Summary
Cancer stem cells (CSCs) are a subpopulation of neoplastic cells with self‐renewal capacity and limitless proliferative potential as well as high invasion and migration capacity. These cells are commonly associated with epithelial‐mesenchymal transition (EMT), which is also critical for tumor metastasis. Recent studies illustrate a direct link between EMT and stemness of cancer cells. Long non‐coding RNAs (lncRNAs) have emerged as important new players in the regulation of multiple cellular processes in various diseases. To date, the role of lncRNAs in EMT‐associated CSC stemness acquisition and maintenance remains unclear. In this study, we discovered that a set of lncRNAs were dysregulated in Twist‐positive mammosphere cells using lncRNA microarray analysis. Multiple lncRNAs‐associated canonical signaling pathways were identified via bioinformatics analysis. Especially, the Shh‐GLI1 pathway associated lncRNA‐Hh, transcriptionally regulated by Twist, directly targets GAS1 to stimulate the activation of hedgehog signaling (Hh). The activated Hh increases GLI1 expression, and enhances the expression of SOX2 and OCT4 to play a regulatory role in CSC maintenance. Thus, the mammosphere‐formation efficiency (MFE) and the self‐renewal capacity in vitro, and oncogenicity in vivo in Twist‐positive breast cancer cells are elevated. lncRNA‐Hh silence in Twist‐positive breast cells attenuates the activated Shh‐GLI1 signaling and decreases the CSC‐associated SOX and OCT4 levels, thus reduces the MFE and tumorigenesis of transplanted tumor. Our results reveal that lncRNAs function as an important regulator endowing Twist‐induced EMT cells to gain the CSC‐like stemness properties. Stem Cells 2016;34:55–66
Keywords: lncRNA, Cancer stem cells, Breast cancer, Hedgehog signaling pathway
Significance Statement.
In this study, we identified a series of long non‐coding RNAs (lncRNAs) and canonical signaling pathways in Twist high‐expressed breast cancer stem cells (CSCs) using lncRNA and mRNA array. Of note, a hedgehog pathway associated lncRNA, named lncRNA‐Hh, was found to directly target GAS1, an enhancer of hedgehog signaling to increase the SOX2 and OCT4 expression, thus played a critical role for Twist‐driven EMT cells and Twist‐positive breast cancer cells to gain the CSCs‐like characteristics. Our study provides a novel insight into the CSCs pathology genesis in Twist‐positive breast cancer cells.
Introduction
Cancer stem cells (CSCs) (or tumor‐initiating cells) comprise only a small minority of neoplastic cells and were first identified in the hematopoietic system 1. They have been since discovered in many solid tumor types, such as breast cancer, liver cancer, lung cancer, and glioma 2, 3, 4, 5. CSCs are capable of self‐renewal and possess limitless proliferative potential and high invasion and migration capacity 6, 7. It is also postulated that CSCs are the “seeds” of tumor recurrence and metastasis 8.
Epithelial‐mesenchymal transition (EMT) is a cellular process commonly associated with CSCs 9, 10. EMT is characterized by a loss of epithelial differentiation and a shift toward mesenchymal phenotype in tumor invasion and metastasis 11. It can be triggered or mediated by several transcription factors, including Snail, Twist, and ZEB1. However, why epithelial tumor cells with the EMT phenotype acquire stem‐like characteristics, such as self‐renewal, drug‐resistance, and cell migration remains unclear.
Several signaling pathways such as Wnt/β‐Catenin, Notch, Hedgehog (Hh), and transforming growth factor‐beta (TGF‐β) signaling are well‐established in the function of CSCs 12, 13, 14, 15. The Hh signaling pathway controls tissue polarity, patterning maintenance, and stem cell renewal during embryonic development 16. Previous studies demonstrated that Hh signaling is also vital in the maintenance of CSCs 17. Canonical Hh signaling pathways are stimulated by the binding of secreted Shh ligand to the transmembrane protein Patched (Ptch), resulting in the activation of another transmembrane protein smoothened (Smo) and its downstream signaling cascade 18. Consequently, the activated Gli family of zinc‐finger transcription factors initiates the transcription of their target genes 19, such as SOX2 and OCT4 14, 20, the crucial genes for stem cell function 21. Gli1 is considered a key mediator of Hh signals 21. Growth arrest‐specific 1 (GAS1), an enhancer of Hh signaling via binding to Shh protein 22, 23, 24, may act as a positive regulator in CSC maintenance.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with little protein‐coding potential 25, 26. Recently, accumulated data show that lncRNAs are critical players in the regulation of multiple cellular processes, such as development, homeostasis, stem cell pluripotency, cell growth and apoptosis, and cancer metastasis 27, 28, 29, 30 via modulating epigenetic, transcriptional, or post‐transcriptional gene regulation mechanisms 31. In our previous work, lncRNAs were demonstrated to play a critical role in Twist‐induced EMT 32. This has led us to hypothesize that lncRNAs could be involved in control of cancer stem‐like properties associated with Twist‐induced EMT.
Using lncRNA and mRNA arrays, we identified a series of lncRNAs and canonical signaling pathways in Twist‐overexpressing breast CSCs. Of note, Hh pathway‐associated lncRNA, lncRNA‐Hh, was found to directly target GAS1 to increase SOX2 and OCT4 expression, endowing Twist‐positive breast cancer cells with stem‐like characteristics. Our study provides a novel insight into the regulation of CSCs in Twist‐positive breast cancer cells.
Materials and Methods
Cell Culture, Plasmids and Reagents
Breast cancer cell lines MCF‐7, Hs578T, BT549, MDA‐MB‐231, human mammary epithelial cell line MCF10A were obtained from ATCC (Manassas, VA, http://www.atcc.org). MCF‐10A cells were maintained as described 32. MCF‐7, Hs578T, BT549, and MDA‐MB‐231 cells were cultured in RPMI‐1640 medium (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) with 10% FBS (Gibco, Grand Island, NY, http://www.invitrogen.com). HEK‐293T cells were cultured in Dulbecco's modified Eagle's media (Invitrogen) with 10% fetal calf serum (FBS) (Gibco). Culture of MCF‐10A/Twist cell line was previously described 32. pCMV‐lncRNA‐Hh was constructed by cloning human lncRNA‐Hh cDNA into pCMV‐HA (Clontech, PaloAlto, CA, http://www.clontech.com). For shRNA plasmids, synthetic DNA inserts were cloned into pGLVH1/GFP+Puro. The target sequences for Twist shRNA are 5′‐AGGTACATCGACTTCCTCTAC‐3′, 5′‐AAGCTGAGCAAGATTCAGACC‐3′, 5′‐ACUCCAAGAUGGCAAGCUG‐3′. The target sequences for lncRNA‐Hh shRNA are 5′‐TGGGATGTTTCAGAAGATGATTG‐3′, 5′‐TGGCTTGAAAATTAAATGATACT‐3′, 5′‐GGGTGACATTTGGATTGATTC‐3′, 5′‐GGTGGCATTTGGCTAGGTTCT‐3′. The control shRNA sequences, which do not match any known human cDNAs, are 5′‐TTCTCCGAACGTGTCACGT‐3′. The lncRNA‐Hh promoter (−2,000 to + 294 region) was inserted into the pGL3‐basic vector (Promega, Madison, WI, http://www.promega.com) at KpnI/XbaI sites to construct pGL3‐lncRNA‐Hh luciferase reporter plasmid (pGL3‐lncRNA‐Hh‐Luc). The Hh signaling‐specific inhibitors GDC‐0449 (Vismodegib, S1082) was bought from Seleckchem (Houston, TX, http://www.selleckchem.com/).
