Abstract
B class MADS-box genes play important roles in petal and stamen development. Some monocotyledonous species, including liliaceous ones, produce flowers with petaloid tepals in whorls 1 and 2. A modified ABCE model has been proposed to explain the molecular mechanism of development of two-layered petaloid tepals. However, direct evidence for this modified ABCE model has not been reported to date. To clarify the molecular mechanism determining the organ identity of two-layered petaloid tepals, we used chimeric repressor gene-silencing technology (CRES-T) to examine the suppression of B function in the liliaceous ornamental Tricyrtis sp. Transgenic plants with suppressed B class genes produced sepaloid tepals in whorls 1 and 2 instead of the petaloid tepals as expected. In addition, the stamens of transgenic plants converted into pistil-like organs with ovule- and stigma-like structures. This report is the first to describe the successful suppression of B function in monocotyledonous species with two-layered petaloid tepals, and the results strongly support the modified ABCE model.
The ABCE model represents the relationship between floral organ development and the expression patterns of three classes of floral homeotic genes, the A, B, and C class genes, most of which encode MADS-box transcription factors1,2. An A class gene alone specifies sepal formation in whorl 1, the combination of A and B class genes determines petal formation in whorl 2, the combination of B and C class genes specifies stamen formation in whorl 3, and a C class gene alone determines carpel formation in whorl 4. B class genes comprise two paralogous genes, DEFICIENS (DEF)/APETALA3 (AP3) and GLOBOSA (GLO)/PISTILLATA (PI); DEF/AP3 and GLO/PI proteins interact directly and form functional complexes3,4. DEF/AP3 and GLO/PI resulted from a duplication event that occurred before the emergence of angiosperms5,6. Multiple DEF/AP3 or GLO/PI homologs, present in the genomes of some plant species, have shown functional division by sub- or neo-functionalization7,8,9,10. Recently, an E class gene has been identified as an additional class of MADS-box genes, which are involved in the specification of all four types of floral organs11,12.
In contrast to higher eudicotyledonous species, such as Arabidopsis thaliana and Antirrhinum majus, some monocotyledonous species, including liliaceous ones, produce two-layered petaloid tepals in whorls 1 and 2. A modified ABCE model has been proposed to explain the molecular mechanisms of organ development of two-layered petaloid tepals (Supplementary Fig. S1)13. In this model, the expression of B class genes extends to whorl 1 in addition to whorls 2 and 3, resulting in the development of two-layered petaloid tepals in whorls 1 and 2. The modified ABCE model has been supported by the expression analysis of B class genes in various plant species with two-layered petaloid tepals14,15,16,17,18,19,20,21,22.
In higher eudicotyledonous species, the mutation of B class genes results in the conversion of petals into sepaloid tepals and stamens into carpeloid organs1,23,24,25,26; similar phenotypes have been induced by suppressing B function in transgenic plants27,28,29. In the liliaceous species Tulipa gesneriana, viridiflora cultivars, which have partially greenish tepals in whorls 1 and 2 and slightly degenerated stamens in whorl 3, show lower expression levels of B class genes than normal cultivars21. However, the suppression of B function in monocotyledonous species with two-layered petaloid tepals has not been reported to date; thus, there is no direct evidence supporting the modified ABCE model.
Tricyrtis spp. are liliaceous ornamental plants with two-layered petaloid tepals. An efficient and reproducible system of Agrobacterium-mediated genetic transformation has been established in Tricyrtis sp.30,31. To clarify the molecular mechanism of two-layered petaloid tepal development, we produced and characterized a transgenic Tricyrtis sp. in which B function was suppressed through chimeric repressor gene-silencing technology (CRES-T).
