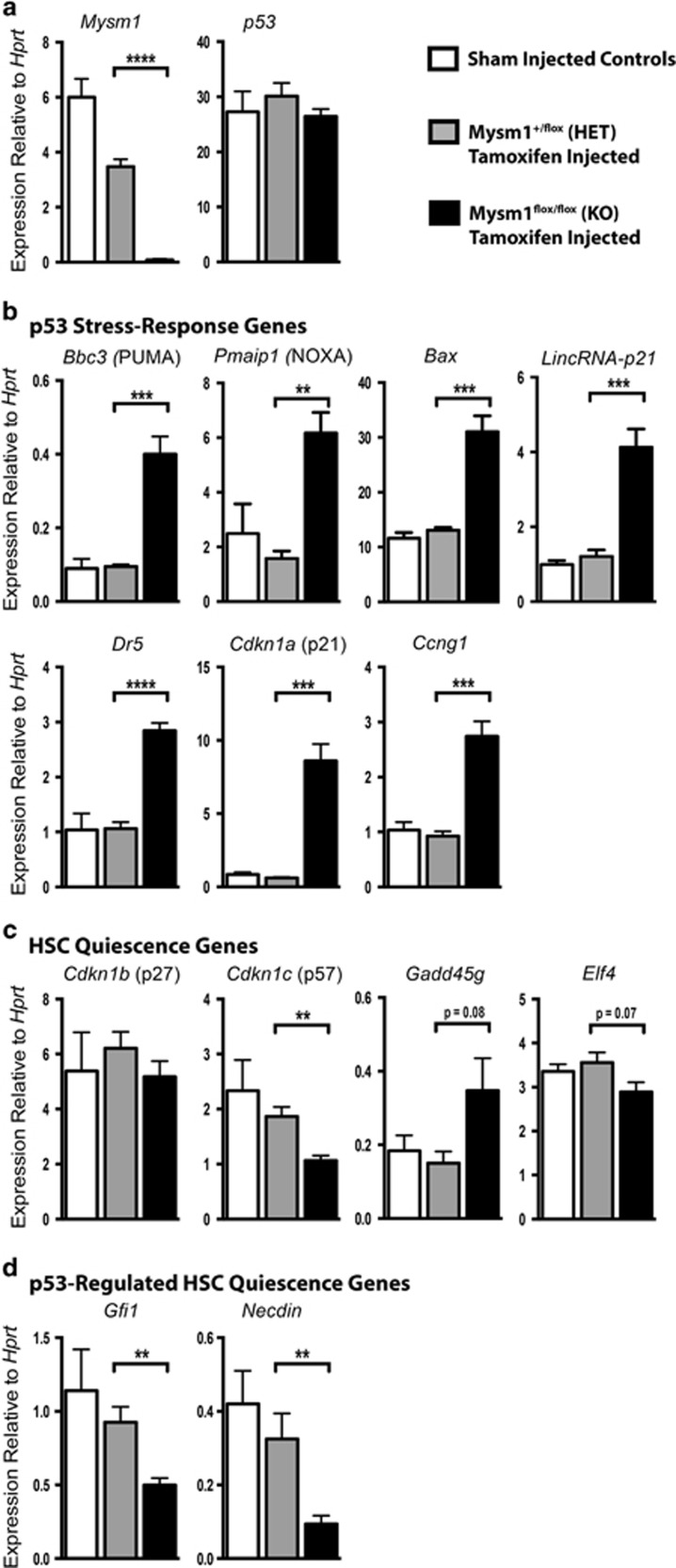
To understand the causes of p53 activation and downstream p53 effector functions leading to hematopoietic failure in Mysm1-deficiency, HSPC gene expression was analyzed. Inducible Mysm1 deletion in the Rosa26CreERT2Mysm1flox/flox model40 was used to focus on direct effects of Mysm1 loss rather than indirect effects of chronic Mysm1 deficiency. HSPCs (Lin−cKit+Sca1+) were sorted from tamoxifen-treated Rosa26CreERT2Mysm1flox/flox mice against control Rosa26CreERT2Mysm1+/flox mice and untreated Mysm1flox/flox animals. qRT-PCR validated loss of Mysm1 expression, without significant changes in p53 transcript (Figure 1a). There was a strong induction of p53 stress-response genes including Bbc3/PUMA, Pmaip1/NOXA, Bax, Cdkn1a/p21, and Ccng1 (Figure 1b).
Figure 1.
Mysm1 deficiency results in dysregulation of p53-target gene expression in HSPCs. qPCR analysis of Lin−cKit+Sca1+ cells sorted from the bone marrow of CreERT2-transgenic Mysm1flox/flox mice following tamoxifen-induced Mysm1 gene inactivation (black bars). CreERT2 Mysm1+/flox mice that received the same tamoxifen treatment (gray bars), and sham-injected Mysm1flox/flox mice (white bars) were used as controls. Transcript levels of (a) Mysm1 and p53, (b) classical p53 stress-response genes, (c) regulators of HSC quiescence, and (d) HSC-specific p53 target genes are presented. All gene expressions were normalized against Hprt expression. Bars represent mean±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, NS, nonsignificant using Student's t-test; data from ≥5 mice per group
We validated the reduced expression of HSC quiescence regulator Gfi1 previously reported in Mysm1−/− HSPCs,37 and demonstrated the downregulation of other HSC quiescence regulators, namely Necdin and Cdkn1c/p5716, 17, 41, 42, 43 (Figure 1c and d). Overall, this indicated severe dysregulation of both p53-mediated stress responses and cell-cycle target genes in HSPCs upon Mysm1 deletion. Importantly, Cdkn2a/p19ARF expression was undetectable in all samples and not elevated in Mysm1-deficient HSPCs (Supplementary Figure S1a). As induction of Cdkn2a/p19ARF was previously implicated in p53 activation in Mysm1−/− thymocytes,39 we conclude that the mechanisms of p53 activation in Mysm1-deficient HSPCs are distinct and p19ARF-independent.
MYSM1 colocalizes with p53 to Bbc3/PUMA and Cdkn1a/p21 gene promoters
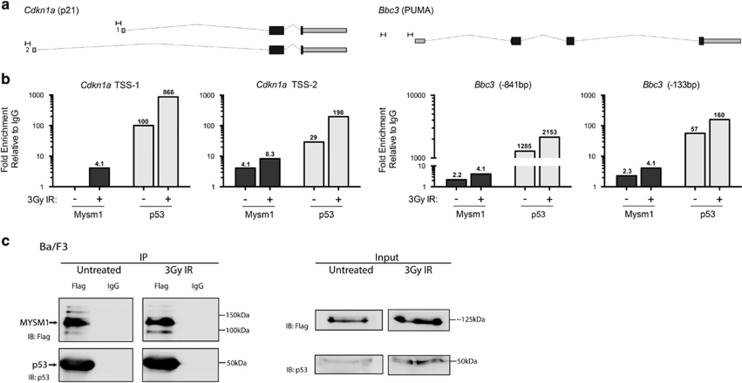
As both p53 and MYSM1 are transcriptional regulators,2, 30 we hypothesized that the increased induction of p53-target genes in Mysm1-deficient HSPCs indicated a direct role of MYSM1 in their regulation. The recruitment of MYSM1 to p53-target gene promoters was tested using chromatin immunoprecipitation (ChIP) in a Ba/F3 hematopoietic progenitor cell line stably expressing FLAG-MYSM1 (Supplementary Figure S2a). MYSM1 binding was detected at known p53-binding sites within Bbc3/PUMA and Cdkn1a/p21 promoters,44 and enhanced with irradiation (Figures 2a and b).
Figure 2.
MYSM1 protein can interact with p53 and colocalize to the promoters of p53-target genes. (a and b) Ba/F3 hematopoietic progenitor cells stably expressing 3xFlag-MYSM1 protein were analyzed for recruitment of MYSM1 and p53 to the promoters of p53 target genes using ChIP-qPCR at a steady state and following 3-Gy irradiation. (a) Genomic structure of Bbc3/PUMA and Cdkn1a/p21 loci according to the UCSC Mouse mm9 assembly with target primer sites and PCR products indicated (to scale). Primers were designed in the vicinity of known p53-binding sites based on ChIP-Seq data from Kenzelmann Broz D et al. (b) Fold enrichment of MYSM1 and p53 near the transcriptional start sites of Bbc3/PUMA and Cdkn1a/p21 genes in untreated and irradiated cells (3 h after 3 Gy). Relative enrichment was normalized relative to a nonspecific IgG ChIP and to a negative control genomic region (Pomc) with normalized values shown above each bar. Data shown are representative of at least three independent experiments with additional replicates in Supplementary Figures S2c and d. (c) Co-immunoprecipitation of endogenous p53 with 3xFlag-MYSM1 stably expressed in the Ba/F3 hematopoietic progenitor cell line. Cell lysates from untreated and irradiated cells (3 h after 3 Gy) were subjected to immunoprecipitation with anti-Flag and IgG control antibodies. Left panel: IP samples were blotted for Flag-MYSM1 (anti-Flag) and p53 showing specific pulldown of both proteins. Right panel: whole-cell lysates (input) were blotted for Flag-MYSM1 (anti-Flag) and p53
Given the recruitment of MYSM1 to genomic p53-binding sites, we tested for interaction between MYSM1 and p53 proteins. In the Ba/F3 line expressing FLAG-MYSM1, endogenous p53 was co-immunoprecipitated with FLAG-MYSM1 (Figure 2c), and similar results were obtained in reciprocal co-immunoprecipitations in HEK293T cells (Supplementary Figure S2b). MYSM1 therefore interacts with p53, either directly or more likely as part of a larger protein complex. The interaction of MYSM1 with p53 and their colocalization to p53-target gene promoters, in addition to p53 transcriptional signature activation in Mysm1−/− HSPCs, suggest a role for MYSM1 in regulating p53 transcriptional activity.