Mammosphere Culture, Self‐Renewal, Cell Growth, and Cell Migration Assay
Mammosphere culture was performed as described 33, 34. Essentially, cells or mammospheres were dissociated into single cells by 0.05% trypsin‐EDTA solution and plated into six‐well plates coated with 2% Poly‐2‐hydroxyethyl methacrylate (poly‐HEMA) (Sigma, http://www.sigmaaldrich.com/) at a density of 1 × 104 viable cells per milliliter in primary culture and 5 × 103 cells per milliliter in following passages 35. Cells were grown in a serum‐free Dulbecco's modified Eagle's medium/F12 medium supplemented with B27 (Invitrogen), 20 ng/mL Epidermal Growth Factor (EGF), 20 ng/mL Basic fibroblast growth factor (b‐FGF), 0.4% Albumin from bovine serum (BSA), 2 μg/mL heparin, and 5 μg/mL insulin. For MCF‐7 cells, mammospheres were kept in culture and passaged every 7 days, whereas Hs578T, BT549, and MDA‐MB‐231mammospheres were passaged every 4 days. The number of secondary generation spheres (at least 50 μm diameter) was counted using a OLYMPUS IX70 microscope (Tokyo, Japan) fitted with a graticule dividing the field into small squares each with an area of 16 mm at × 100 magnification). The percentage (%) of MFE was calculated as previously described 36. The average size of mammospheres (N = 30 spheres) was calculated. The percentage of MFE (%) from primary generation to tertiary generation indicated the self‐renewal capacity of each kind of mammosphere. Mammosphere cell growth was determined by cell numbers contained within each generation of mammospheres, which were disaggregated using 0.05% trypsin‐EDTA solution, and single cells were counted. Cell migration abilities were measured by Transwell assay as described previously 37. All experiments were performed at least three times.
Immunofluorescence
For MCF‐7/Vector and MCF‐7/Twist cells, immunofluorescence analysis was performed as previously described 37. For mammosphere cells, secondary generation spheres were collected and resuspended with PBS. Mammospheres were placed on coverslips coated with 0.1% poly‐l‐lysine and then fixed within 4% paraformaldehyde for 20 minutes at room temperature. After washing with PBS, cells were treated by 0.1% Triton X‐100 for 15 minutes at room temperature and incubated with 5% goat serum solution for 30 minutes at 37°C. Cells were stained with primary antibodies against Twist (1:50), SOX2 (1:150), and OCT4 (1:150) at 4°C overnight, followed by a fluorescein isothiocyanate (FITC)‐labeled goat anti‐rabbit or mouse secondary antibody (1:200, Sigma) for 1 hour at 37°C in a humidified incubator. Sections were mounted in aqueous medium containing 4′,6‐diamidino‐2‐phenylindole (DAPI) as a nuclear counterstain (Vector Labs, Burlingame, CA, http://www.vectorlabs.com). A negative control was used to ensure the specificity of fluorescent immunostaining by replacing the primary antibody with a normal rabbit IgG. A Nikon Eclipse 80i microscope (Eclipse 80i, Tokyo, Japan) was used to take immunofluorescent images.
Quantitative Real‐Time Polymerase Chain Reaction Assay and Western Blot Analysis
Total RNA was isolated from secondary generation mammospheres and fresh tumor tissues using Trizol (Invitrogen) according to the instruction of the manufacturer. RNA quantity was determined by agarose gel electrophoresis and spectrophotometry. Quantitative real‐time polymerase chain reaction (PCR) was performed using SYBR Premix Ex Taq (Takara Bio, Japan, http://www.takara.co.jp) according to the manufacturer's protocol. The primers used are listed in Supporting Information Tables 1 and 2. All experiments were performed at least three times.
Table 1.
The associated signaling pathways of lncRNAs and mRNAs. The underlined pathways are reported to be associated with CSC features
| lncRNA target gene | mRNA | ||
|---|---|---|---|
| Signaling pathway | PValue | Signaling pathway | PValue |
| Hedgehog signaling pathway | 6.4E‐02 | Cell adhesion molecules (CAMs) | 1.5E‐02 |
| Apoptosis | 1.2E‐01 | mTOR signaling pathway | 2.9E‐02 |
| Wnt signaling pathway | 2.3E‐01 | Androgen and estrogen metabolism | 3.3E‐02 |
| MAPK signaling pathway | 2.3E‐01 | Drug metabolism | 3.4E‐02 |
| TGF‐beta signaling pathway | 2.5E‐01 | Calcium signaling pathway | 6.7E‐02 |
| Tight junction | 2.6E‐01 | Histidine metabolism | 9.4E‐02 |
| p53 signaling pathway | 2.7E‐01 | Jak‐STAT signaling pathway‐ | 1.0E‐01 |
| Regulation of actin cytoskeleton | 4.5E‐01 | Insulin signaling pathway | 1.7E‐01 |
| Drug metabolism | 5.0E‐01 | B cell receptor signaling pathway | 1.8E‐01 |
| ABC transporters | 5.1E‐01 | Hedgehog signaling pathway | 1.9E‐01 |
| Chemokine signaling pathway | 5.9E‐01 | Apoptosis | 2.0E‐01 |
| ECM‐receptor interaction | 6.4E‐01 | NOD‐like receptor signaling pathway | 2.7E‐01 |
| Gap junction | 6.8E‐01 | ABC transporters | 3.2E‐01 |
| Cell cycle | 7.2E‐01 | GnRH signaling pathway | 3.2E‐01 |
| PPAR signaling pathway | 7.5E‐01 | Focal adhesion | 3.4E‐01 |
| Notch signaling pathway | 8.5E‐01 | T cell receptor signaling pathway | 4.3E‐01 |
| mTOR signaling pathway | 8.7E‐01 | VEGF signaling pathway | 4.5E‐01 |
| Cell adhesion molecules (CAMs) | 8.9E‐01 | MAPK signaling pathway | 5.0E‐01 |
| Insulin signaling pathway | 9.0E‐01 | PPAR signaling pathway‐ | 5.4E‐01 |
| Toll‐like receptor signaling pathway | 9.1E‐01 | Drug metabolism | 7.2E‐01 |
| Jak‐STAT signaling pathway | 9.4E‐01 | Wnt signaling pathway | 7.5E‐01 |
| VEGF signaling pathway | 9.5E‐01 | ErbB signaling pathway‐ | 7.6E‐01 |
| Adherens junction | 9.5E‐01 | Gap junction | 7.8E‐01 |
| Focal adhesion | 9.6E‐01 | Adherens junction | 7.9E‐01 |
| ErbB signaling pathway | 9.7E‐01 | Tight junction | 8.1E‐01 |
| GnRH signaling pathway | 9.8E‐01 | ECM‐receptor interaction | 8.5E‐01 |
| Toll‐like receptor signaling pathway | 8.7E‐01 | ||
| TGF‐beta signaling pathway | 9.5E‐01 | ||
| Notch signaling pathway | 9.8E‐01 | ||
| Cell cycle | 1.0E+00 | ||
Table 2.