Results
Phylogenetic relationship of MADS-box genes from Tricyrtis sp
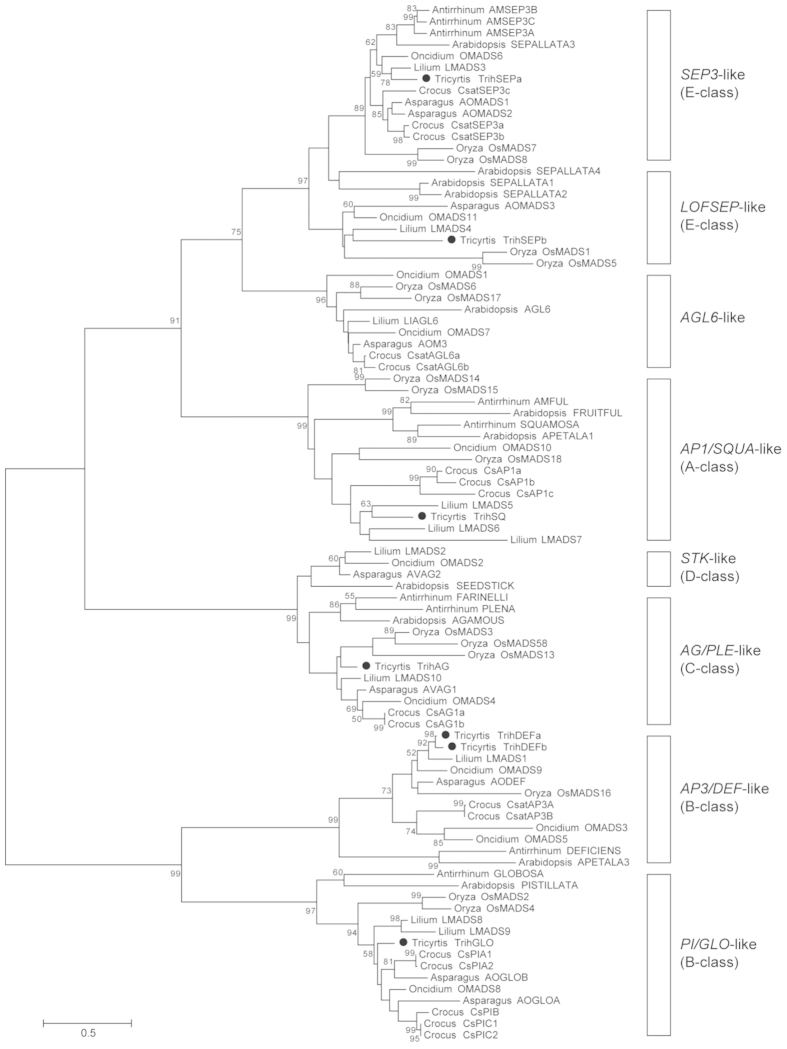
Seven MADS-box genes were isolated from Tricyrtis sp. Phylogenetic analyses classified these genes into major MADS-box gene lineages: AP1/SQUA-like (TrihSQ), AP3/DEF-like (TrihDEFa, TrihDEFb), PI/GLO-like (TrihGLO), AG/PLE-like (TrihAG), and SEP-like (TrihSEPa, TrihSEPb) lineages (Fig. 1). Each Tricyrtis sp. gene clustered closely with homologous genes from Lilium. The deduced amino acid sequences of TrihDEFa and TrihDEFb were similar, with only seven differences in amino acid residues. Conversely, the deduced amino acid sequences of TrihSEPa and TrihSEPb differed greatly; phylogenetic tree analysis assigned TrihSEPa to the SEP3 clade and TrihSEPb to the LOFSEP clade.
Figure 1. Phylogenetic relationships of MADS-box protein sequences.
A phylogenetic tree was constructed using the maximum likelihood method with the JTT + G model. Node values indicate bootstrap support greater than 50% from 1,000 replicates.
Expression of floral organ identity genes in Tricyrtis sp
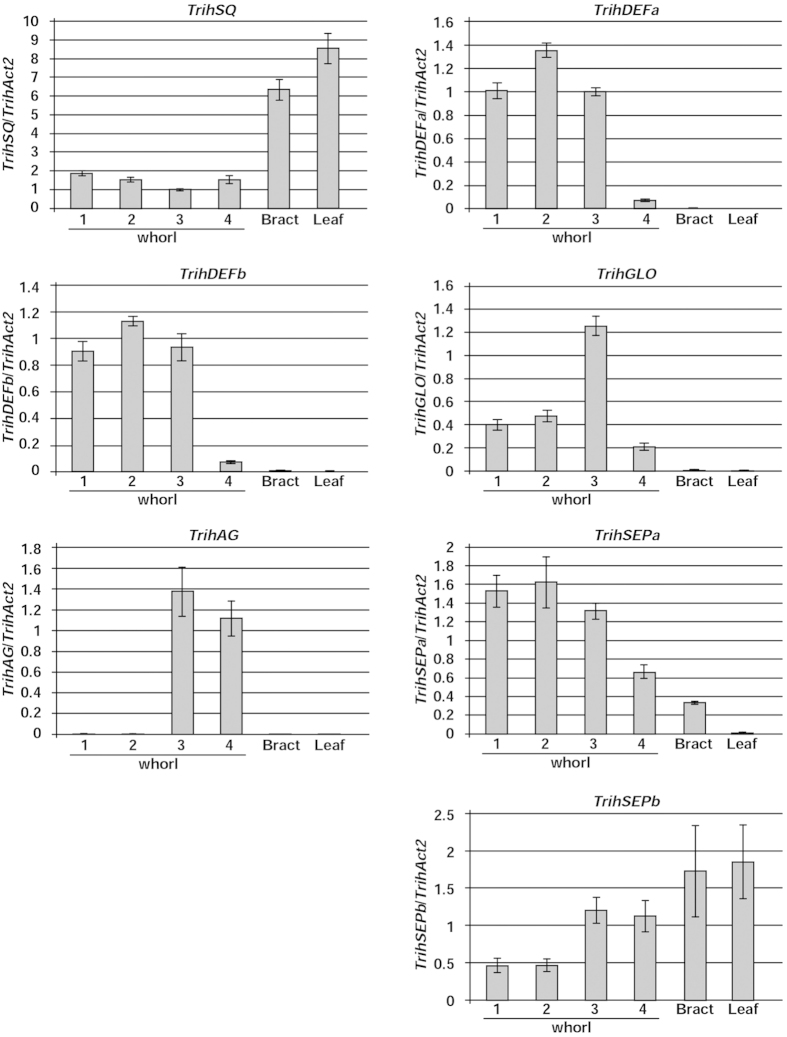
To investigate the spatial expression pattern of floral organ identity genes in a tetraploid variant of Tricyrtis sp. ‘Shinonome’, the expression levels of ABCE class genes in the floral organs (i.e., whorl 1, outer tepals; whorl 2, inner tepals; whorl 3, stamens; and whorl 4, carpels), bracts, and leaves of non-transgenic plants were quantified with real-time reverse transcription polymerase chain reaction (RT-PCR) analysis (Fig. 2). The expression of the A class gene TrihSQ was detected in all floral organs, bracts, and leaves. Although three B class genes, TrihDEFa, TrihDEFb, and TrihGLO, were expressed in all floral organs, their expression levels were lower in the whorl 4 organs. The C class gene TrihAG was strongly expressed in whorls 3 and 4. One E class gene, TrihSEPa, was expressed in all floral organs and bracts, whereas the other E class gene, TrihSEPb, was strongly expressed in all floral organs and bracts and in the leaves.
Figure 2. Real-time RT-PCR analysis of endogenous ABCE model gene transcripts in floral organs, bracts, and leaves of wild-type, non-transgenic Tricyrtis sp. plants.
Relative amounts of transcripts for each gene were normalized to TrihAct2. Values represent the means ± standard error of triplicates.
NMorphological characterization of transgenic Tricyrtis sp