MYSM1 regulates p53-target gene expression in hematopoietic progenitors
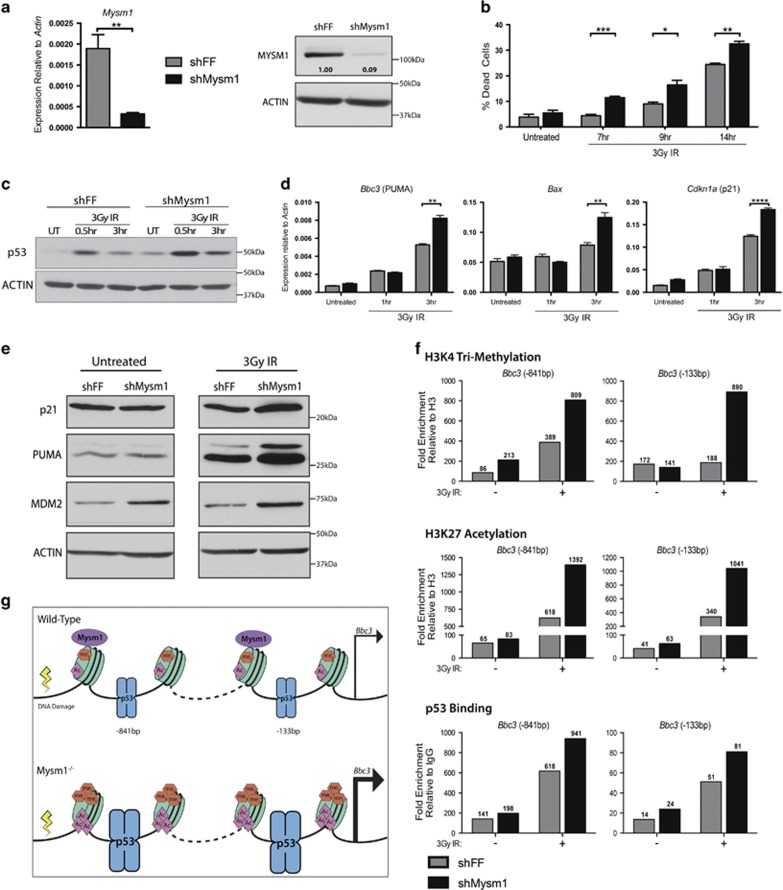
To overcome the limitations of the severe depletion of primary HSPCs in Mysm1−/− mice, Mysm1 shRNA knockdown in Ba/F3 hematopoietic progenitor cells was employed as a model for further analysis of MYSM1 functions in p53-target gene regulation. The shMysm1-knockdown lines showed a 10-fold reduction in the MYSM1 protein as compared with firefly luciferase shRNA (shFF) control lines (Figure 3a). To validate the model's relevance, shMysm1 lines were tested for the phenotypes observed in primary Mysm1−/− HSPCs, including activation of the p53 stress response and apoptosis. Following irradiation, shMysm1 cells displayed increased susceptibility to cell death, and slightly increased levels of p53 protein. Furthermore, shMysm1 cells showed elevated induction of p53 stress-response genes such as Bbc3/PUMA and Cdkn1a/p21 at both transcript and protein levels (Figures 3b–e; Supplementary Figure S3a). The knockdown Ba/F3 cells therefore reproduced all the major phenotypic and transcriptional features of Mysm1−/− HSPCs, despite having only partial reduction in MYSM1 protein. This confirms that shMysm1-Ba/F3 cells are an appropriate model for assessing the role of MYSM1 in regulating p53-target gene expression.
Figure 3.
MYSM1 regulates p53-target gene expression in hematopoietic progenitors. (a) Validation of reduced Mysm1 transcript and protein levels in the Mysm1-knockdown Ba/F3 hematopoietic progenitor cells (shMysm1, black bars), relative to control lines expressing shFF (gray bars). Gene expression normalized to Actin. (b) Measurement of cell death in Ba/F3 cells at 7, 9, and 14 h following 3-Gy irradiation. (c) Immunoblot showing levels of total p53 protein with ACTIN-loading control following irradiation. (d) Expression of p53-stress-response genes in knockdown shMysm1 cells and control shFF cells over a 3-h time course following 3-Gy irradiation. Normalization against Actin expression. (e) Immunoblots showing protein levels of p21, PUMA, MDM2, and ACTIN-loading control in knockdown shMysm1 and control shFF cells at a steady state and following 3-Gy irradiation. All data in a–e are representative of three independent shMysm1 and shFF lines. Bars in a, b, d represent mean±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 using Student's t-test. (f) Effects of MYSM1 on chromatin modification and p53 recruitment to the Bbc3/PUMA gene promoter in untreated and 3-Gy-irradiated cells (3 h). Fold enrichment of histone H3K4 tri-methylation (H3K4me3), histone H3K27 acetylation (H3K27ac), and p53 at the Bbc3/PUMA promoter in Mysm1 knockdown (shMysm1) and control (shFF) Ba/F3 cells analyzed using ChIP-qPCR. Enrichment values (shown above each bar) were calculated relative to control IgG for p53, and relative to total histone H3 for the histone marks using Pomc as a negative genomic region. Genomic structure of the locus with target primer sites is provided in Figure 2a. Data are representative of two independent experiments, with extra replicates in Supplementary Figure S3c. (g) Schematic summary of mechanism: MYSM1 antagonizes p53-induced Bbc3/PUMA gene expression in hematopoietic progenitors by inhibiting p53 recruitment and chromatin modification (H3K4me3 and H3K27ac)
We analyzed p53 recruitment and histone modification at the Bbc3/PUMA and Cdkn1a/p21 promoters in untreated and irradiated Mysm1-knockdown cells. At the Bbc3/PUMA promoter, Mysm1 knockdown resulted in increased p53 recruitment and striking increases in histone H3 lysine 27 acetylation (H3K27ac) and histone H3 lysine 4 trimethylation (H3K4me3) histone modifications (Figures 3f and g; Supplementary Figure S3b). At the Cdkn1a/p21 locus, increased H3K27ac and H3K4me3 levels were also observed, but without increased p53 binding (Supplementary Figure S3b), which indicates that MYSM1 can have an impact on H3K27ac levels independently of effects on p53 recruitment. Increased levels of H3K27ac and H3K4me3, hallmarks of promoter activity, are known to correlate with increased transcription. Therefore, even partial Mysm1 knockdown results in significant changes in chromatin dynamics at its binding sites within p53 target gene promoters. As the chromatin state represents a propensity for transcription, we conclude that MYSM1 acts as a transcriptional cofactor that restricts p53 stress-response transcriptional activity by modulating the epigenetic state of activation at p53-target gene regulatory elements (Figures 3f and g; Supplementary Figure S3b).
Testing the roles of PUMA and p21 as mediators of Mysm1-deficiency phenotypes
Our data implicate MYSM1 as a transcriptional regulator of p53-target genes Bbc3/PUMA and Cdkn1a/p21, with MYSM1 loss resulting in increased p53 transcriptional activity in vitro and in vivo (Figures 1–3). Considering our recent demonstration that Mysm1-deficiency phenotypes are p53-mediated,38 we tested the roles of PUMA and p21 as specific mediators of Mysm1−/− phenotypes by generating Mysm1−/−Puma−/− and Mysm1−/−Cdkn1a−/− mouse lines.
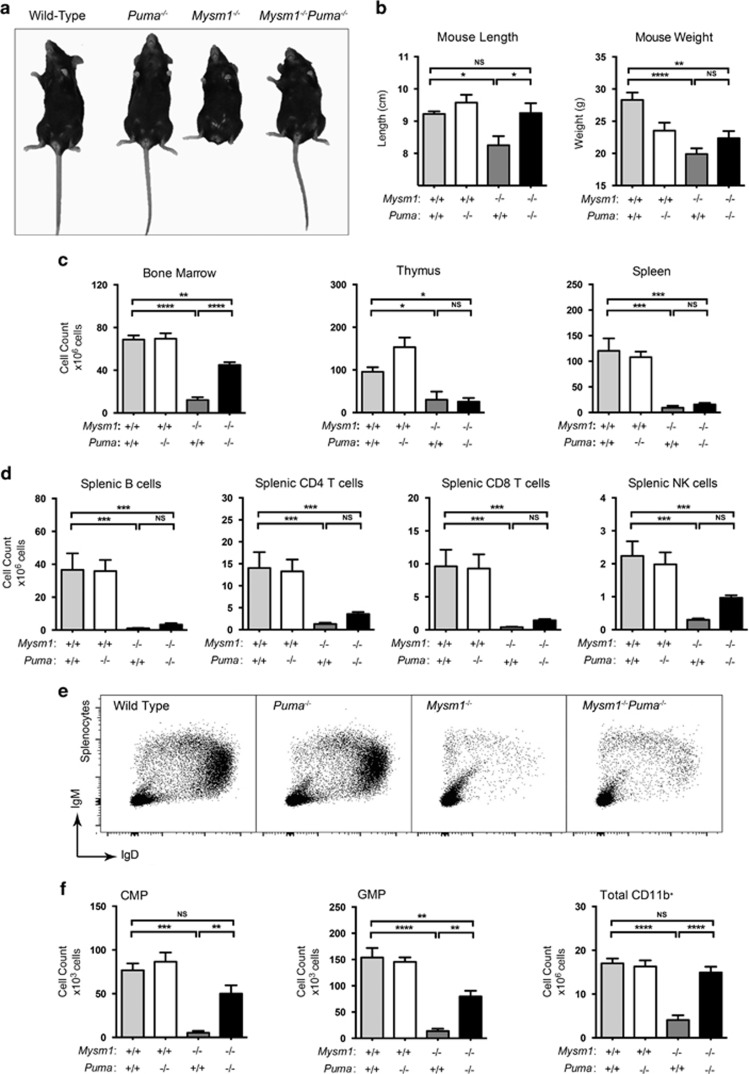
Mysm1−/−Puma−/− mice showed rescue of Mysm1−/− coat pigmentation defects, hindlimb and tail dysmorphology, and partial rescue of growth retardation (Figures 4a and b). There was a significant increase in bone marrow cellularity in Mysm1−/−Puma−/− relative to Mysm1−/− controls, suggesting some rescue of hematopoiesis; however, thymus and spleen cellularity remained low (Figure 4c). In contrast, Mysm1−/−Cdkn1a−/− mice showed high levels of embryonic and neonatal mortality, and the few surviving animals had no observable rescue of Mysm1-deficiency phenotypes (Supplementary Figures S5 and 6). Overall, this implicated PUMA, but not p21, in mediating developmental and hematopoietic abnormalities in Mysm1 deficiency.
Figure 4.