The potential lncRNAs associated with Hedgehog signaling pathway lncRNAs were named according to “chromosome number of the location, the plus lncRNA position from the start to the end in the chromosome, and the same (+) or the opposite (−) strand direction of prediction related to 5′ terminus of RNA”. The potential targets neighboring genes were also showed
| Pathway | lncRNA(chr,chr start‐end,strand)(target gene) |
|---|---|
| Hedgehog signaling pathway | IncRNA (chrl5, 37090798_37110707,−) (CSNK1A1) |
| IncRNA (chrl9,438420_2083745,−) (CSNK1G2) | |
| IncRNA (chr9,89563614_89616948,+) (GAS1) | |
| IncRNA (chrl, 84630386_84644921,+) (PRKACB) | |
| IncRNA (chrl2, 5090327_5090604,+) (BMP7) | |
| IncRNA (chr20, 18768614_18774626,−) (FBXW11) |
Western blot analysis was performed as described previously 32. Briefly, the secondary generation mammospheres and fresh tumor tissues were lyzed with RIPA buffer (Boster, China), and cell lysates were separated by SDS‐PAGE. Proteins were incubated with specifically primary antibodies, and β‐actin was used as a loading control. The primary antibodies against Myc‐tagged‐Twist, Twist, β‐catenin, p‐β‐catenin ALDH1, sHh, GAS1, and GLI1 were purchased from Abcam (Cambridge, MA, http://www.abcam.com); OCT4, SOX2, E‐cadherin, Histone H3, Vimentin, and fibronectin antibodies were from Bioworld (St Louis Park, MN); p‐SMAD3, SMAD3, p‐SMAD1/5/8, SMAD1/5/8, p‐Erk, Erk, and p‐β‐catenin antibodies were from Cell Signaling Technology (Beverly, MA, http://www.cellsignal.com); and β‐Actin antibodies were from Zhongshan Golden Bridge (Beijing, China http://www.zsbio.com/). The appropriate horseradish peroxidase‐conjugated secondary antibodies were applied, and the proteins were visualized by using the enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ, http://www.amersham.com).
LncRNA and mRNA Microarray Analysis
LncRNA and mRNA microarray analysis was performed as described previously 32. Total RNA was separately isolated from secondary generation MCF‐7/Vector mammospheres, MCF‐7/Twist mammospheres, MCF‐10A/Vector mammospheres, and MCF‐10A/Twist mammospheres using Trizol reagent as above described. The Agilent LncRNA and mRNA arrays (4 × 180K) contained more than 20,000 lncRNAs and 40,000 cDNA were used in this analysis. Dysregulated lncRNA or mRNA was filtered by at least fold change ≥5 or fold change ≥ 1.5, respectively. In our study, we named lncRNA according to “chromosome number of the location, the plus lncRNA position from the start to the end in the chromosome, and the same (+) or the opposite (−) strand direction of prediction related to 5′ terminus of RNA”.
lncRNA Target Prediction
Differentially expressed lncRNAs (fold change ≥ 5) were selected for potential target‐gene prediction 38. Two independent algorithms were used. The first algorithm searches for target genes acting in cis. Using gene annotations at UCSC (http://genome.ucsc.edu/), lncRNAs and potential target genes were paired and visualized using UCSC genome browser. The genes transcribed within a 10 kb region upstream or downstream of lncRNAs were considered as cis target genes. The second algorithm is based on mRNA sequence complementarity and RNA duplex energy prediction, assessing the impact of lncRNA binding on complete mRNA molecules. It uses the BLAST software for first round screening. Finally, the RNA plex software was used to choose trans‐acting target genes 39.
Gene Ontology and KEGG Pathway Analysis
To better understand the large number of differentially expressed genes in the microarray data and identify specific signaling pathways, we performed Gene Ontology (GO) and KEGG pathway analyses 40 in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.7 with the dysregulated lncRNAs targets genes and differentially expressed mRNA (fold change ≥ 1.5).
Luciferase Reporter Assay
Transient transfection and luciferase assays were performed as described previously 41. HEK293, MCF‐7/Twist, Hs578T, and BT549 cells were transiently cotransfected with the pGL3‐lncRNA‐Hh‐Luc promoter luciferase plasmid and Twist expression plasmids using Lipofectamine 2000 (Sigma). After 48 hours transfection, the cell lysates were prepared, and luciferase activity was assayed according to the instruction of the manufacturer (Promega). Luciferase activity was showed as fold change. Three independent experiments were taken.
Tumor Formation Assay In Vivo
The MCF‐7/lncRNA‐Hh, MDA‐MB‐231‐shRNA/lncRNA‐Hh mammosphere cells and their control mammosphere cells were injected subcutaneously into the groin of 5 weeks‐old athymic nude mice (1 × 105 cells per mouse). The longest diameter and its widest vertical width of tumor were measured each 3 days with a dial‐caliper. Tumor volume was calculated using the equation L × W 2 × 1/2. The mice were euthanized at fifth week, and tumors were surgically isolated. All animal experiments were approved by the Animal Care Committee of the Chongqing medical University.
Immunohistochemistry Staining
Tumor tissues were fixed with 4% paraformaldehyde. Paraffin‐embedded specimens were then sectioned at 4 mm thickness. Immunohistochemistry (IHC) staining was performed as previously described 37. The sections were incubated with rabbit anti‐GAS1 polyclonal antibody (Abcam) (1/50)) overnight at 4°C and then 1 hour at room temperature. Then, the sections were sequentially incubated with polymer helper solution (ZSBiO, Beijing, China) for 20 minutes, polyperoxidase‐anti‐mouse/rabbit IgG (ZSBiO) for 30 minutes at 37°C, and stained with diaminobenizidine. A Nikon Eclipse 80i microscope (Eclipse 80i, Tokyo, Japan) was used to take immunohistochemistry images.
Statistical Analysis
Statistical analysis was done using SPSS standard version 13.0 software. The independent Student's t test was used to compare the continuous variables between two groups. The data were expressed as means ± SD at least three independent determination. Values of p < .05 were considered statistically significant.
Results
Twist‐Induced EMT Endows Cells with a CSC‐Like Characteristics
Previous reports showed that Twist is capable of inducing EMT in human mammary epithelial cells 32. Using retrovirus infection, constitutive expression of Myc‐tagged Twist in MCF‐7 human breast cancer cells (MCF‐7/Twist) or MCF10A human mammary epithelial cells (MCF10A/Twist) resulted in a mesenchymal morphology in MCF‐7 (Supporting Information Fig. 1A) and MCF10A cells 32. EMT was confirmed by reduced expression of epithelial cell markers and increased expression of mesenchymal cell markers (Supporting Information Fig. 1B, 1C). Previous studies indicated that EMT can endow cells with stem cell‐like phenotypes 9. MFE assays were conducted using MCF‐7/Twist, MCF10A/Twist, and their controls. Larger size of mammospheres and higher MFE were found in MCF‐7/Twist and MCF10A/Twist cells than the corresponding control cells (Fig. 1A, 1B). The OCT4, SOX2, NANOG, ALDH1 mRNA levels and OCT4, SOX2, ALDH1 protein levels were significantly increased in MCF‐7/Twist (Fig. 1C, 1D) and MCF10A/Twist cells (data not shown) compared with their control cells. The CD44+/High/CD24−/Low cells are regarded as CSCs in breast cancer 42. Thus, CD44 and CD24 expression was determined by quantitative real‐time PCR (qRT‐PCR). Higher levels of CD44 and lower levels of CD24 were identified in MCF‐7/Twist cells than those in MCF‐7/Vector cells (Fig. 1C). In line with this, Twist overexpression enhanced SOX2 and OCT4 expression as shown by immunofluorescence staining (Fig. 1E). The self‐renewal capacity of each mammosphere‐generating cell can be determined by mammosphere growth 33. MCF‐7/Twist formed more mammospheres than MCF‐7/Vector in primary, secondary, and tertiary mammosphere culture (Fig. 1F). More cells were found in MCF‐7/Twist mammospheres than in MCF‐7/Vector mammospheres (Fig. 1G). Similar results were observed for the cell migration ability of MCF‐7/Twist mammosphere cells and MCF‐7/Vector mammosphere cells (Fig. 1H). These data suggest that Twist‐overexpressing breast cancer cells have the stem‐like characteristics.