To clarify the role of B class genes in the development of two-layered petaloid tepals, transgenic Tricyrtis sp. plants, in which B function was suppressed using CRES-T, were produced via Agrobacterium-mediated transformation. Forty-one independent transgenic plants carrying an artificial chimeric repressor of TrihDEFa (TrihDEFa-SRDX; Supplementary Fig. S2) were obtained and termed CrB strains. The presence of the transgene (HPT) was confirmed by PCR analysis with the primer set hpt290-F and hpt290-R (Fig. S3). Morphological characterization was performed one year after cultivation in pots during the flowering season. Vector control plants, transformed with A. tumefaciens strain EHA101/pIG121Hm32, exhibited no phenotypic alterations compared with non-transgenic plants (Fig. 3a–c). CrB strains could be classified into three types according to the degree of phenotypic alteration. Type I CrB strains (CrB1, CrB13, and CrB82) showed significant morphological alterations in floral organs (Fig. 3a,b,d). These strains produced greenish tepals in whorls 1 and 2 instead of petaloid tepals and pistil-like organs in whorl 3 instead of stamens (Fig. 3d,e). The pistil-like organs had a stigma-like structure on the apical part and ovary-like structures on the basal to middle parts (Fig. 3d,e). Furthermore, ectopic ovules were formed on the inside of the ovary-like structures (Fig. 3e). Because the ovary-like structures did not fuse with each other, these ectopic ovules were exposed (Fig. 3d,e). The flowers of Type I CrB strains did not open (Fig. 3b). Although whorl 4 organs (pistil) of Type I CrB strains showed no phenotypic alterations, they were strongly flexed, owing to the unopened state of the flowers (Fig. 3c,d). Type II CrB strains (CrB21, CrB32, CrB45, CrB53, and CrB67) showed moderate morphological alterations to the floral organs. They produced partially greenish tepals in whorl 1, and their flowers were slightly opened (Fig. 3b). In contrast to Type I CrB strains, Type II CrB strains showed no morphological alterations in whorl 2 and whorl 3 organs (Fig. S4). Thirty-three strains were classified as Type III CrB strains and showed no morphological alterations in any floral organs (Fig. 3b). The soil-plant analysis development (SPAD) values of whorl 1 and whorl 2 organs and leaves were measured using a chlorophyll meter. Whorl 1 and whorl 2 organs of both Type I and Type II CrB strains showed higher mean SPAD values than those observed for Type III CrB strains and vector control plants (Supplementary Fig. S5).