The developmental and hematopoietic phenotypes of Mysm1−/− mice are partially mediated by Bbc3/PUMA. The presented data are from mice of the following genotypes: wild-type, Mysm1+/+Puma−/−, Mysm1−/−Puma+/+, and Mysm1−/−Puma−/−. (a) Representative photograph showing overall appearance of the mice. (b) Lengths and weights of the mice. (c) Absolute cell counts of the bone marrow, thymus, and spleen of the mice. (d) Absolute numbers of splenic B cells (B220+), CD4 T cells, CD8 T cells, and NK cells (CD3−NK1.1+) in the mice. (e) Representative flow cytometry plots of the spleen stained for B-cell markers IgM and IgD. (f) Absolute numbers of common myeloid progenitors (CMP, Lin−cKit+Sca1−IL7R−CD34+CD16/CD32−), granulocyte–macrophage progenitors (GMP, Lin−cKit+Sca1−IL7R−CD34+CD16/CD32+), and total CD11b+ cells in the bone marrow of the mice. Bars represent mean±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, NS, nonsignificant using ANOVA with Bonferroni post hoc test; data from four to five mice per group and reproduced in two independent experiments
Lymphocyte development arrest in Mysm1 −/− mice is PUMA-independent
B cells, CD4 T cells, and CD8 T cells remained strongly diminished in Mysm1−/−Puma−/− spleens (Figures 4d and e). There was also no significant rescue of common lymphoid progenitor cells (CLPs), bone marrow B-cell precursors, or thymic T-cell precursors in Mysm1−/−Puma−/− mice relative to Mysm1−/− controls (Supplementary Figure S4). We conclude that the lymphoid-lineage differentiation arrest in Mysm1−/− mice is independent of Bbc3/PUMA, and is mediated through other p53 effector functions, as it was rescued in Mysm1−/−p53−/− mice.38, 39
PUMA drives MPP cell depletion in Mysm1 deficiency
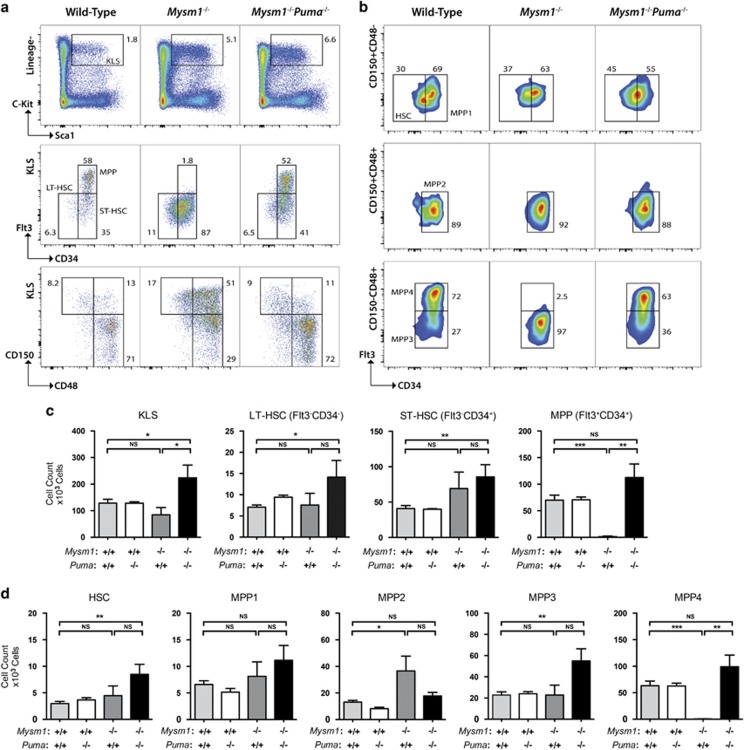
Surprisingly, the severe loss of lymphoid-primed MPPs (MPP4s; cKit+Lin−Sca1+CD150−CD48+Flt3+CD34+) characteristic of Mysm1 deficiency was fully rescued in Mysm1−/−Puma−/− mice.31, 33, 37 This implicates PUMA-mediated apoptosis in the depletion of this cell population (Figure 5). Restricted myeloid progenitor cell numbers were also rescued, further demonstrating that the p53/PUMA axis mediates Mysm1−/− hematopoietic dysfunction (Figure 4f). Moreover, we observed an expansion of HSC numbers in Mysm1−/−Puma−/− mice not only relative to the Mysm1−/− group but also significantly above wild-type and Puma−/− levels (Figure 5; Supplementary Figure S7). This suggested that the loss of HSC quiescence previously reported in the Mysm1−/− line33 persists in Mysm1−/−Puma−/− mice. We conclude that PUMA is the essential non-redundant mediator of p53-driven depletion of MPP4s and myeloid lineage cells in Mysm1 deficiency.
Figure 5.
Bbc3/PUMA deletion rescues the MPP cell deficiency of Mysm1-knockout mice. (a) Representative flow cytometry plots showing Lin−cKit+Sca1+ (KLS) HSPCs within the bone marrow of wild-type, Mysm1−/−, and Mysm1−/−Puma−/− mice (top panel) and further characterizing these cells based on the expression of CD34 and Flt3 markers (middle panel), or CD150 and CD48 markers (bottom panel). (b) Representative flow cytometry plots of the bone marrow from wild-type, Mysm1−/−, and Mysm1−/−Puma−/− mice gated on Lin−cKit+Sca1+ KLS cells and showing HSCs (CD150+CD48−CD34−Flt3−, top panel), MPPs MPP1 (CD150+CD48−CD34+Flt3−, top panel), MPP2 (CD150+CD48+CD34+Flt3−, middle panel), MPP3 (CD150−CD48+CD34+Flt3−, bottom panel), and MPP4 (CD150−CD48+CD34+Flt3+, bottom panel) cell populations. (c) Absolute cell counts of long-term stem cells (LT-HSCs, KLS CD34−Flt3−), short-term stem cells (ST-HSCs, KLS CD34+Flt3), and MPPs (KLS CD34+Flt3+). (d) Absolute cell counts of HSC, and MPP1–4 populations, as defined in Figure 5b. Bars represent mean±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, NS, nonsignificant using Student's t-test. Bone marrow cell numbers are presented per two tibias and femurs. Data are from ≥4 mice per group and reproduced in at least two independent experiments
Partial rescue of Mysm1−/−Puma−/− HSC function
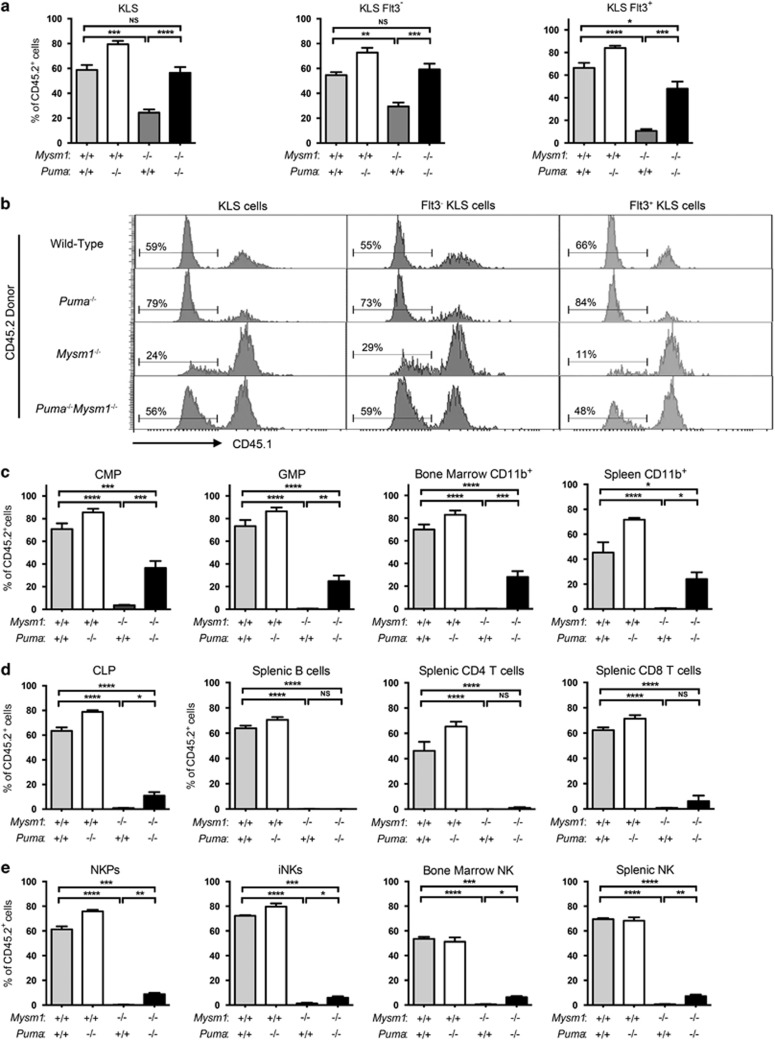
Bone marrow chimeras were employed to further assess the role of Bbc3/PUMA in Mysm1-deficient HSPC dysfunction. B6.SJL-CD45.1 bone marrow was mixed with CD45.2 bone marrow from wild-type, Mysm1−/−, Puma−/−, or Mysm1−/−Puma−/− mice, and were transplanted into four groups of lethally irradiated recipients. As previously reported, Mysm1−/− donors were strongly impaired in hematopoietic reconstitution capacity.31, 33 However, the performance of Mysm1−/−Puma−/− donors was significantly enhanced in comparison (Figure 6). Specifically, Mysm1−/−Puma−/− bone marrow was rescued in HSPC compartment reconstitution (Figures 6a and b), indicating partial rescue of HSC functions. There was a strongly improved contribution to the myeloid cell lineage, including progenitors and total CD11b+ cells of the bone marrow, spleen, and blood (Figure 6c, Supplementary Figure S9). Lymphoid-lineage reconstitution, however, remained severely impaired, with no rescue of B- and T-cell reconstitution, and only a minor increase in the reconstitution of CLP and NK cells (Figures 6d and e,Supplementary Figures S8 and 9). The results show a significant yet incomplete rescue of Mysm1−/−Puma−/− HSC function and further establish that Bbc3/PUMA is an important mediator of the hematopoietic failure in Mysm1 deficiency.
Figure 6.