Figure 1.
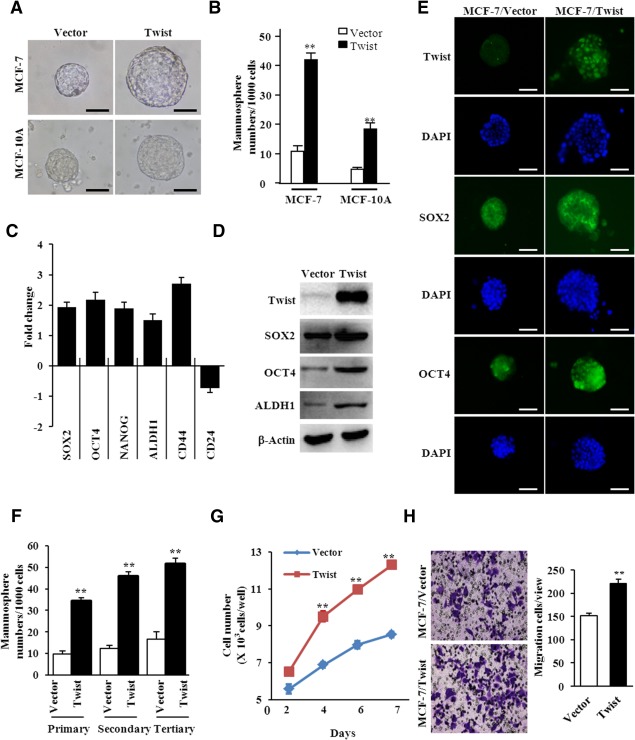
Epithelial‐mesenchymal transition (EMT) induced by Twist enhances the enrichment of cancer stem cells (CSCs). (A): Reprehensive images of mammospheres formed from MCF‐7/Twist, MCF‐10A/Twist, and its controls (magnification of × 200). (B): The quantification of mammosphere forming efficiency of MCF‐7/Twist, MCF‐10A/Twist, and its controls in independent experiments. The data are shown as the mammospheres number per 1,000 seeded cells ± SEM (**, p ≤ .01). (C–E): The expression of CSC‐associated genes in MCF‐7/Vector mammosphere and MCF‐7/Twist mammosphere cells were determined by quantitative real‐time polymerase chain reaction (C, data shown as mean ± SD), Western blot analysis (D), and immunofluorescence staining (E, magnification of × 200). (F): The mammosphere forming efficiency was quantitatively analyzed for MCF‐7/Vector mammospheres and MCF‐7/Twist mammospheres from primary generation to tertiary generation (data showed as means ± SD, **, p ≤ .01). (G): The growth curve of MCF‐7/Vector mammosphere and MCF‐7/Twist mammosphere cells was tested by cell counting. The data represent means ± S.D. from three different experiments (**, p ≤ .01). (H): Migration ability of mammosphere cells was examined by Transwell chamber. The migrated cells were showed as mean ± SD (N > 3,**, p ≤ .01). Abbreviation: DAPI, 4′,6‐diamidino‐2‐phenylindole.
A Series of Dysregulated lncRNAs are Identified in Twist Positive Mammospheres
Our previous study has shown that lncRNAs are involved in Twist‐induced EMT in mammary epithelial cells 32. We wondered whether lncRNAs are associated with CSC properties. LncRNAs and mRNA expression profiles were analyzed between MCF‐7/Twist mammosphere cells and MCF‐7/Vector mammosphere cells using Agilent Human lncRNA/mRNA microarrays. Compared with MCF‐7/Vector mammosphere cells, 270 upregulated lncRNAs and 192 downregulated lncRNAs of more than fivefold changes were identified. A heat‐map of the significantly changed lncRNAs was generated by unsupervised hierarchical clustering analysis (Supporting Information Fig. 2A). These results were validated by qRT‐PCR using 10 randomly chosen lncRNAs and 10 mRNAs (Supporting Information Fig. 2B, 2C).
In order to identify their target genes and the corresponding pathways of these CSC‐related lncRNAs, we predicted the potential target genes of the 462 differentially expressed lncRNAs (fold change ≥ 5) using cis‐ or trans‐regulatory mechanism‐based algorithms 43. A total of 797 target genes were found. These target genes and 2,065 differentially expressed genes identified by mRNA arrays (fold change ≥ 1.5) were then classified via DAVID v6.7 using Gene Ontology and KEGG pathway databases. Several well‐known canonical pathways such as Hedgehog, Wnt, mitogen‐activated protein kinase (MAPK), TGF‐β and Notch signaling pathways, which are related with CSCs were identified in both the lncRNAs array dataset and the mRNA array dataset (Table 1). Interestingly, these canonical pathways (e.g., Hedgehog, Wnt, MAPK, TGF‐β were also found in the MCF‐10A/Twist mammospheres (Supporting Information Table 3). These data indicate that lncRNAs‐regulated Hedgehog, Wnt, MAPK, and TGF‐β signaling may play a role in Twist and EMT‐induced CSC properties.
Hedgehog Pathway Plays a Crucial Role in Mammospheres
In order to determine the actually activated signaling pathway in Twist‐positive mammosphere cells, these canonical pathways (e.g., Hedgehog, Wnt/β‐Catenin, MAPK, and TGF‐β) conducted by bioinformatics analysis were evaluated by immunoblotting assay. Interestingly, almost all of these pathways were detected to be activated in MCF‐7/Twist mammosphere cells (Fig. 2A). Hh signaling, which was the top changed pathway in lncRNA/mRNA array assays, was found to be increased sHh (sonic hedgehog protein) and Gli‐1 (Zinc finger protein) expression in MCF10A‐Twist cells (Fig. 2A). Because sHh and GLI1 are critical components of the Hh pathway and involved in regulating the proliferation, migration, and differentiation of CSCs 14, we proceeded to determine the role of the Hh pathway in regulating CSC properties of Twist‐positive breast cancer cells. To this end, Twist was verified to be efficiently knocked down by shRNAs in BT549 breast cancer cells (Supporting Information Fig. 3A). Twist knockdown reduced the MFE (Fig. 2B upper panel; Supporting Information Fig. 3B) and led to smaller mammospheres (Fig. 2B down panel; Supporting Information Fig. 3C) in Hs578T‐sh/Twist cells and BT549‐sh/Twist cells compared with parent cells and shRNA/vector control cells. Knockdown of endogenous Twist expression in Hs578T and BT549 cells also decreased GLI1, SOX2, and OCT4 expression in mammosphere cells (Fig. 2C). So, loss of Twist expression may impede corresponding CSC self‐renewal and maintenance via the Hh signaling pathway in breast cancer cells.