Figure 3. Morphological characterization of transgenic plants containing TrihDEFa-SRDX.
(a) Vector control plants and the Type I CrB strain (CrB1) during the flowering season. Bar = 10 cm. (b) Flowers in vector control plants and Type I (CrB1), Type II (CrB53), and Type III (CrB19) CrB strains. Bar = 1 cm. (c,d) Floral organs of (c) vector control plants and (d) CrB1 plants. Bar = 1 cm. (e) Close-up of pistil-like whorl 3 organs of a CrB1 plant. White arrows indicate stigma-like structures. Yellow arrowheads indicate heterotopic ovules. Bar = 5 mm.
The surface of each floral organ of the vector control and Type I CrB strain CrB1 plants was observed through scanning electron microscopy (SEM). In the greenish areas of whorl 1 and whorl 2 organs in CrB1, dome-shaped cells were observed on the adaxial surface; these cells were also observed on petaloid tepals in vector control plants, although the cell size was smaller in CrB1 than in vector control plants (Fig. 4a,b,e,f). The abaxial surface of whorl 1 organs in vector control plants mainly consisted of flat and complex irregular-shaped cells, whereas that in CrB1 plants mainly consisted of slightly swelled and more simple-shaped cells (Fig. 4c,d). The abaxial epidermis of whorl 2 organs in vector control plants mainly consisted of flat cells, whereas that in CrB1 plants mainly consisted of slightly swelled cells (Fig. 4g,h). The surface of the middle part of whorl 3 organs in vector control plants (filaments) contained only rectangular cells (Fig. 4i), whereas CrB1 plants (ovary-like structures) had many irregular-shaped cells that were similar to the ovaries in vector control plants (Fig. 4j,k). Ectopically formed ovules in the middle part of whorl 3 organs in CrB1 showed a structure similar to that of the ovules in vector control plants (Fig. 4l,m). The surfaces of the apical part of the whorl 3 organs in CrB1 (stigma-like structures) had typical papillae (Fig. 4n,p) and were morphologically similar to the stigmas in vector control plants (Fig. 4n–q).
Figure 4. SEM observations of epidermal cells from floral organs of transgenic plants containing TrihDEFa-SRDX.
(a,b) Adaxial surfaces of whorl 1 organs (outer tepals) of (a) vector control plants and (b) the Type I CrB strain (CrB1). (c,d) Abaxial surfaces of whorl 1 organs (outer tepals) of (c) vector control and (d) CrB1 plants. (e,f) Adaxial surfaces of whorl 2 organs (inner tepals) of (e) vector control and (f) CrB1 plants. (g,h) Abaxial surfaces of whorl 2 organs (inner tepals) of (g) vector control and (h) CrB1 plants. (i) Surface of the middle part of whorl 3 organs (filaments) in vector control plants. (j) Surface of the middle part of whorl 3 organs (ovary-like structures) in CrB1 plants. (k) Surface of the basal part of whorl 4 organs (ovaries) in vector control plants. (l) Ectopic ovules in the middle part of whorl 3 organs (ovary-like structures) in CrB1 plants. (m) Longitudinal section of the basal part of whorl 4 organs (ovaries) in vector control plants. (n,o) Surface of the apical part of whorl 4 organs (stigmas) in vector control plants. (p,q) Surface of the apical part of whorl 3 organs (stigma-like structures) in CrB1 plants.