Partial rescue of hematopoietic stem and progenitor cell functions in Mysm1−/−Puma−/− mice. Competitive bone marrow transplantations were set up; B6.SJL-CD45.1 bone marrow was mixed with CD45.2 bone marrow from wild type, Puma−/−, Mysm1−/−, or Mysm1−/−Puma−/− mice, and was transplanted into four independent groups of lethally irradiated recipients. Recipients were analyzed for the relative contribution of CD45.2 donor cells to hematopoiesis at 20 weeks after reconstitution. (a) Percentage contribution of wild-type, Puma−/−, Mysm1−/−, or Mysm1−/−Puma−/− donors to Lin−cKit+Sca1+ (KLS) cells, Flt3− KLS HSCs, and Flt3+ KLS MPP cells. (b) Flow cytometry histograms of the bone marrow of the chimeric mice gated on Lin−cKit+Sca1+ cells (KLS, left), Flt3− KLS HSCs (middle), or Flt3+ KLS MPP cells (right). Histogram gates and percentages indicate CD45.2+CD45.1− donor cells. (c–e) Percentage contribution of wild type, Puma−/−, Mysm1−/−, or Mysm1−/−Puma−/− donors to (c) CMPs (Lin−cKit+Sca1−IL7R−CD34+CD16/32−), granulocyte-macrophage progenitors (GMP, Lin−cKit+Sca1−IL7R−CD34+CD16/32+), and CD11b+ myeloid lineage cells in the bone marrow and spleen; (d) CLPs (Lin−cKit+Sca1+IL7R+), splenic B cell (B220+), splenic CD4 T cell, and splenic CD8 T-cell populations; (e) NK cell progenitors (NKPs, Lin−CD122+NK1.1−CD11b−), immature bone marrow NK cells (iNKs, Lin−CD122+NK1.1+CD11b−), and mature NK cells in the bone marrow (Lin−CD122+NK1.1+CD11b+) and spleen (CD3−NK1.1+). Bars show means±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, NS, nonsignificant using ANOVA with Bonferroni post-hoc test
PUMA mediates apoptosis and other stress phenotypes in Mysm1−/− HSPCs
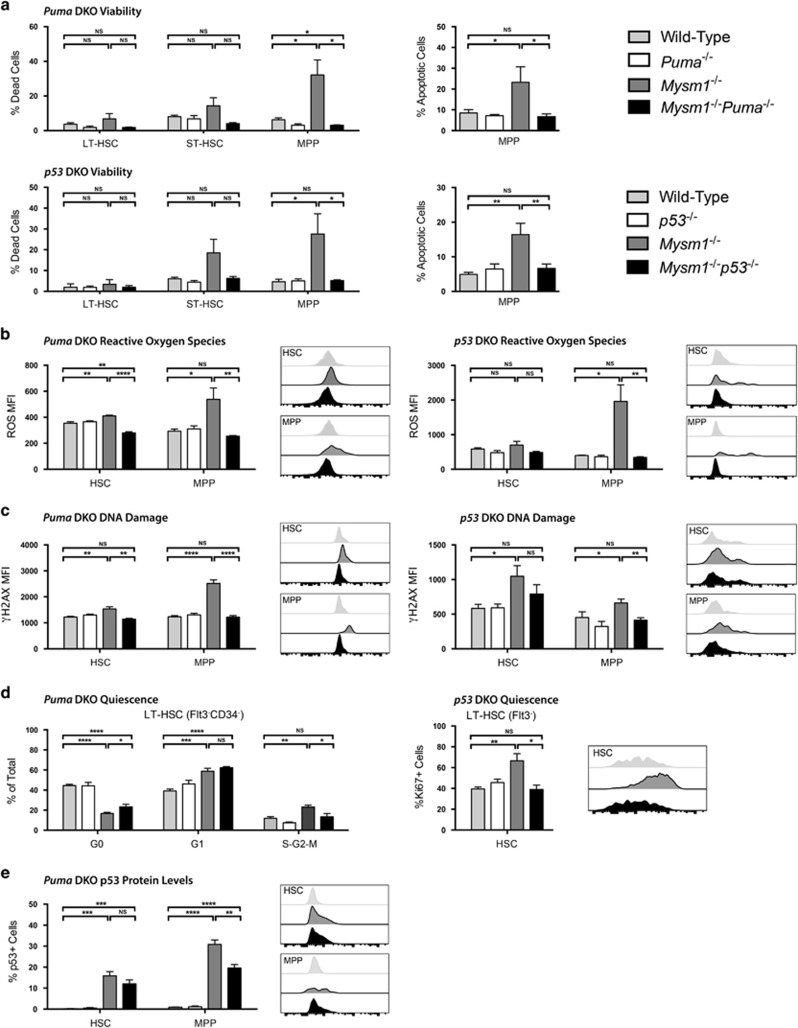
Our data indicate that PUMA is an important mediator of MPP4 depletion and HSC dysfunction in Mysm1 deficiency. To further explore the p53/PUMA axis as the mediator of Mysm1−/− HSPC stress phenotypes,31, 33 we analyzed Mysm1−/−Puma−/− and Mysm1−/−p53−/− HSPCs for oxidative stress, DNA damage, and apoptosis. Increased levels of apoptosis in Mysm1−/− MPP4 cells were fully rescued upon deletion of either p53 or Bbc3/PUMA (Figure 7a), indicating that Mysm1−/− MPP4s are depleted via p53-dependent apoptosis, with PUMA as its primary non-redundant mediator. Increases in reactive oxygen species (ROS) and DNA damage (Phospho-Serine 139 Histone H2A Variant X, γH2AX) in Mysm1−/− HSPCs were also restored to normal levels in Mysm1−/−p53−/− and Mysm1−/−Puma−/− mice (Figure 7b and c). This indicates that they are neither direct products of MYSM1 loss nor primary inducers of p53, but rather downstream results of p53-PUMA activation and apoptosis.45, 46
Figure 7.
p53 and PUMA as mediators of cell death and other stress phenotypes in Mysm1-deficient HSPCs. All data presented are either from Puma- double knockout (DKO) mice: wild-type (gray), Puma−/− (white), Mysm1−/− (charcoal), and Mysm1−/−Puma−/− (black); or from p53-DKO mice: wild-type (gray), p53−/− (white), Mysm1−/− (charcoal), and Mysm1−/−p53−/− (black). Cell gating was as follows: HSCs (Lin−cKit+Sca1+Flt3−) and MPPs (Lin−cKit+Sca1+Flt3+); or LT-HSCs (Lin−cKit+Sca1+Flt3−CD34−), ST-HSCs (Lin−cKit+Sca1+Flt3−CD34+) and MPPs (Lin−cKit+Sca1+Flt3+CD34+). (a) Cell death was assessed by the uptake of an amine-reactive fixable viability dye, and apoptosis was assessed with Annexin V staining. (b) Reactive oxygen species levels were quantified using the H2DCFDA dye. (c) DNA damage was measured by staining for γH2AX protein levels. (d) HSC quiescence for Puma-DKO mice was evaluated by staining for Ki67 protein levels and DNA content with 7-AAD (left panel) with G0 (Ki67− 2 N DNA), G1 (Ki67+ 2 N DNA), and S-G2-M (Ki67− >2 N DNA) phase shown. HSC quiescence for p53-DKO mice was evaluated by staining for Ki67 protein levels only (right panel) with G0 (Ki67−) and G1-S-G2-M (Ki67+) phases shown. (e) p53 levels in Puma-DKO mice were assessed using p53 antibody (1C12) staining. Data for viability and reactive oxygen species represent total cells; data for γH2AX, Ki67, and p53 represent live cells only. MFI is the mean fluorescence intensity; percentage of positive cells refers to cells with fluorescence intensity above FMO control levels. Histogram overlays show representative FACS plots of mice from wild-type, Mysm1−/−, and DKO groups only. Bars show the mean±S.E.M.; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, NS nonsignificant using Student's t-test; data are from five to nine mice per group and are representative of at least two independent experiments
Previous studies reported reduced quiescence and increased proliferation of Mysm1−/− HSCs.33 Using Ki67 as a marker of non-quiescent cells, we show that Mysm1−/− HSC loss of quiescence is rescued in Mysm1−/−p53−/− HSCs (Figure 7d, right). This is consistent with the full rescue of Mysm1−/−p53−/− HSC functions reported in our recent work.38 Cell-cycle analysis was also performed in Mysm1−/−Puma−/− HSCs by staining for Ki67 and DNA content to resolve G0, G1, and S–G2–M cell-cycle phases. This revealed a partial rescue, with normalized percentages of proliferating Mysm1−/−Puma−/− HSCs in the S–G2–M phases of the cell cycle, but persistent loss of quiescence with a significant reduction in G0-phase cells and increase in G1-phase cells (Figure 7d, left). The significant but incomplete rescue of HSC quiescence in the Mysm1−/−Puma−/− model is consistent with the aforementioned partial rescue of Mysm1−/−Puma−/− HSC functions (Figure 6).
Finally, we assessed p53 protein levels in Mysm1−/−Puma−/− mice and found that they were still significantly elevated in both HSC and MPP populations, although partially reduced in the latter (Figure 7e). Overall, this suggests that continued dysregulation of p53 effector functions beyond the upregulation of Bbc3/PUMA contributes to persistent Mysm1−/− HSC loss of quiescence, and also to the lymphoid differentiation arrest of Mysm1−/−Puma−/− MPP4s.