Figure 2.
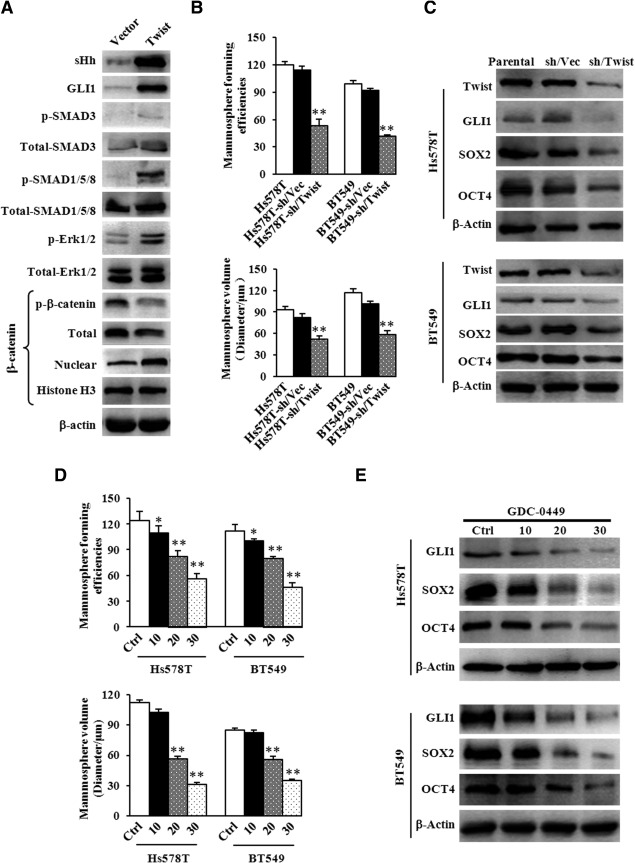
Hedgehog pathway plays a crucial role in mammospheres enrichment for Twist‐positive breast cancer. (A): The activation of a series of classic signaling pathways in MCF‐7/Twist mammosphere cells was determined by Western blotting analysis. β‐Actin was used as a loading control. (B): The mammosphere‐formation efficiency (MFE) and mammosphere sizes were quantified for mammospheres formed by Hs578T, Hs578T‐sh/Vector, Hs578T‐sh/Twist, BT549, BT549‐sh/Vector, and BT549‐sh/Twist cells. The data represent means ± SD (N = 3, **, p ≤ .01). (C): Immnoblotting analysis was used to examine the activation of Hedgehog signaling and expression of cancer stem cell (CSC) markers in the cells described in (B). β‐Actin was used as a loading control. (D): The MFE and mammosphere sizes were quantitatively analyzed for Hs578T and BT549 under treatment with or without GDC‐0449 (specific inhibitor of Hedgehog pathways). The data represent means ± SD from three different experiments (*, p ≤ .05; **, p ≤ .01). (E): The activation of Hedgehog pathways and CSC markers were tested by immunoblotting analysis in the cells described in (C). β‐Actin was used as a loading control.
To consolidate our study, a Hh specific inhibitor GDC‐0449 was used. Accordingly, GDC‐0449 dramatically reduced MFE (Fig. 2D upper panel) and resulted in the smaller mammosphere formation capacity (Fig. 2D down panel) of Hs578T and BT549 cells in a dose‐dependent manner. Correspondingly, the expression of GLI1, SOX2, and OCT4 was decreased in GDC‐0449‐treated Hs578T and BT549 cells (Fig. 2E). Together, these data strongly indicate that the Hh signaling pathway plays a crucial role in mammosphere enrichment in Twist‐positive breast cancer cells.
Hedgehog Signaling Pathway Associated‐LncRNA (LncRNA‐Hh) is Regulated by Twist
We next analyzed the potential of dysregulated lncRNAs involved in regulating the Hh pathway. As shown in Table 2, the Hh pathway may be regulated by lncRNA (chr15, 37090798‐37110707, −), lncRNA (chr19, 438420‐2083745, −), lncRNA (chr9, 89563614‐89616948, + ), lncRNA (chr1, 84630386‐84644921, + ), lncRNA (chr12, 5090327‐5090604, + ), and lncRNA (chr20, 18768614‐18774626, −) as indicated by bioinformatics analysis. The expression of these lncRNAs was determined in MCF‐7/Twist mammospheres and MCF‐7/Vector mammospheres. Only lncRNA (chr9, 89563614‐89616948, + ) (named as lncRNA‐Hh here after) was remarkably upregulated in MCF‐7/Twist mammospheres (Fig. 3A), whereas the levels of other lncRNAs were only mildly upregulated in MCF‐7/Twist mammosphere cells (Supporting Information Fig. 4). In line with this finding, the level of lncRNA‐Hh was found to be higher in Twist‐positive parental Hs578T and BT549 mammosphere cells than that in Twist‐negative parental MCF‐7 mammosphere cells (Fig. 3B). In addition, knockdown of Twist in MCF‐7/Twist, Hs578T, and BT549 by shRNAs decreased the lncRNA‐Hh expression in their mammosphere cells, which was comparable to that in MCF‐7/Vector mammosphere cells (Fig. 3C). These data indicate the possible association between the expression of lncRNA‐Hh and Twist.
Figure 3.
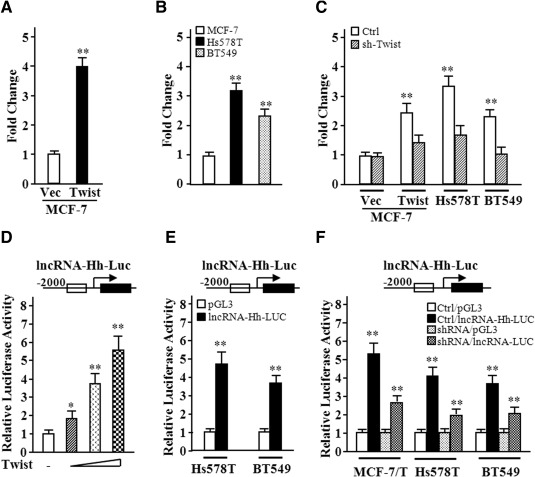
LncRNA‐Hh levels are correlated with endogenous expression of Twist in breast cancer cells. (A–C): Levels of the lncRNA‐Hh in mammospheres formed from different breast cancer cells were analyzed using quantitative real‐time polymerase chain reaction. (A): lncRNA‐Hh level in ectopic Twist expressed MCF‐7 and its control mammospheres; (B) comparison of the lncRNA‐Hh levels in mammospheres from MCF‐7, Hs578T, and BT549 cells; (C) lower levels of the lncRNA‐Hh in breast cancer cells with knocking‐down Twist by siRNA. (D–F): The relative transcript activity of lncRNA‐Hh was examined by luciferase analysis. (D): lncRNA‐Hh promoter luciferase plasmid and Twist expression plasmid were cotransfected to EK293 cells. (E): lncRNA‐Hh promoter luciferase plasmid was transfected into MCF‐7, Hs578T, and BT549 cells. (F): Luciferase activity of lncRNA‐Hh in MCF‐7/Twist, Hs578T, and BT549 with or without sh/Twist. Data are the means ± SD of three independent experiments performed in triplicate (*, p ≤ .05; **, p ≤ .01). Abbreviations: lncRNA‐Hh, hedgehog pathway associated lncRNA; lncRNA‐Hh‐Luc, shRNA, short hairpin RNA.