Expression analysis of endogenous B class genes in transgenic Tricyrtis sp
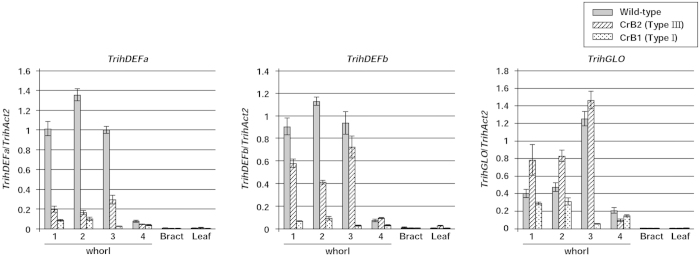
Figure 5 shows the expression levels of endogenous B class genes in floral organs, bracts, and leaves of Type I (CrB1) and Type III (CrB2) CrB strains and in non-transgenic plants. Although the expression of all three endogenous B class genes (TrihDEFa, TrihDEFb, and TrihGLO) was detected in whorls 1, 2, and 3 in CrB1 and CrB2, their expression levels were significantly lower than those observed in non-transgenic plants. The relative amounts of TrihDEFa transcript in organs of whorls 1, 2 and 3 in CrB1 decreased to 8.3%, 7.5%, and 2.5% of non-transgenic plant levels, respectively, whereas those in CrB2 decreased to 19.3%, 12.1%, and 29.4% of the levels observed in non-transgenic plants, respectively. The relative amounts of the TrihDEFb transcript in organs of whorls 1, 2, and 3 in CrB1 decreased to 7.4%, 8.2%, and 3.1% of non-transgenic plant levels, respectively, and those in CrB2 decreased to 63.9%, 36.5%, and 77.2% of the levels seen in non-transgenic plants, respectively. The relative amounts of TrihGLO transcript in organs of whorls 1, 2, and 3 in CrB1 plants decreased to 72.3%, 66.1%, and 4.4% of the levels observed in non-transgenic plants, respectively. In contrast, no apparent differences in the relative amounts of TrihGLO transcript in the organs of whorls 1, 2, and 3 were observed between CrB2 and non-transgenic plants. These results indicate that the expression levels of endogenous B class genes in CrB strains are correlated with the degree of morphological alteration.
Figure 5. Real-time RT-PCR analysis of endogenous B class MADS-box genes (TrihDEFa, TrihDEFb, and TrihGLO) in floral organs, bracts, and leaves of transgenic plants containing TrihDEFa-SRDX.
Type I (CrB1) and Type III (CrB2) CrB strains showed significant or no morphological alterations, respectively, in floral organs. Relative amounts of transcripts of each gene were normalized to TrihAct2. Values represent the means ± standard error of triplicates.
Discussion
ABCE model-related MADS-box genes were isolated from Tricyrtis sp., and their expression levels were analyzed in non-transgenic plants. Of the B class genes, two AP3/DEF-like genes (TrihDEFa and TrihDEFb) and one PI/GLO-like gene (TrihGLO) were isolated. Phylogenetic tree analyses indicated that the duplication of DEF-like genes in Tricyrtis sp. occurred recently, after the evolutionary split between Lilium and Tricyrtis. TrihDEFa, TrihDEFb, and TrihGLO were strongly expressed in whorls 1, 2 and 3 (Fig. 2; Supplementary Fig. S6), indicating that the modified ABCE model7 is applicable to Tricyrtis sp. The A class gene (TrihSQ) was expressed not only in whorls 1 and 2 but also in whorls 3 and 4, bracts and leaves (Fig. 2; Supplementary Fig. S6). Similar extended expression patterns of A class genes have been reported in monocotyledonous species33.
In the Arabidopsis ap3 mutant, petals and stamens are transformed to sepals and carpels, respectively23. Transgenic tomato (Solanum lycopersicum) plants, in which the B class gene SlGLO1 is repressed by RNA interference (RNAi), develop greenish petals in whorl 2 and greenish stamens in whorl 3, and the expression of chlorophyll biosynthetic genes is significantly up-regulated in these organs34. In the present study, Type I CrB strains produced greenish tepals in whorls 1 and 2 instead of petaloid tepals, and pistil-like organs with ovule- and stigma-like structures were produced in whorl 3 instead of stamens (Figs 3 and 4). Abaxial epidermal cells of the organs of whorls 1 and 2 showed different shapes between Type I CrB strains and the vector control plants (Fig. 4c,d,g,h). In addition, chlorophyll accumulation was observed in organs of whorls 1 and 2 of Type I CrB strains (Supplementary Fig. S5). Thus, there were apparent changes in floral organ identity in whorls 1 and 2 in Type I CrB strains. Because Tricyrtis sp. plants do not intrinsically develop sepals, it is difficult to clarify the identity of whorl 1 and whorl 2 organs in Type I CrB strains. In T. gesneriana, no apparent morphological differences in the surface structures of whorls 1 and 2 organs were observed between viridiflora cultivars and normal cultivars, although the expression of B class genes in whorls 1 and 2 organs of viridiflora cultivars is lower than in normal cultivars21. Thus, greenish tepals caused by the mutation of B class genes might not always exhibit phenotypic alterations in epidermal cells. However, the conversion of stamens into pistil-like organs in whorl 3 of Type I CrB strains, which is one of the typical phenotypes of B class gene mutants such as the Arabidopsis ap3 mutant23, strongly supports that whorl 1 and 2 organs of Type I CrB strains were converted into sepaloid organs. In Type I CrB strains, the flowers did not open (Fig. 3), and the cell sizes of the adaxial surface of whorl 1 and 2 organs were smaller than in the vector control plants (Fig. 4a,b,e,f). Because flower opening is mainly caused by cell expansion at the adaxial side of tepals35, unopened flowers of Type I CrB strains might result from the insufficient expansion of epidermal cells in whorl 1 and 2 organs.