Mysm1−/−Puma−/− MPP transcriptome reveals persistent p53 stress-response activation
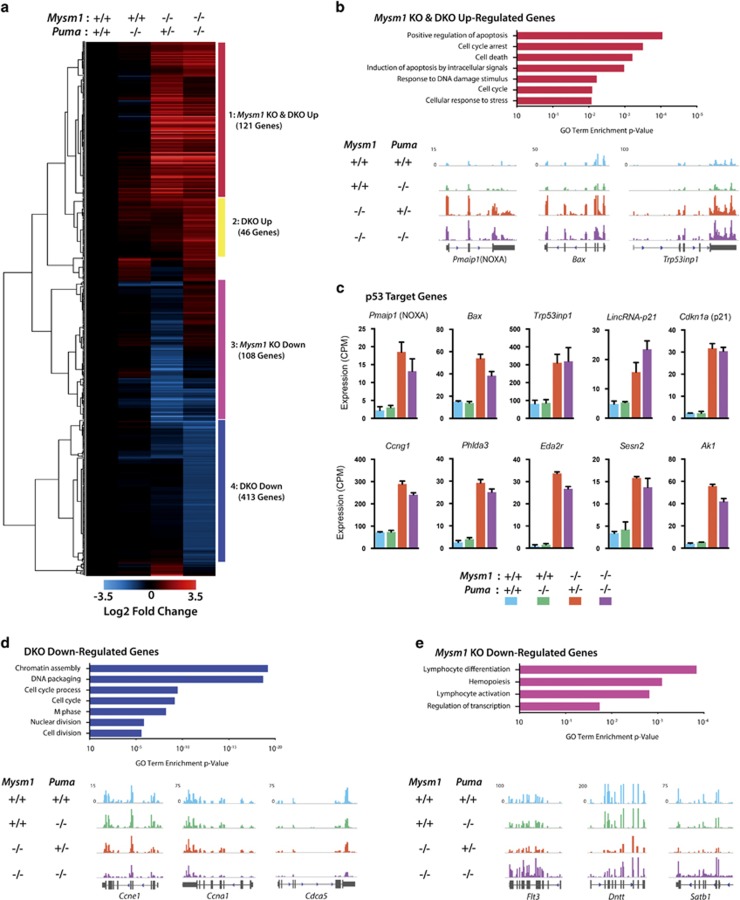
We establish PUMA as the primary effector of p53-mediated depletion of MPP4 and myeloid cells, but not CLPs and lymphoid cells in Mysm1 deficiency. To gain insight into the functional defects in Mysm1−/−Puma−/− MPP4s contributing to cell death and downstream lymphopoiesis failures, we performed RNA-sequencing (RNA-Seq) of MPPs (Lin−cKit+Sca1+CD150−) from Mysm1−/−Puma−/−, Mysm1−/−Puma+/−, Puma−/−, and wild-type mice (Mysm1−/−Puma+/− phenocopies Mysm1−/−). Unbiased clustering analysis of differentially regulated genes yielded four main groups (Figure 8a). Puma−/− cells displayed no significant gene expression differences as compared with wild-type controls.
Figure 8.
Transcriptome analysis of Mysm1−/−Puma−/− MPPs reveals widespread p53 stress-response activation and rescue of lymphoid-priming genes. Data shown represent RNA-Seq analysis of FACS-sorted MPPs from wild-type, Puma−/−, Mysm1−/−Puma+/− (phenocopy Mysm1−/−), and Mysm1−/−Puma−/− mice. (a) Hierarchical clustering of differentially regulated genes (defined as statistically significant based on adjusted P-values <0.05, and with ≥2-fold change in expression over wild type in at least one group). Four major clusters are labeled and color-coded. Heatmap scale represents Log2 of fold change. All representative RNA-Seq tracks and bar charts in other panels show genes included in heat map. (b) Mysm1 KO and DKO upregulated gene cluster 1: genes upregulated in Mysm1−/−Puma−/− and Mysm1−/− as compared with wild type. Gene ontology enrichment analysis and RNA-seq tracks representative of expression trends in this cluster. (c) p53 target genes from heatmap cluster 1 that were significantly upregulated (more than twofold change) in both Mysm1−/−Puma−/− (purple bars) and Mysm1−/− (orange bars) as compared with wild-type (blue bars) MPPs. All expression values shown as normalized counts per million reads (CPM). (d) DKO downregulated cluster 4: genes downregulated in Mysm1−/−Puma−/− and unchanged in Mysm1−/− as compared with wild type. Gene ontology enrichment analysis and RNA-seq tracks representative of expression trends in this cluster. (e) Mysm1 KO downregulated cluster 3: genes downregulated in Mysm1−/− and unchanged in Mysm1−/−Puma−/− as compared with wild type. Gene ontology enrichment analysis and RNA-seq tracks representative of expression trends in this cluster. Note: Flt3 was significantly differentially expressed between Mysm1−/− and wild-type groups but had less than twofold change, so did not pass the heatmap threshold. Data from three mice per group; RNA-Seq tracks represent composite files of all three data sets within each genotype
In cluster 1, the strong p53 stress-response signature of Mysm1−/−Puma+/− MPPs, and Mysm1−/− HSPCs (Figure 1), was fully retained in Mysm1−/−Puma−/− MPPs (Figures 8a–c; Supplementary Figures S11a–c). Gene ontology analysis revealed that genes implicated in ‘positive regulation of apoptosis' and ‘cell-cycle arrest' were strongly enriched (Figure 8b), including p53-target genes such as Pmaip1/NOXA, Bax, and Cdkn1a/p21 (Figure 8c, Supplementary Figures S11a–c), and downstream apoptotic mediators including Casp12 and Bcl2l11/BIM (Supplementary Figure S11f). This indicates a persistent activation of p53-mediated stress-response pathways in Mysm1−/−Puma−/− MPPs.
In cluster 4, Mysm1−/−Puma−/− MPPs showed a striking downregulation of genes required for cell-cycle progression, such as Ccne1, Ccna1, Cdca5, histone genes,47 and many others, compared with control and Mysm1−/−Puma+/− levels (Figure 8d). This was associated with a reduced proportion of cells in the S–G2–M cell-cycle phases (Supplementary Figure S10). However, HSC-specific quiescence regulators were either unchanged or undetectable in expression in MPPs across all genotypes (Supplementary Figure S11i; Figure 1). The downregulation of cell-cycle progression genes, together with the upregulation of p53 effectors in Mysm1−/−Puma−/− MPPs further supports persistent stress-response activation in these cells. Importantly, as with HSPC qPCR analysis (Supplementary Figure S1), Cdkn2a was not expressed in Mysm1-deficient MPPs (Supplementary Figure S11d), further distinguishing MYSM1 functions in HSPCs and thymocytes.39
In cluster 3, the downregulation of many lymphoid-lineage genes observed in Mysm1−/−Puma+/− cells was rescued in Mysm1−/−Puma−/− cells (Figure 8e; Supplementary Figure S11h). Interestingly, Flt3, Satb1, Dntt, Il7r, and most other genes in this cluster were recently shown to be upregulated in the differentiation of lymphoid-primed MPP4 cells.48 This indicates that their downregulation in Mysm1 deficiency represents PUMA-dependent death of MPP4s, rather than an intrinsic requirement for MYSM1 to induce their expression. Notably, all the genes implicated as direct MYSM1 targets in previous studies, including Gfi1, Flt3, Id2, and Ebf1,32, 33, 36, 37 did not differ in expression between Mysm1−/−Puma−/− and control MPPs (Supplementary Figure S11g). This indicates that, at least in MPPs, Mysm1-deficiency phenotypes are independent of these genes, and instead are p53 stress-response-mediated, as supported by all our other analyses.
We conclude that the rescued Mysm1−/−Puma−/− MPP4 viability, despite a strong upregulation of other apoptotic mediators such as Pmaip1/NOXA, Bax, and others, demonstrates the key non-redundant role of PUMA as the mediator of p53-dependent apoptosis within the MPP4 population. The other upregulated p53 stress-response genes identified in the transcriptome analysis likely contribute to the p53-dependent but PUMA-independent phenotypes in Mysm1 deficiency.
Discussion
Our work demonstrates that MYSM1 represses p53 stress-response gene expression by localizing to p53-binding sites within Bbc3/PUMA and Cdkn1a/p21 promoters and antagonizing local histone modification (H3K27ac and H3K4me3) and p53 recruitment (Figure 3g). We further establish that PUMA, but not p21, is an important mediator of the p53-driven hematopoietic dysfunction in Mysm1 deficiency, with Mysm1−/−Puma−/− mice showing full rescue of MPP4 cell viability, partial rescue of HSC functions, but persistent depletion of CLPs and lymphopenia. We demonstrate that PUMA is an essential non-redundant effector of p53-induced MPP4 apoptosis, and through global transcriptome analysis of Mysm1−/−Puma−/− MPPs identify other p53 effectors that remain induced and may contribute to the other p53-dependent but PUMA-independent Mysm1−/− phenotypes. Whereas recent studies established p53 activation as the common mediator of all Mysm1−/− hematopoietic phenotypes,38, 39 our current work provides novel mechanistic insight into the distinct outcomes of p53 activation in different hematopoietic populations. This highlights the Mysm1−/− and Mysm1−/−Puma−/− lines as unique models for the study of p53 hyperactivity within different hematopoietic populations without exogenous stress. We conclude that MYSM1 is an essential regulator of p53 transcriptional programs in hematopoiesis, acting to maintain hematopoietic progenitor viability by antagonizing p53-driven Bbc3/PUMA expression.
Although PUMA is a major mediator of p53-dependent apoptosis, its functions overlap significantly with other BH3-only proteins, for example, with NOXA in lymphocyte responses to irradiation, in B-cell lymphomagenesis, and in non-hematopoietic cells such as fibroblasts and oocytes.22, 23, 49 Similarly, BIM contributes to apoptosis during lymphocyte development50 and exposure to mutagens.24, 25 In contrast, we demonstrate an essential non-redundant role of PUMA in p53-mediated apoptosis of MPP4s but not CLPs in the context of Mysm1 deficiency. Transcriptional profiling of Mysm1−/−Puma−/− MPPs provides valuable insights into the p53 transcriptional network of this hematopoietic population, and identifies upregulation of many p53-target genes, including pro-apoptotic Pmaip1/NOXA, Bax, Trp53inp1, and others. The full rescue of Mysm1−/−Puma−/− MPP4 viability, despite strong upregulation of other apoptotic mediators, further demonstrates that PUMA is the key effector of MPP4 apoptosis. Considering that Mysm1−/−p53−/− mice displayed complete rescue of hematopoietic dysfunction,38 the other p53 effectors identified in our Mysm1−/−Puma−/− transcriptome analysis may contribute to the PUMA-independent but p53-dependent Mysm1−/− phenotypes.
Lymphoid cell depletion in Mysm1 deficiency was previously attributed to several mechanisms, involving either MYSM1-mediated Cdkn2a/p19ARF repression to restrict p53 activation in thymocytes,39 or MYSM1-mediated induction of lymphoid-lineage genes, such as Ebf1 and Id2.32, 36 Importantly, Cdkn2a expression was undetectable in Mysm1-deficient HSPCs or MPPs, demonstrating the distinct mechanism of p53 activation in Mysm1−/− HSPCs, as compared with thymocytes.39 Furthermore, the full-transcriptome analysis of Mysm1−/−Puma−/− MPPs showed no global changes in lymphoid-lineage priming, and normal expression of all genes previously implicated as MYSM1 targets.32, 36, 33, 37 Although lymphoid genes were downregulated in Mysm1−/− MPPs, this reflected PUMA-mediated death of lymphoid-primed MPP4 cells, as the gene expression was restored in Mysm1−/−Puma−/− MPPs together with restoration of MPP4 viability. This shows that MYSM1 has no direct role in transcriptional regulation of MPP lymphoid priming, but instead maintains MPP4 survival by regulating p53 transcriptional programs. Lymphoid cell depletion via p53-dependent38, 39 but PUMA-independent mechanisms suggests the involvement of other p53-induced effectors. The upregulation of p53-regulated pro-apoptotic mediators, together with downregulation of many genes essential for normal cell-cycle progression in Mysm1−/−Puma−/− MPPs, may contribute to the p53-mediated depletion of downstream CLPs and lymphopenia in Mysm1 deficiency.