Next, we asked whether lncRNA‐Hh expression is directly regulated by Twist. The transcription of lncRNA‐Hh was measured by luciferase reporter assays. After the lncRNA‐Hh promoter luciferase plasmid (pGL3‐lncRNA‐Hh‐Luc) and the constructor encoding Twist were transfected into HEK293 cells, a dose‐dependent lncRNA‐Hh promoter transactivation by Twist was observed (Fig. 3D). Transcriptional activity of lncRNA‐Hh was high in Hs578T and BT549 breast cancer cells with high Twist levels (Fig. 3E). The reporter activity was notably repressed in Twist‐knockdown cells (MCF‐7/Twist‐sh/Twist, Hs578T‐sh/Twist, BT549‐sh/Twist) compared with their control cells (Fig. 3F). These results show that Twist is involved in lncRNA‐Hh transcription.
lncRNA‐Hh Maintains Mammosphere Enrichment in Breast Cancer Cells In Vitro
The above data suggest that lncRNA‐Hh is a regulator of the Hh pathway and may be vital for mammosphere enrichment. To address this question, a construct encoding lncRNA‐Hh was stably transfected into MCF‐7 cells (Fig. 4A) using lentivirus. LncRNA‐Hh overexpression enhanced mammosphere formation and induced a larger mammosphere size in MCF‐7 (Fig. 4B). In addition, expression of GAS1 (the lncRNA‐Hh potential‐target), GLI1, and Hh target genes SOX2, OCT4 was increased in MCF‐7/lncRNA‐Hh mammosphere cells compared with control mammosphere cells (Fig. 4C). Knockdown of the endogenous lncRNA‐Hh expression in Hs578T and BT549 cells (Fig. 4D; Supporting Information Fig. 5A, 5B) using lentivirus‐mediated RNA interference attenuated MFE and mammosphere volume (Fig. 4E; Supporting Information Fig. 5C, 5D). Correspondingly, expression of GAS1, GLI1, SOX2, and OCT4 was also reduced in Hs578T‐sh/lncRNA‐Hh and BT549‐sh/lncRNA‐Hh mammosphere cells compared with their control mammospheres (Fig. 4F). Consistence with these findings, knockdown of endogenous Twist and overexpression of lncRNA‐Hh in Hs578T and BT549 did not alter MFE, mammospheres volume (Supporting Information Fig. 6A, 6B), and expression of GAS1, GLI1, SOX2, and OCT4 (Supporting Information Fig. 6E) compared with their parent cells. Similar data were acquired in MCF7 with ectopic Twist overexpression and lncRNA‐Hh knockdown and parent MCF7 cells (Supporting Information Fig. 6C, 6D, 6F). These data demonstrate that lncRNA‐Hh is essential for mammosphere enrichment in Twist‐positive breast cancer cells.
Figure 4.
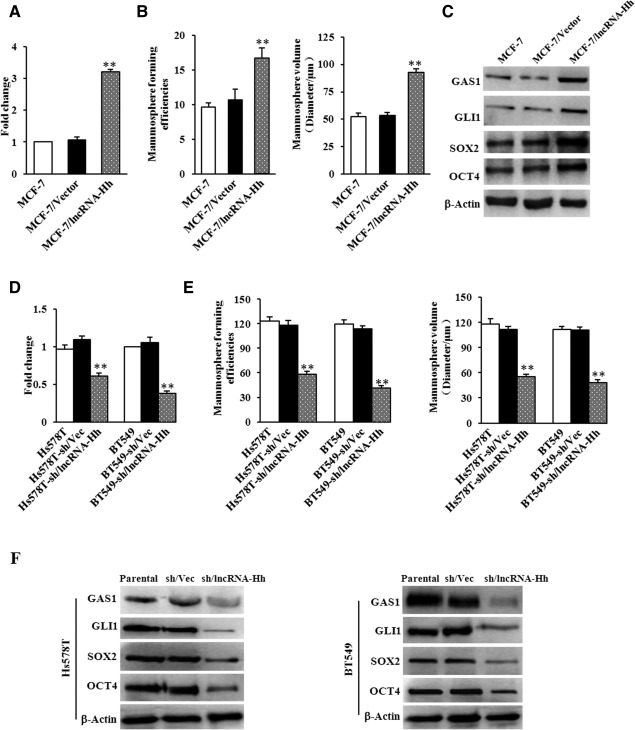
lncRNA‐Hh promotes mammosphere enrichment in breast cancer cells. (A): Levels of the lncRNA‐Hh in MCF‐7/lncRNA‐Hh mammosphere and their control mammosphere cells were analyzed using quantitative real‐time polymerase chain reaction (qRT‐PCR). (B): The quantification of mammosphere forming efficiency (MFE) and mammosphere sizes formed by MCF‐7/lncRNA‐Hh and their control cells. (C): Immunoblotting analysis of GAS1, GLI1, and cancer stem cell (CSC) markers in the mammosphere cells described in (B). β‐Actin was used as a loading control. (D): qRT‐PCR was used to test the endogenous lncRNA‐Hh in Hs578T‐sh/lncRNA‐Hh mammospheres and BT549‐sh/lncRNA‐Hh mammosphere cells and their control mammosphere cells. (E): MFE and mammosphere sizes were quantitatively analyzed for mammospheres formed by Hs578T‐sh/lncRNA‐Hh and BT549‐sh/lncRNA‐Hh and their control cells. (F): Immunoblotting analysis of GAS1, GLI1, and CSC markers in the mammosphere cells described in (D). β‐Actin was used as a loading control. Data are the means ± SD of three independent experiments in (D) and (E) (**, p ≤ .01). Abbreviations: GAS1, growth arrest‐specific 1; lncRNA‐Hh, hedgehog pathway associated long noncoding RNA.
lncRNA‐Hh Promotes Breast Cancer Tumorigenesis In Vivo
To investigate the functional role of lncRNA‐Hh in tumorigenesis, breast cancer xenograft models were used. As shown in Figure 5A, lncRNA‐Hh markedly enhanced the subcutaneous tumor growth in MCF‐7 mammosphere xenografts compared with the control mice after 5 weeks (upper panel). Meanwhile, lncRNA‐Hh knockdown significantly reduced tumor growth in MDA‐MB‐231 mammosphere xenografts compared with the control group (Fig. 5A, down panel). As expected, MCF‐7/lncRNA‐Hh mammosphere‐derived tumors grew faster than MCF‐7/Vector tumors in nude mice. Tumors derived from lncRNA‐Hh knockdown MDA‐MB‐231 mammospheres grew slower than those from MDA‐MB‐231‐sh/Vec mammospheres (Fig. 5B). The expression levels of lncRNA‐Hh (Fig. 5C) and GAS1, GLI1, SOX2, and OCT4 (Fig. 5D, 5E) in xenografts were confirmed by qRT‐PCR, Western blotting, and IHC. Taken together, these data suggest that lncRNA‐Hh is required to promote tumorigenesis in vivo.
Figure 5.