B class genes have a positive autoregulatory feedback system that is important for the maintenance of high expression levels36. Although there were no differences in the relative amounts of endogenous A, C and E class genes (TrihSQ, TrihAG, TrihSEPa and TrihSEPb) transcripts between CrB1 and non-transgenic plants (Supplementary Fig. S6), the expression levels of endogenous B class genes in CrB strains decreased compared with those observed in non-transgenic plants, which might be due to the inhibition of the autoregulatory feedback system. In CrB strains, the expression levels of endogenous B class genes correlated with the degree of morphological alteration in floral organs (Fig. 5), indicating that morphological alterations may have resulted from the suppression of the function of endogenous B class genes. In A. majus, the threshold expression level of B class genes required to specify petal identity was approximately 11% of that in the wild-type37. The expression levels of endogenous TrihDEFa and TrihDEFb in organs of whorls 1 and 2 in Type I CrB strains decreased to 7.5–8.3% and 7.4–8.2% of those in non-transgenic plants, respectively (Fig. 5). Thus, the phenotypic alterations observed in organs of whorls 1 and 2 in Type I CrB strains might be caused by a transition in the identity from petaloid to sepaloid organs. Although indirect evidence for the modified ABCE model has been obtained by the expression analysis of B class genes in various plant species with two-layered petaloid tepals14,15,16,17,18,19,20,21,22, the results obtained in the present study directly support the modified ABCE model via the suppression of B class gene function in Tricyrtis sp.
Methods
Plant materials and growth conditions
Potted plants of Tricyrtis sp. ‘Shinonome’ and a tetraploid variant of this cultivar were cultivated in a greenhouse without heating. Tepal-derived embryogenic calli of a tetraploid variant were induced as previously described38 and used for transformation. This tetraploid variant showed high transformation efficiency and vigorous growth of transgenic plants compared with the original diploid cultivar ‘Shinonome’.
Isolation of MADS-box genes
Total RNA was extracted from young flower buds (5–7 mm in length) with an RNeasy Plant Mini Kit (QIAGEN Sciences, Maryland, USA) in accordance with the manufacturer’s instructions. Poly(A)+ RNA was separated from total RNA using DYNABEADS (DYNAL, Oslo, Norway), and cDNA was synthesized using AMV reverse transcriptase (Roche Diagnostics GmbH, Mannheim, Germany). Partial cDNAs were isolated by 3′-rapid amplification of cDNA ends (RACE)39 using a 5′/3′-RACE Kit (Roche) and four MADS-box degenerate primers: P038, P041, AD, and SP314. Upstream sequences overlapping the 3′ fragments were isolated by 5′-RACE using a 5′/3′-RACE Kit. cDNA clones with complete open reading frames (ORFs) were isolated via PCR using primers located in the 5′- and 3′-UTR regions with cDNA pools as templates. DNA sequencing was performed using a BigDye Terminator Cycle Sequencing Premix Kit (Applied Biosystems, Foster City, CA, USA) with an automated sequencer (Model 310, Applied Biosystems) according to the manufacturer’s protocol.
Phylogenetic analysis
The phylogenetic classification of MADS-box genes from Tricyrtis sp. was analyzed using MEGA 6.06 software40. Genes from Arabidopsis (Brassicaceae), Antirrhinum (Plantaginaceae), Oryza (Poaceae), Asparagus (Asparagaceae), Crocus (Iridaceae), Lilium (Liliaceae), and Oncidium (Orchidaceae) were used for the phylogenetic analysis. Accession numbers for these genes are listed in Supplementary Table S1. A molecular phylogenetic tree of the deduced amino acid sequences was constructed using ClustalW implemented in MEGA. To find the most appropriate evolutionary models, maximum likelihood fits for amino acid substitution models were tested using the program included in MEGA. Based on the Bayesian Information Criterion (BIC) score, we selected the JTT + G model and constructed a maximum-likelihood tree with 1,000 bootstrap replicates.
RNA isolation and gene expression analysis by real-time RT-PCR
Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and was then treated with DNase I (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. For cDNA synthesis, 500 ng of total RNA was reverse transcribed using a PrimeScript™ RT reagent Kit (Takara, Shiga, Japan) in accordance with the manufacturer’s instructions.