Our study also provides important insights into the unique effects of p53 stress responses in HSCs, where p53 activation in Mysm1 deficiency does not result in apoptosis, but promotes loss of quiescence. Our findings support the conclusions of several studies, which indicated that HSCs are more resistant to apoptosis compared with downstream progenitors, and have lower induction of pro-apoptotic genes in response to exogenous stress such as irradiation.10, 26, 27, 28, 51 We also demonstrate that Mysm1−/− HSC hyperproliferation is in part a compensatory effect driven by PUMA-dependent depletion of downstream progenitors, similar to the compensatory mechanisms operating following irradiation-induced MPP depletion.27, 51
Although the rescue of HSC quiescence and function in Mysm1−/−Puma−/− mice was significant, it was incomplete. This represents an important distinction from the complete rescue in Mysm1−/−p53−/− mice and indicates a cell-intrinsic role of MYSM1 in HSC homeostasis in crosstalk with p53. p53 is known to promote homeostatic HSC quiescence by regulating its HSC-specific targets Gfi1 and Necdin.11, 16, 17 We confirm downregulation of Gfi1 in Mysm1−/− HSPCs,33 and also show downregulation of Necdin. This suggests that MYSM1 may have distinct effects at different p53-target genes, repressing classical stress-response genes such as Bbc3/PUMA to promote survival while supporting expression of HSC-specific targets such as Gfi1 and Necdin to maintain quiescence (Supplementary Figure S12). Although MYSM1 was initially characterized as a transcriptional activator acting via histone H2AK119 deubiquitination,30, 52 recent studies linked H2AK119 deubiquitination by other proteins to transcriptional repression.53 Our work shows that MYSM1 represses p53 stress-response genes by antagonizing other histone modifications (H3K27ac and H3K4me3). Further understanding of the mechanisms through which MYSM1 regulates p53 transcriptional activity in HSCs to maintain HSC quiescence and function is an important focus for future work.
In conclusion, we establish MYSM1 as an essential regulator of p53 transcriptional activity and stress-response programs in HSPCs. We demonstrate that PUMA is an essential non-redundant mediator of p53-induced apoptosis in MPPs but not lymphoid cells in the context of Mysm1 deficiency. Together with the characterization of p53 stress-response transcriptional signatures in MPPs, this provides valuable novel insights into the cell-type-specific roles of p53 in hematopoiesis. Our work advances the understanding of human bone marrow failures resulting from MYSM1 mutations,34, 35 and highlights MYSM1 as a potential drug target for the activation of p53 and PUMA in hematopoietic progenitors for myeloablation or leukemia treatment.7, 8, 54
Materials and Methods
Mouse lines
Mysm1tm1a(KOMP)WTSI mice derived from EPD0019_1_A05 ESC clone were previously described31, 55 and have over 100-fold reduction in the Mysm1 transcript in a homozygous state (referred here as Mysm1−/−). Mysm1flox/flox line was generated by crossing Mysm1tm1a mice to the Gt(ROSA)26 Sortm1(FLP1)Dym/Wtsi line with ubiquitous expression of Flp recombinase,56 and subsequently bred to Gt(ROSA)26Sortm1(cre/ERT2) mice for tamoxifen-induced Mysm1 inactivation.40 The p53 knockout and Bbc3/PUMA knockout lines were obtained from The Jackson Laboratory (JAX002101 and JAX011067). All lines were on C57BL/6 background. The mice were maintained under specific pathogen-free conditions; all experiments were in accordance with the guidelines of the Canadian Council on Animal Care, and protocols approved by the McGill Animal Care Committee.
Mouse procedures
For competitive bone marrow transplantation, recipient B6.SJL-PtprcaPepcb/Boy mice (JAX002014, congenic for CD45.1) were irradiated with two doses of 4.5 Gy, delivered 3 h apart, in the RS2000 irradiator (Rad Source, Brentwood, TN, USA). The mice were intravenously injected with 2.5 × 106 bone marrow cells from the following donor mice in a 1 : 1 ratio: B6.SJL cells mixed with C57BL/6 cells from either wild-type, Mysm1−/−, Puma−/−, or Mysm1−/−Puma−/− mice. The mice were kept on neomycin in drinking water (2 g/l, BioShop, Burlington, ON, Canada) for 3 weeks, and were killed at 20 weeks for immunophenotypic analysis. Bleeds were performed at 12.5 weeks post transplantation.
For tamoxifen-induced Mysm1 gene inactivation in the Mysm1flox/flox Gt(ROSA)26Sortm1(cre/ERT2) mouse line, the mice were injected intraperitoneally with tamoxifen (T5648, Sigma, St. Louis, MO, USA) in sterilized corn oil (Sigma C8267) at 0.15 mg/g per injection, with eight doses in total administered over 16 days. The mice were analyzed 16 weeks after the treatment. Successful deletion of Mysm1 exon 3 was validated by genotyping on genomic DNA extracted from hematopoietic and lymphoid organs with primers Mysm1_wt_F5 5′-CCACAGTGATTCCTGGCTG-3′, Mysm1_wt_Rv6 5′-CTAGGCTTCAGGCATTTTGC-3′, Mysm1_wt_R7 5′-CCACATGTTCTTGACCTTGC-3′,40 and the loss of Mysm1 transcript ENSMUST00000075872 was further confirmed using qRT-PCR analysis of HSPCs FACS-sorted from the mouse bone marrow. Mysm1flox/+ heterozygous mice expressing the same Gt(ROSA)26Sortm1(cre/ERT2) transgene and undergoing identical tamoxifen treatment, as well as sham-mice treated with corn-oil only, were used as control groups.
Flow cytometry
Cell suspensions of mouse tissues were prepared in RPMI-1640 (Wisent, Jean-Baptiste, QC, Canada) with 2% (v/v) fetal calf serum (FCS), 100 μg/ml streptomycin, and 100 U/ml penicillin (Wisent). Cells were stained for surface markers in PBS with 2% FCS and 0.2% (w/v) sodium azide (VWR, Amresco, Solon, OH, USA) for 20 min on ice, with the following fluorophore-conjugated antibodies: FITC-conjugated antibody against CD34 (clone RAM34), CD48 (HM48-1, both from eBioscience, San Diego, CA, USA) or CD127 (A7R34, BioLegend, San Diego, CA, USA); PE-conjugated antibodies against CD11b (M1/70, BD Biosciences, Franklin Lakes, NJ, USA), CD150 (mShad150, eBioscience), IgM (II/41, eBioscience), or CD135/FLT3 (A2F10, BioLegend); PerCP/Cy5.5 antibodies against: CD45R/B220 (RA3–6B2), CD11b (M1/70), TER119 (TER119), CD4 (RM4–5), CD8a (53-6.7, all from BioLegend); APC antibodies against CD27 (LG3A10), CD49b (DX5), IgD (11–26c.2a), Sca-1 (E13–161.7, all from BioLegend), and Sca-1 (D7, eBioscience); Alexa Fluor 647 anti-CD150 (TC15-12F12.2, BioLegend); Pacific Blue antibodies against CD8a (53-6.7), CD11b (M1/70), cKit/CD117 (2B8, all from BioLegend); eFluor450 antibodies against CD48 (HM48-1) or CD122 (TM-b1, both from eBioscience); BV421 anti-CD34 (RAM34, BD Biosciences); PE-Cy7 antibodies against CD4 (RM4–5), CD19 (6D5), CD45.2 (104), CD71 (RI7217, all from BioLegend), and cKit/CD117 (2B8, BD Biosciences); APC-Cy7 antibodies against: CD45 (30-F11), CD45.1 (A20), or Sca1 (D7, all from BioLegend). Fixable Viability Dye eFluor 506 (eBioscience) was used to discriminate live and dead cells. Annexin V PeCy7 (eBioscience) was used for detection of apoptotic cells. Compensation was performed with BD CompBeads (BD Biosciences). The data were acquired on FACS Canto II flow cytometer (BD Biosciences) and analyzed with FACS Diva (BD Biosciences) or FlowJo (Tree Star, Ashland, OR, USA) software.
For intracellular staining, samples were pre-stained with Fixable Viability Dye eFluor506 (eBioscience), APC anti-Sca1 (D7, eBiosciences), Pacific Blue anti-cKit/CD117 (2B8, BioLegend), and PE anti-CD135/Flt3 (A2F10, BioLegend). The cells were fixed in 2% paraformaldehyde in PBS with 2% FCS at 37 °C for 10 min, and permeabilized in 90% methanol for 30 min on ice. The cells were then stained with PerCP/Cy5.5-conjugated antibodies against lineage markers and either with FITC anti-H2AX phospho-Ser139 (clone 20E3, Cell Signaling), or Alexa Fluor 488 anti-p53 (clone 1C12, Cell Signaling), or FITC anti-Ki67 (clone SolA15, eBioscience), or FITC Ki67 Set (clone B56, BD Pharmingen), or appropriate isotype controls. For ROS measurements the cells were loaded with carboxy-H2DCFDA (Life Technologies, Carlsbad, CA, USA) at 5 μM in PBS for 30 min at room temperature, and were washed twice before analyses.
Cell sorting protocols (FACS)
Mouse bone marrow was flushed from femurs, tibiae, humeri, and ilia of mice with a 26-G needle in PBS supplemented with 0.1% BSA and 2 mM EDTA, filtered through 40-μm cell strainers, and subjected to red blood cell lysis in ACK buffer (0.15 M NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA). The cells were stained with Biotin anti-mouse Lineage Panel (BioLegend) and lineage-positive cells depleted using biotin-binding Dynabeads (Life Technologies). The lineage-negative fraction was stained with PeCy7-cKit/CD117 (clone 2B8, BD Biosciences), APC-Sca1 (E13–161.7, BioLegend), FITC-CD48 (HM48-1, eBioscience), PE-CD150 (mShad150, eBioscience), and streptavidin-PECy5 (BioLegend). Propidium iodide or DAPI was added immediately before sorting for dead cell exclusion. Cell sorting was performed on the FACSAria instrument and was analyzed with FACS Diva software (BD Biosciences).