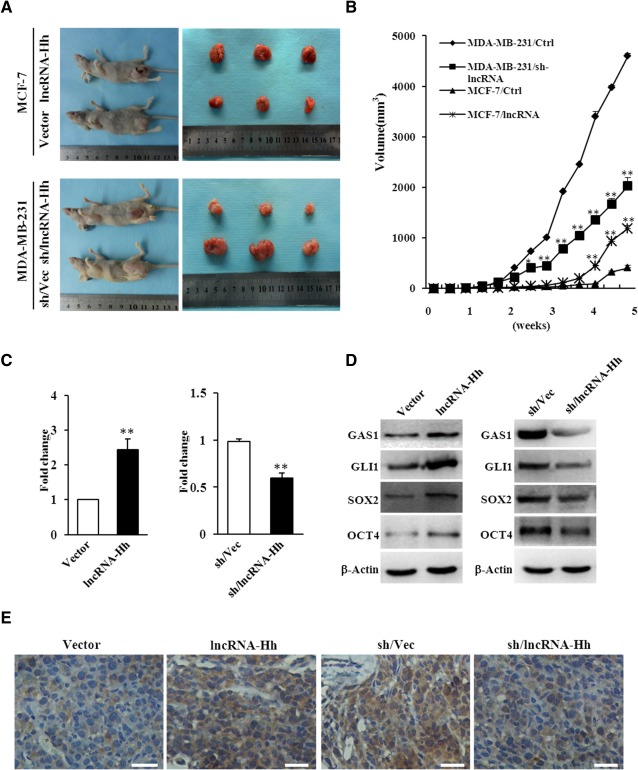
lncRNA‐Hh promotes breast cancer tumorigenesis in nude mice. (A): Photos of the tumor‐bearing BALB/c female nude mice and the xenograft tumors dissected from the nude mice at 5 weeks after being subcutaneously injected with MCF‐7/lncRNA‐Hh, MDA‐MB‐231/shRNA‐lncRNA‐Hh mammosphere cells and their control mammosphere cells. (B): Tumor growth curve. The data were generated from three mice in four different groups described in (A). Data are the means ± SD of three independent experiments performed in triplicate (*, p ≤ .05; **, p ≤ .01). (C): Quantitative real‐time polymerase chain reaction was used to test levels of lncRNA‐Hh in MDA‐MB‐231‐sh/lncRNA‐Hh tumor tissue and MCF‐7/lncRNA‐Hh tumor tissue and their control tumor tissues. (D): Immunoblotting analysis of GAS1, GLI1, SOX2, and OCT4 in the tumor tissues described in (C). β‐Actin was used as a loading control. Data are the means ± SD of three independent experiments in (C) and (D). (E): Representative photomicrographs of tumor tissue sections stained with SOX2‐specific polyclonal antibody (magnification of 400×). Scale bars = 100 μm. Abbreviations: GAS1, growth arrest‐specific 1; lncRNA‐Hh, hedgehog pathway associated long noncoding RNA.
Discussion
It has been demonstrated that stem‐like characteristics are associated with EMT 9. As previous studies have suggested that EMT induction through ectopic expression of Snail, Twist, or ZEB1 provides mammary epithelial cells with stem‐like properties 44, 45, 46. However, the underlying molecular mechanism is still not well‐known. In this study, we found that the ectopic Twist overexpression endows breast cancer cells with strong mammosphere‐formation potential and self‐renewal capacity. More interestingly, several dysregulated lncRNAs and the multiple aberrant CSC‐associated canonical signal pathways (such as Hedgehog, Wnt/β‐Catenin, and TGF‐β signaling pathways) were identified in Twist‐overexpressing mammosphere cells. In particular, a Hedgehog signaling pathway‐associated lncRNA, named lncRNA‐Hh, was found to promote CSC enrichment and self‐renewal abilities via activation of Shh‐GLI1 signaling. Our data highlights a novel lncRNA regulator in Twist‐induced EMT associated CSC properties.
Some classical signaling pathways, such as Hedgehog 14, TGF‐β 15, Notch 13, WNT/β‐Catenin 12, and Hippo 47 signaling are critical in the maintenance of CSC characteristics. For example, Wnt/β‐catenin signaling has been found to regulate the self‐renewal of head and neck squamous cell carcinoma cells with stem cell characteristics, and this phenomenon occurs partly through direct Oct4 regulation by β‐catenin 12. In addition, Hedgehog signaling pathway promotes thyroid CSC self‐renewal in ATC cell lines by inducing Snail expression 48 and CSC regulatory factors, SOX2, OCT4, NANOG, and GLI1 49, 50. Despite the current understanding of signaling in CSCs, how EMT induction can drive CSC generation remains poorly understood.
Increasing evidence has confirmed that lncRNAs are involved in many cellular processes and play a core role in controlling the abnormal activation pathways and various disease 32. HOTAIR plays a critical role in cancer through epigenetic regulation mechanisms 51. H19 lncRNA can function as an oncogene 52, and EFNA3 lncRNA can promote metastatic dissemination in human tumors 53. In this work, we found that a set of dysregulated lncRNAs are in the EMT‐associated breast CSCs, suggesting that lncRNAs play an important role in cancer cell stemness maintenance.
We provided the direct evidence that lncRNAs are a novel regulator in modulating the CSC characteristics in breast cancer cells. A set of lncRNAs were dysregulated and multiple lncRNAs‐associated canonical signal pathways (such as Hedgehog, Wnt/β‐Catenin, and TGF‐β signaling pathways) were abnormally activated in Twist‐overexpressing mammosphere cells. Of note, the lncRNA‐Hh directly targets GAS1 to stimulate the activation of SHH‐GLI1 pathway, thus upregulating expression of CSC‐associated genes in Twist‐positive mammospheres. This lncRNA is essential for Twist‐induced mammosphere‐formation ability, the CSC self‐renewal capacity, and tumorigenesis in breast cancer cells. LncRNA‐Hh silence by RNA interference attenuated the generation of CSCs in breast cancer cells. Consistently, rescue of lncRNA‐Hh in MCF‐7 cells increased CSCs and tumor formation.
Conclusion
In conclusion, our results show that lncRNA (e.g., lncRNA‐Hh) plays a key role in mediating Twist‐induced CSC properties via regulating the Hedgehog signaling pathway. Our data indicate that lncRNAs may be a new type of regulators for CSC function. We hope that this study could provide a new avenue for targeting CSC in breast cancer.
Author Contributions
M.Z. and Y.H. contributed equally to this work.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article
Supporting Information
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (NSFC 31171336, NSFC 81472476, NSFC 81402180); the Doctoral Fund of Ministry of Education, China (20125503110001); the Scientific Research Foundation for the Returned Overseas Chinese Scholars, Ministry of Education, China ([2011]508). X.C. was supported by National Institutes of Health (CA151610), the Avon Foundation (02‐2014‐063), David Salomon Translational Breast Cancer Research Fund, the Fashion Footwear Charitable Foundation of New York, and the Margie and Robert E. Petersen Foundation.