RT-PCR analysis was performed using SYBR® Premix Ex Taq™ II (Takara, Shiga, Japan) on a DNA Engine Opticon System (MJ Research, Waltham, MA, USA) as previously described31,41. The primer sets used in the analysis are listed in Supplementary Table S2. Each PCR was performed in three replicates under the following conditions: initial denaturation at 95 °C for 30 s; 45 cycles of 5 s at 95 °C and 30 s at 60 °C; and plate reading (detection of fluorescent product). To characterize the PCR products, melting curve analysis was performed by slowly increasing the temperature from 60 to 95 °C, and recording fluorescence data at 0.5 °C intervals42. The relative amounts of transgene transcripts were calculated using the comparative cycle threshold method, and results were normalized to the actin gene of Tricyrtis sp. (TrihAct2; AB196260 in the GenBank/EMBL/DDBJ databases).
Plasmid construction and production of transgenic Tricyrtis sp
The A. tumefaciens strain EHA101/pIG-CrB was used for transformation. The full-length coding region of TrihDEFa was fused with the ERF-associated amphiphilic repression (EAR) motif repression domain (SRDX)43. The T-DNA region of the binary vector pIG-CrB contained TrihDEFa-SRDX under the control of the cauliflower mosaic virus (CaMV) 35S promoter, the neomycin phosphotransferase II gene (NPTII) under the control of the nopaline synthase (NOS) promoter, and the hygromycin phosphotransferase gene (HPT) under the control of the CaMV35S promoter (Supplementary Fig. S2). Co-cultivation of embryogenic calli with Agrobacterium, selection of transgenic cells and tissues, and regeneration of transgenic plants were performed as previously described24. The presence of HPT in transgenic plants was confirmed by PCR analysis with the primer set hpt290-F and hpt290-R (Supplementary Table S2).
Transgenic plants were transplanted to pots and cultivated in a growth chamber as previously described25. One year after cultivation, morphological characterization was performed during the flowering season. The mean SPAD value, which expresses the relative amount of chlorophylls and has a high correlation with the chlorophyll concentration, of whorl 1 and whorl 2 organs was measured during the flowering season using a chlorophyll meter (SPAD-502; Fujiwara Scientific Co., Tokyo, Japan) as previously described44. SEM observation of the surfaces of floral organs was performed as previously described45.
Additional Information
How to cite this article: Otani, M. et al. Suppression of B function strongly supports the modified ABCE model in Tricyrtis sp. (Liliaceae). Sci. Rep. 6, 24549; doi: 10.1038/srep24549 (2016).
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Nobutaka Mitsuda, National Institute of Advanced Industrial Science and Technology (AIST), Japan, for constructing and providing the CRES-T vector. We thank Ms. Yumi Kajino, Tohoku University, Japan, for technical support in the cloning and DNA sequencing. This work was supported in part by Grants-in-Aid for Scientific Research (Nos. 23580037, 16380024 and 15K07287) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Author Contributions M.O., S.K. and A.K. wrote the manuscript. M.O., A.S., S.K., K.O., F.U. and M.H. performed the gene isolation and characterization and M.O. performed the transgenic experiments. Y.H. provided valuable suggestions. M.N. and A.K. designed and integrated the project.
References
- Bowman J. L., Smyth D. R. & Meyerowitz E. M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20 (1991). [DOI] [PubMed] [Google Scholar]
- Coen E. S. & Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991). [DOI] [PubMed] [Google Scholar]
- Goto K. & Meyerowitz E. M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560 (1994). [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Krizek B. A. & Meyerowitz E. M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93, 4793–4798 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 9, 2102–2118 (2004). [DOI] [PubMed] [Google Scholar]
- Hernández-Hernández T., Martínez-Castilla L. P. & Alvarez-Buylla E. R. Functional diversification of B MADS-box homeotic regulators of flower development: Adaptive evolution in protein-protein interaction domains after major gene duplication events. Mol. Biol. Evol. 24, 465–481 (2007). [DOI] [PubMed] [Google Scholar]
- de Martino G. et al. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18, 1833–1845 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K. & Irish V. Hidden variability of floral homeotic B genes in Solanaceae provides a molecular basis for the evolution of novel functions. Plant Cell 22, 2562–2578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B. & Kramer E. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae). New Phytol. 197, 949–957 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J. S. et al. Deciphering the Physalis floridana Double-Layered-Lantern1 mutant provides insights into functional divergence of the GLOBOSA duplicates within the Solanaceae. Plant Physiol. 164, 748–764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S. et al. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000). [DOI] [PubMed] [Google Scholar]