RNA isolation and qPCR
RNA was isolated from Ba/F3 cells using the EZ-10 DNAaway RNA mini-prep kit (Biobasic, Markham, ON, Canada) according to the manufacturer's protocol. Quality was assessed with nanodrop spectrophotometry and gel electrophoresis. cDNA was prepared using the M-MLV reverse-transcription kit (Invitrogen, Life Technologies).
RNA was isolated from FACS-sorted hematopoietic cells using the MagMAX total RNA kit (Ambion, Life Technologies) according to the manufacturer's protocol. Quality was assessed on Bioanalyzer Pico chip (Agilent, Santa Clara, CA, USA). KLS cDNA was prepared using the qScript cDNA Supermix (Quanta, Gaithersburg, MD, USA) with 5-ng input RNA.
All qPCR analyses were performed on a StepOnePlus instrument with Power SYBR master mix (Applied Biosystems, Life Technologies). Primers were purchased from IDT Technologies (Coralville, IA, USA), and primer sequences are provided in Supplementary Tables S1 and S2.
RNA-Seq
RNA integrity was assessed on a Bioanalyzer RNA pico chip. RNA (10 ng) was rRNA-depleted and used for library preparation with the SMARTer Stranded RNA-Seq kit (Takara Clontech, Mountain View, CA, USA) designed for low input. The RNA-seq libraries were sequenced on an Illumina HiSeq 2500 sequencer in a paired-end 50-bp configuration. The number of reads obtained for each sample ranged between 50 and 95 millions.
The high quality of sequence reads was confirmed using the FastQC tool (Babraham Bioinformatics, Cambridge, UK). To ensure high mapability, the few low-quality bases were trimmed from read extremities using Trimmomatic v.0.33.57 Reads were then mapped to the mouse UCSC mm9 reference assembly using TopHat v2.0.9 in conjunction with the Bowtie 1.0.0 algorithm,58, 59, 60 and gene expression was quantified by counting the number of uniquely mapped reads with featureCounts using default parameters.61
Normalization and differential gene expression analysis was conducted using the edgeR Bioconductor package.62 We retained genes that had a minimum expression level of five counts per million reads in at least 3 of the 12 samples. Pairwise differential gene expression analysis was performed by comparing samples from mutant mice (Puma−/−, Mysm1−/−Puma+/−, Mysm1−/−Puma−/−) relative to wild-type samples. Genes with changes in expression ≥|2| and an adjusted P-value of <0.05 were considered significant.
For figure preparation and RNA-seq data visualization in IGV genome browser,63 triplicate data were combined and bigwig scaled per 10 million reads. Mapping into gene exons was generated using a succession of genomeCoverageBed and wigToBigWig tools.64 Unbiased clustering analysis of relative gene expression changes (Log2 fold change; Figure 8) and heatmap visualization were performed using Pearson correlation within MeV software.65
Tissue culture and transfection
HEK293T cells were maintained in DMEM (Wisent) with 10% FCS and 2 mM L-Glutamine (Wisent). Cells were passaged every 2–3 days and all transfections were performed within 10 passages from thawing. The cells were transiently transfected using Lipofectamine 2000 (Life Technologies) with pcDNA3.1(+) vector encoding N-terminal triple-Flag-tagged mouse Mysm1 (NM177239, Life Technologies), and/or a pMiG vector encoding mouse wild-type untagged p53.
Ba/F3 cells were maintained in RPMI-1640 (Wisent) with 10% FCS, 5% WEHI-conditioned media, 2 mM L-Glutamine, 100 μg/ml streptomycin, and 100 U/ml penicillin (Wisent). The cells were maintained at 0.5–2 × 106 cells/ml at all times. Ba/F3 line stably expressing triple-Flag-tagged MYSM1 was generated by electroporation of 107 cells with 2.5 μg of BglII-linearized Mysm1-pcDNA3.1(+) vector (Life Technologies) in GenePulser II (Bio-Rad, Hercules, CA, USA). Transfected cells were selected and maintained in G418-containg media (0.8 mg/ml, EMD Millipore, Billerica, MA, USA).
shRNA knockdown
For Mysm1 knockdown in Ba/F3 cells, six shRNA sequences were designed to target Mysm1 transcript as described by Robert FW et al.66 Oligonucleotides were purchased from BioCorp (DDO, QC, Canada), PCR-amplified, and cloned into the pMSCV-mir30-PIG (MLP) vector.67 MLP-shFF vector was used for off-target knockdown controls. Retrovirus was produced in Phoenix cells (ATCC, Manassas, VA, USA).68 Ba/F3 cells were infected, selected, and further maintained in 2 μg/ml puromycin (Wisent). On the basis of knockdown efficiency, the following shRNA was chosen 5′-TGCTGTTGACAGTGAGCGACCGGGAAATGATGAAAGTACATAGTGAAGCCACAGATGTATGTACTTTCATCATTTCCCGGCTGCCTACTGCCTCGGA-3′, and six independent infections performed to generate six knockdown lines for experiments.
Ba/F3 irradiation assays
Independently infected shFF and shMysm1 lines (six each) were plated at 1 × 106 cells/ml in fresh media without IL-3 at the beginning of each experiment. As IL-3 has been shown to block apoptotic programs in this cell line,69, 70 it was excluded from the experiments to avoid masking of p53-mediated stress responses. Cells were irradiated with 3 Gy in a RS2000 irradiator (Rad Source) at indicated time points and always harvested alongside untreated cells to control for endogenous stress. Cell death was assessed usingtrypan blue dye exclusion.
ChIP
ChIP was performed as described previously,71 with minor modifications. Briefly, cells were fixed by addition of formaldehyde in the culture media to a final concentration of 1%, and was incubated for 11 min at room temperature, followed by addition of 0.125 M of glycine to stop fixation. Nuclei were then extracted with 5 min lysis in 0.25% Triton buffer (10 mM Tris-HCl pH 8, 10 mM EDTA, 0.5 mM EGTA), followed by 30 min lysis in 200 mM NaCl buffer (10 mM Tris-HCl pH 8, 1 mM EDTA, 0.5 mM EGTA). Nuclei were resuspended in sonication buffer (10 mM Tris pH 8, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% SDS, 0.5% Triton, and 0.05% NaDOC) and sonicated 12 times for 30 s in S450D Sonifier (Crystal Electronics, Newmarket, ON, Canada) at 80%, with 30 s rest in a cooling water bath.
Beads were prepared overnight with 20 μl of Dynabeads Protein G (Invitrogen, Life Technologies) conjugated with 2–3 μg of test antibody (listed in Supplementary Table S3) or control IgG (Santa Cruz, Dallas, TX, USA). Immunoprecipitation was performed by overnight incubation of antibody–bead matrices with sheared chromatin from the equivalent of 5 × 106 cells. For transcription factor ChIP, six washes were performed with low stringency buffers, whereas four medium-stringency washes were used for histone ChIP. Samples were de-crosslinked by overnight incubation at 65°C in 1% SDS buffer (50 mM Tris pH 8, 10 mM EDTA), and following RNaseA and Protienase K enzymatic treatments, ChIP DNA was purified using the Qiaquick PCR cleanup kit (Qiagen, Venlo, Netherlands). ChIP enrichment was quantified using real-time qPCR analysis (primer sequences provided in Supplementary Table S2). All CT values were normalized to those of the pro-opiomelanocortin (Pomc) gene, which serves as a negative binding region. Enrichment was calculated relative to control IgG for transcription factors, and relative to total histone H3 for H3K27ac and H3K4me3.
Co-immunoprecipitation and western blotting
For co-immunoprecipitation, cells were lysed in B450 buffer (25 mM Tris pH 7.5, 2 mM EDTA, 450 mM NaCl, 0.2% NP40, 5% glycerol) 24–48 h after transfection. Samples were dounce-homogenized and cleared using centrifugation. Cleared lysates were diluted 1/3 to 150 mM NaCl and 0.1% NP40 and their protein content quantified with the BCA assay (Thermo Scientific, Waltham, MA, USA). Antibody-conjugated Dynabeads Protein G (Invitrogen, Life Technologies) was prepared by incubation of 25 μl bead slurry with 2.5 μg of Flag M2 (Sigma), p53 1C12 (Cell Signaling, Danvers, MA, USA), or control IgG (Santa Cruz) for at least 4 h. Immunoprecipitation of the protein lysate with antibody-conjugated Dynabeads was performed with rotation at 4°C for 2 h. For HEK co-transfection IP, 500 μg total protein was used; for Ba/F3 endogenous IP, 5 mg total protein was used. After four washes with IP buffer (25 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% NP40), protein complexes were eluted in 1 × SDS sample buffer with boiling for 10 min at 95°C. Eluates were then separated from the beads and reduced by addition of 50 mM DTT, followed by heating for further 10 min at 95 °C. For western blot analysis, cells were lysed in a modified RIPA buffer supplemented with DTT, EDTA, and protease and phosphatase inhibitors (Thermo Scientific). Protein concentration was assessed using the BCA assay (Thermo Scientific), and samples were prepared by boiling in Laemmli buffer. Commercial antibodies used for western blot analysis are listed in Supplementary Table S4.
Production of anti-MYSM1 rabbit polyclonal antibody
This was performed according to previously described protocols.72 Briefly, mouse Mysm1 cDNA fragment encoding amino acids 162–363 was cloned into pGEX-2TK vector (GE Healthcare, Buckinghamshire, UK), expressed as a GST fusion protein in Escherichia coli strain BL21 (New England Biolabs, Ipswich, MA, USA) and purified using affinity chromatography against glutathione sepharose 4B (GE Healthcare). Rabbit immunization was performed according to SOP 406 of the McGill Comparative Medicine and Animal Resource Centre (CMARC, http://www.mcgill.ca/research/researchers/compliance/ animal/sop), with 0.1 mg of protein injected subcutaneously in incomplete Freund's adjuvant, boosting every 4 weeks. For antibody purification, the same Mysm1 cDNA fragment was cloned into plasmid pQE40 (Qiagen), expressed as a DHFR fusion protein in E. coli strain M15(pREP4) (Qiagen), and was isolated using affinity chromatography against Ni-NTA agarose (Qiagen). Antibody purification from rabbit serum was performed by preparative immunoblotting, as previously described.72
Statistical analyses
Statistical comparisons were performed with Prism 5.0 (GraphPad, La Jolla, CA, USA), using Student's t-test for two data sets and ANOVA for multiple comparisons.