References
- 1. Dominique B, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–737. [DOI] [PubMed] [Google Scholar]
- 2. Charafe‐Jauffret E, Ginestier C, Iovino F et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 2009;69:1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang ZF, Ho DW, Ng MN et al. Significance of CD90 + cancer stem cells in human liver cancer. Cancer Cell 2008;13:153–166. [DOI] [PubMed] [Google Scholar]
- 4. Eramo A, Lotti F, Sette G et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008;15:504–514. [DOI] [PubMed] [Google Scholar]
- 5. Yuan X, Curtin J, Xiong Y et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 2004;23:9392–9400. [DOI] [PubMed] [Google Scholar]
- 6. Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: First steps into uncharted territory. Cell Stem Cell 2007;1:241–242. [DOI] [PubMed] [Google Scholar]
- 7. Sottoriva A, Verhoeff JJ, Borovski T et al. Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res 2010;70:46–56. [DOI] [PubMed] [Google Scholar]
- 8. Adorno‐Cruz V, Kibria G, Liu X et al. Cancer stem cells: Targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res 2015;75:924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mani SA, Guo W, Liao MJ et al. The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu M, Casimiro MC, Wang C et al. p21CIP1 attenuates Ras‐ and c‐Myc‐dependent breast tumor epithelial mesenchymal transition and cancer stem cell‐like gene expression in vivo. Proc Natl Acad Sci USA 2009;106:19035–19039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thiery JP: Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442–454. [DOI] [PubMed] [Google Scholar]
- 12. Lee SH, Koo BS, Kim JM et al. Wnt/beta‐catenin signalling maintains self‐renewal and tumourigenicity of head and neck squamous cell carcinoma stem‐like cells by activating Oct4. J Pathol 2014;234:99–107. [DOI] [PubMed] [Google Scholar]
- 13. Abel EV, Kim EJ, Wu J et al. The Notch pathway is important in maintaining the cancer stem cell population in pancreatic cancer. PloS One 2014;9:e91983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clement V, Sanchez P, de Tribolet N et al. HEDGEHOG‐GLI1 signaling regulates human glioma growth, cancer stem cell self‐renewal, and tumorigenicity. Curr Biol 2007;17:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Yu Y, Tsuyada A et al. Transforming growth factor‐beta regulates the sphere‐initiating stem cell‐like feature in breast cancer through miRNA‐181 and ATM. Oncogene 2011;30:1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev 2001;15:3059–3087. [DOI] [PubMed] [Google Scholar]
- 17. Zhao C, Chen A, Jamieson CH et al: Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature 2009;458:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rohatgi R, Scott M P. Patching the gaps in Hedgehog signalling. Nat Cell Biol 2007;9:1005–1009. [DOI] [PubMed] [Google Scholar]
- 19. Osterlund T, Kogerman P. Hedgehog signalling: How to get from Smo to Ci and Gli. Trends Cell Biol 2006;16:176–180. [DOI] [PubMed] [Google Scholar]
- 20. Gopinath S, Malla R, Alapati K et al. Cathepsin B and uPAR regulate self‐renewal of glioma‐initiating cells through GLI‐regulated Sox2 and Bmi1 expression. Carcinogenesis 2013;34:550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leis O, Eguiara A, Lopez‐Arribillaga E et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012;31:1354–1365. [DOI] [PubMed] [Google Scholar]
- 22. Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev 2007;21:1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee CS, Buttitta L, Fan CM. Evidence that the WNT‐inducible growth arrest‐specific gene 1 encodes an antagonist of sonic hedgehog signaling in the somite. Proc Natl Acad Sci USA 2001;98:11347–11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinelli DC, Fan C‐M. The role of Gas1 in embryonic development and its implications for human disease. Cell Cycle 2014;6:2650–2655. [DOI] [PubMed] [Google Scholar]
- 25. Frith MC, Bailey TL, Kasukawa T et al. Discrimination of non‐protein‐coding transcripts from protein‐coding mRNA[J]. RNA Biol 2006;3:40–48. [DOI] [PubMed] [Google Scholar]
- 26. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loewer S, Cabili MN, Guttman M et al. Large intergenic non‐coding RNA‐RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010;42:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guttman M, Donaghey J, Carey BW et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta RA, Shah N, Wang KC et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang KC, Yang YW, Liu B et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–641. [DOI] [PubMed] [Google Scholar]
- 32. Hu P, Yang J, Hou Y et al. LncRNA expression signatures of twist‐induced epithelial‐to‐mesenchymal transition in MCF10A cells. Cell Signal 2014;26:83–93. [DOI] [PubMed] [Google Scholar]
- 33. Dontu G, Abdallah WM, Foley JM et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003;17:1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ponti D, Costa A, Zaffaroni N et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506–5511. [DOI] [PubMed] [Google Scholar]
- 35. Han M, Wang Y, Liu M et al. MiR‐21 regulates epithelial‐mesenchymal transition phenotype and hypoxia‐inducible factor‐1α expression in third‐sphere forming breast cancer stem cell‐like cells. Cancer Sci 2012;103:1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shaw FL, Harrison H, Spence K et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia 2012;17:111–117. [DOI] [PubMed] [Google Scholar]
- 37. Wang L, Hou Y, Sun Y et al. c‐Ski activates cancer‐associated fibroblasts to regulate breast cancer cell invasion. Mol Oncol 2013;7:1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han L, Zhang K, Shi Z et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol 2012;40:2004–2012. [DOI] [PubMed] [Google Scholar]
- 39. Tafer H, Hofacker IL. RNAplex: A fast tool for RNA‐RNA interaction search. Bioinformatics 2008;24:2657–2663. [DOI] [PubMed] [Google Scholar]
- 40. Ashburner M, Ball CA, Blake JA et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uramoto H, Izumi H, Ise T et al. p73 Interacts with c‐Myc to regulate Y‐box‐binding protein‐1 expression. J Biol Chem 2002;277:31694–31702. [DOI] [PubMed] [Google Scholar]
- 42. Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem‐like cells that self‐renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 2008;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petruk S, Sedkov Y, Riley KM et al. Transcriptional elongation of non‐coding bxd RNAs promoted by the Trithorax TAC1 complex represses Ubx by a transcriptional interference mechanism. Cell 2006;127:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou W, Lv R, Qi W et al. Snail contributes to the maintenance of stem cell‐like phenotype cells in human pancreatic cancer. PloS One 2014;9:e87409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li J, Zhou BP. Activation of beta‐catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell‐like characters. BMC Cancer 2011;11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wellner U, Schubert J, Burk UC et al. The EMT‐activator ZEB1 promotes tumorigenicity by repressing stemness‐inhibiting microRNAs. Nat Cell Biol 2009;11:1487–1495. [DOI] [PubMed] [Google Scholar]
- 47. Cordenonsi M, Zanconato F, Azzolin L et al. The Hippo transducer TAZ confers cancer stem cell‐related traits on breast cancer cells. Cell 2011;147:759–772. [DOI] [PubMed] [Google Scholar]
- 48. Heiden KB, Williamson AJ, Doscas ME et al: The sonic hedgehog signaling pathway maintains the cancer stem cell self‐renewal of anaplastic thyroid cancer by inducing Snail expression. J Clin Endocrinol Metab 2014;99:E2178–E2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santini R, Vinci MC, Pandolfi S et al. Hedgehog‐GLI signaling drives self‐renewal and tumorigenicity of human melanoma‐initiating cells. Stem Cells 2012;30:1808–1818. [DOI] [PubMed] [Google Scholar]
- 50. Mimeault M, Batra SK. Molecular biomarkers of cancer stem/progenitor cells associated with progression, metastases, and treatment resistance of aggressive cancers. Cancer Epidemiol Biomarkers Prev 2014;23:234–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rinn JL, Kertesz M, Wang JK et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non‐coding RNAs. Cell 2007;129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lottin S, Adriaenssens E, Dupressoir T et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 2002;23:1885–1895. [DOI] [PubMed] [Google Scholar]
- 53. Gómez‐Maldonado L, Tiana M, Roche O et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 2015;34:2609–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article
Supporting Information