- Ditta G. et al. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940 (2004). [DOI] [PubMed] [Google Scholar]
- Van Tunen A. J., Eikelboom W. & Angenent G. C. Floral organogenesis in Tulipa. Flowering Newslett. 16, 33–37 (1993). [Google Scholar]
- Tzeng T. Y. & Yang C. H. A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) In Arabidopsis thaliana. Plant Cell Physiol. 42, 1156–1168 (2001). [DOI] [PubMed] [Google Scholar]
- Kanno A. et al. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52, 831–841 (2003). [DOI] [PubMed] [Google Scholar]
- Nakamura T. et al. The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. orientalis (Agapanthaceae) flowers. Plant Mol. Biol. 58, 435–445 (2005). [DOI] [PubMed] [Google Scholar]
- Nakada M. et al. Isolation of MaDEF from Muscari armeniacum and analysis of its expression using laser microdissection. Plant Sci. 170, 143–150 (2006). [Google Scholar]
- Xu Y. et al. Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46, 54–68 (2006). [DOI] [PubMed] [Google Scholar]
- Tsaftaris A., Polidoros A. N., Pasentsis K. & Kalivas A. Tepal formation and expression pattern of B-class paleoAP3-like MADS-box genes in crocus (Crocus sativus L.). Plant Sci. 170, 238–246 (2006). [Google Scholar]
- Hirai M., Kamimura T. & Kanno A. The expression patterns of three class B genes in two distinctive whorls of petaloid tepals in Alstroemeria ligtu. Plant Cell Physiol. 48, 310–321 (2007). [DOI] [PubMed] [Google Scholar]
- Hirai M., Ochiai T. & Kanno A. The expression of two DEFICIENS-like genes was reduced in the sepaloid tepals of viridiflora tulips. Breed. Sci. 60, 110–120 (2010). [Google Scholar]
- Su C. L. et al. A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. Plos ONE 8, e80462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R. & Meyerowitz E. M. Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W. et al. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11, 4693–4704 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent G. C. et al. Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 4, 101–112 (1993). [DOI] [PubMed] [Google Scholar]
- Lange M. et al. The seirena B class floral homeotic mutant of California Poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. Plant Cell 25, 438–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R. et al. Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula. Plant J. 60, 102–111 (2009). [DOI] [PubMed] [Google Scholar]
- Yan X. et al. Functional identification and characterization of the Brassica napus transcription factor gene BnAP2, the ortholog of Arabidopsis thaliana APETALA2. Plos ONE 7, e33890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque E. et al. Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 73, 663–675 (2013). [DOI] [PubMed] [Google Scholar]
- Adachi Y., Mori S. & Nakano M. Agrobacterium-mediated production of transgenic plants in Tricyrtis hirta (Liliaceae). Acta Hort. 673, 415–419 (2004). [Google Scholar]
- Mori S. et al. Stability of β-glucuronidase gene expression in transgenic Tricyrtis hirta plants after two years of cultivation. Biol. Plant 52, 513–516 (2008). [Google Scholar]
- Hiei Y., Ohta S., Komari T. & Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. 43, 724–744 (2005). [DOI] [PubMed] [Google Scholar]
- Guo X. et al. The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development. Sci. Rep. 6, 20454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W. G. & Van Meeteren U. Flower opening and closure: a review. J. Exp. Bot. 54, 1801–1812 (2003). [DOI] [PubMed] [Google Scholar]
- Vandenbussche M. et al. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16, 741–754 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado-Rojo M. et al. Quantitative levels of Deficiens and Globosa during late petal development show a complex transcriptional network topology of B function. Plant J. 72, 294–307 (2012). [DOI] [PubMed] [Google Scholar]
- Nakano M. et al. Somatic embryogenesis and plant regeneration from callus cultures of several species in the genus Tricyrtis. In Vitro Cell Dev. Biol. Plant 40, 274–278 (2004). [Google Scholar]
- Frohman M. A., Dush M. K. & Martin G. R. Rapid production of full length cDNAs from rare transcripts: Amplification using a single gene specific oligonucleotide primer. Proc. Natl Acad. Sci. USA 85, 8998–9002 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiishi Y. et al. Flower color alteration in the liliaceous ornamental Tricyrtis sp. by RNA interference-mediated suppression of the chalcone synthase gene. Mol. Breed. 30, 671–680 (2012). [Google Scholar]
- Ririe K., M. Rasmussen R. P. & Wittwer C. T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245, 154–160 (1997). [DOI] [PubMed] [Google Scholar]
- Ohta M. et al. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike Y., Hoshino Y., Mii M. & Nakano M. Horticultural characterization of Angelonia salicariifolia plants transformed with wild-type strains of Agrobacterium rhizogenes. Plant Cell Rep. 21, 981–987 (2003). [DOI] [PubMed] [Google Scholar]
- Nonaka T. et al. Chromosome doubling of Lychnis spp. by in vitro spindle toxin treatment of nodal segments. Sci. Hort. 129, 832–839 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.