Acknowledgments
This work was funded by the Canadian Institutes of Health Research (CIHR, grant # 123403 to AN), the Canadian Foundation for Innovation (CFI, grant # 29838 to AN), Merck, Sharp & Dohme and McGill Faculty of Medicine Grant for Translational Research (grant #238371 to AN), and Startup Funds from the McGill Faculty of Medicine. AN is a Canada Research Chair Tier 2 in Hematopoiesis and Lymphocyte Differentiation (grant # 950–228977). DL was supported by a postdoctoral fellowship from the CIHR Neuroinflammation training program and from the Fonds de Recherche du Québec—Santé. We thank Eric Massicotte, Julie Lord, and Julien Leconte of the IRCM flow cytometry facility for cell sorting; Dr. Odile Neyret and Myriam Rondeau of the IRCM Molecular Biology and Functional Genomics Facility for whole-genome library preparation and RNA-Seq; Su-Jin Park, Karen Hope Stone, and McGill CMARC for technical assistance with anti-MYSM1 antibody production; Dr Anatoli Onichtchenko and Isabella Albanese for the production of FLAG-MYSM1 Ba/F3 cells; Dr. Peiman Shooshtarizadeh, and Professor Tarik Möröy for advice on HSPC transcriptional analysis; Tanya Koch, Geneviève Perreault, Patricia D'Arcy, and other staff of McGill CMARC for their technical assistance; Shiyang Shen and Su-Jin Park for genotyping.
Author contributions
Experiments were performed by JIB (Figures 1–8), JCP (Figures 1 and 4–8), SS (Figures 4 and 5), and AN (Figures 4–7). Experiments were designed by JIB (Figures 1–8), AN (Figures 1 and 4–8), and DL (Figures 2, 3, and 8). JIB, AN, DL, and JCP analyzed the data. DL performed RNA-Seq bioinformatics analysis. AN, JIB, and DL wrote the paper, with assistance from JCP in editing and revisions. AN, DL, FR, RC, PG, and JP supervised the work.
Glossary
- Bbc3
BCL2 binding component 3
- ChIP
chromatin immunoprecipitation
- CLP
common lymphoid progenitor
- DKO
double knockout
- H2AK119
Histone H2A Lysine 119
- H3K27ac
histone H3 lysine 27 acetylation
- H3K4me3
histone H3 lysine 4 trimethylation
- HSC
hematopoietic stem cell
- HSPCs
hematopoietic stem and progenitor cells
- MPP
multipotent progenitor
- MYSM1
Myb-like SWIRM and MPN Domains 1
- PUMA
p53 upregulated modulator of apoptosis
- RNA-Seq
RNA-sequencing
- ROS
reactive oxygen species
- shFF
firefly luciferase shRNA
- shMysm1
Mysm1 shRNA
- ϒH2AX
Phospho-Serine 139 Histone H2A Variant X
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer 2009; 9: 724–737. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW et al. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell 1994; 76: 1013–1023. [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009; 9: 862–873. [DOI] [PubMed] [Google Scholar]
- Saha MN, Qiu L, Chang H. Targeting p53 by small molecules in hematological malignancies. J Hematol Oncol 2013; 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Quintas-Cardama A, Lozano G. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 2012; 120: 5118–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Mohrin M, Sotiropoulou PA, Passegue E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell 2011; 8: 16–29. [DOI] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Huang G, Di Giandomenico S et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 2009; 4: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 2007; 109: 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 2012; 148: 1001–1014. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000; 287: 1804–1808. [DOI] [PubMed] [Google Scholar]
- van Os R, Kamminga LM, Ausema A, Bystrykh LV, Draijer DP, van Pelt K et al. A limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells 2007; 25: 836–843. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Osawa M, Jakt LM, Yoshikawa K, Nishikawa S. Necdin restricts proliferation of hematopoietic stem cells during hematopoietic regeneration. Blood 2009; 114: 4383–4392. [DOI] [PubMed] [Google Scholar]
- Asai T, Liu Y, Di Giandomenico S, Bae N, Ndiaye-Lobry D, Deblasio A et al. Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood 2012; 120: 1601–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Sun Y, Zhang Z, Feng W, Gao Y, Cai Z et al. Deletion of proapoptotic Puma selectively protects hematopoietic stem and progenitor cells against high-dose radiation. Blood 2010; 115: 4707–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 2005; 123: 641–653. [DOI] [PubMed] [Google Scholar]
- Liu D, Ou L, Clemenson GD Jr, Chao C, Lutske ME, Zambetti GP et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol 2010; 12: 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Shen H, Yuan Y, XuFeng R, Hu X, Garrison SP et al. Deletion of Puma protects hematopoietic stem cells and confers long-term survival in response to high-dose gamma-irradiation. Blood 2010; 115: 3472–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ 2008; 15: 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ 2009; 16: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005; 106: 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happo L, Cragg MS, Phipson B, Haga JM, Jansen ES, Herold MJ et al. Maximal killing of lymphoma cells by DNA damage-inducing therapy requires not only the p53 targets Puma and Noxa, but also Bim. Blood 2010; 116: 5256–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 2010; 7: 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev 2010; 24: 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 2014; 15: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T et al. Structural and functional differences of SWIRM domain subtypes. J Mol Biol 2007; 369: 222–238. [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 2007; 27: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Clare S, Hale C, Raisen C, McIntyre RE, Yusa K et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood 2012; 119: 1370–1379. [DOI] [PubMed] [Google Scholar]
- Jiang XX, Nguyen Q, Chou Y, Wang T, Nandakumar V, Yates P et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity 2011; 35: 883–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Nandakumar V, Jiang XX, Jones L, Yang AG, Huang XF et al. The control of hematopoietic stem cell maintenance, self-renewal, and differentiation by Mysm1-mediated epigenetic regulation. Blood 2013; 122: 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsultan A, Shamseldin HE, Osman ME, Aljabri M, Alkuraya FS. MYSM1 is mutated in a family with transient transfusion-dependent anemia, mild thrombocytopenia, and low NK- and B-cell counts. Blood 2013; 122: 3844–3845. [DOI] [PubMed] [Google Scholar]
- Le Guen T, Touzot F, Andre-Schmutz I, Lagresle-Peyrou C, France B, Kermasson L et al. An invivo genetic reversion highlights the crucial role of Myb-Like, SWIRM, and MPN domains 1 (MYSM1) in human hematopoiesis and lymphocyte differentiation. J Allergy Clin Immunol 2015; e-pub ahead of print 25 July 2015; doi:10.1016/j.jaci.2015.06.008. [DOI] [PubMed]
- Nandakumar V, Chou Y, Zang L, Huang XF, Chen SY. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. Proc Natl Acad Sci USA 2013; 110: E3927–E3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Nandakumar V, Yates P, Sanchez S, Jones L, Huang XF et al. Epigenetic control of dendritic cell development and fate determination of common myeloid progenitor by Mysm1. Blood 2014; 124: 2647–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle JI, Langlais D, Petrov JC, Pardo M, Jones RG, Gros P et al. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1 deficiency. Blood 2015; 125: 2344–2348. [DOI] [PubMed] [Google Scholar]
- Gatzka M, Tasdogan A, Hainzl A, Allies G, Maity P, Wilms C et al. Interplay of H2A deubiquitinase 2A-DUB/Mysm1 and the p19/p53 axis in hematopoiesis, early T-cell development and tissue differentiation. Cell Death Differ 2015; 22: 1451–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster M, Belle JI, Petrov JC, Ryder EJ, Clare S, Nijnik A. Deubiquitinase mysm1 is essential for normal fetal liver hematopoiesis and for the maintenance of hematopoietic stem cells in adult bone marrow. Stem Cells Dev 2015; 24: 1865–1877. [DOI] [PubMed] [Google Scholar]
- Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 2011; 9: 247–261. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Takeishi S, Kanie T, Susaki E, Onoyama I, Tateishi Y et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 2011; 9: 262–271. [DOI] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 2004; 431: 1002–1007. [DOI] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 2013; 27: 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med 2008; 44: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid Redox Signal 2012; 16: 1285–1294. [DOI] [PubMed] [Google Scholar]
- Zhao J. Coordination of DNA synthesis and histone gene expression during normal cell cycle progression and after DNA damage. Cell Cycle 2004; 3: 695–697. [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014; 15: 507–522. [DOI] [PubMed] [Google Scholar]
- Kerr JB, Hutt KJ, Michalak EM, Cook M, Vandenberg CJ, Liew SH et al. DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol Cell 2012; 48: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med 2006; 203: 2939–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge AR, Wu L, Scott CL, Adams JM et al. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev 2010; 24: 1608–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle JI, Nijnik A. H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. Int J Biochem Cell Biol 2014; 50: 161–174. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010; 465: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbridge AR, Strasser A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ 2015; 22: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W, Rosen B, West A, Koutsourakis M, Bushell W, Iyer V et al. A conditional knockout resource for genome-wide analysis of mouse gene function. Nature 2011; 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 2000; 28: 106–110. [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013; 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 2010; 26: 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C et al. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc Natl Acad Sci USA 2014; 111: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 2010; 12: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Malina A, Lee T, Di Paola D, Larsson O, Miething C et al. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Emu-myc/Bcl-2 mouse model. Blood 2013; 121: 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez del Arroyo A, Gil-Lamagniere C, Lazaro I, de Marco MC, Layunta I, Silva A. Involvement of p53 and interleukin 3 in the up-regulation of CD95 (APO-1/Fas) by X-ray irradiation. Oncogene 2000; 19: 3647–3655. [DOI] [PubMed] [Google Scholar]
- Canman CE, Gilmer TM, Coutts SB, Kastan MB. Growth factor modulation of p53-mediated growth arrest versus apoptosis. Genes Dev 1995; 9: 600–611. [DOI] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell 2012; 47: 38–49. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Canonne-Hergaux F, Gauthier S, Hackam DJ, Grinstein S, Gros P. The iron transport protein NRAMP2 is an integral membrane glycoprotein that colocalizes with transferrin in recycling endosomes. J Exp Med 1999; 189: 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.