Abstract
Hematopoietic stem cells (HSCs) maintain blood cell production life‐long by their unique abilities of self‐renewal and differentiation into all blood cell lineages. Growth arrest and DNA‐damage‐inducible 45 alpha (GADD45A) is induced by genotoxic stress in HSCs. GADD45A has been implicated in cell cycle control, cell death and senescence, as well as in DNA‐damage repair. In general, GADD45A provides cellular stability by either arresting the cell cycle progression until DNA damage is repaired or, in cases of fatal damage, by inducing apoptosis. However, the function of GADD45A in hematopoiesis remains controversial. We revealed the changes in murine HSC fate control orchestrated by the expression of GADD45A at single cell resolution. In contrast to other cellular systems, GADD45A expression did not cause a cell cycle arrest or an alteration in the decision between cell survival and apoptosis in HSCs. Strikingly, GADD45A strongly induced and accelerated the differentiation program in HSCs. Continuous tracking of individual HSCs and their progeny via time‐lapse microscopy elucidated that once GADD45A was expressed, HSCs differentiate into committed progenitors within 29 hours. GADD45A‐expressing HSCs failed to long‐term reconstitute the blood of recipients by inducing multilineage differentiation in vivo. Importantly, γ‐irradiation of HSCs induced their differentiation by upregulating endogenous GADD45A. The differentiation induction by GADD45A was transmitted by activating p38 Mitogen‐activated protein kinase (MAPK) signaling and allowed the generation of megakaryocytic‐erythroid, myeloid, and lymphoid lineages. These data indicate that genotoxic stress‐induced GADD45A expression in HSCs prevents their fatal transformation by directing them into differentiation and thereby clearing them from the system. Stem Cells 2016;34:699–710
Keywords: Hematopoietic stem cells, Differentiation, Self‐renewal, Cell fate decisions, GADD45 family, Single cell tracking, In vivo transplantations
Significance Statement.
Since there have been a lot of controversies about the function of the DNA‐damage induced tumor suppressor gene Growth arrest and DNA‐damage‐inducible 45 alpha (GADD45A) in hematopoiesis, we revisited the role of GADD45A in hematopoietic stem cell fate choice control at single cell resolution. We show here that neither cell cycle arrest nor cell death was induced by GADD45A in HSCs, but a strong differentiation program via the initiation of the p38 MAPK signaling pathway. Forcing terminal differentiation of damaged hematopoietic stem and progenitor cells (HSPCs) might be a prominent function of GADD45A in the hematopoietic system, besides providing DNA repair and genomic stability.
Introduction
Hematopoietic stem cells (HSCs) serve for the life‐long production of all blood cell lineages 1, 2. Their unique ability of renewing themselves maintains the HSC pool. Since HSCs are long‐lived they might be susceptible for acquiring genomic alterations leading to blood disorders. The genomic integrity of HSCs is instrumental to avoid detrimental mutations that may cause clonal dominance and leukemia initiation 3, 4. GADD45A belongs to the growth arrest and DNA damage‐inducible 45 family consisting of three genes Gadd45a, Gadd45b, and Gadd45g 5, 6, 7. Various cell fate decisions and functions are regulated by individual GADD45 family members including cell cycle progression and arrest, survival and death, DNA repair and epigenetic modifications 8. Because of their pleiotropic action, GADD45 proteins regulate basic cell biological mechanisms with wide implications in development, regeneration, aging, and disease 8, 9. We previously elucidated the function of the family member GADD45 Gamma as a hematopoietic cytokine‐induced mediator of differentiation and lineage choice control in HSCs 10. Different stimuli activate the expression of individual GADD45 family members which execute distinct and redundant functions in a cell type and context‐dependent manner 8, therefore, their individual functional contribution needs further attention.
The importance of GADD45A in DNA excision repair through the interaction with proliferating cell nuclear antigen (PCNA) 11, 12 becomes apparent in mice lacking Gadd45a (Gadd45a −/−) that show increased genome instability, reduced nucleotide excision repair, a higher rate of mutations, and consequently, a higher incidence of carcinogenesis 13. The absence of GADD45A in long‐term repopulating HSCs (LT‐HSCs) resulted in a delayed DNA repair and increased leukemogenesis in aged animals 14. GADD45A expression is initiated by ultra violet irradiation (UV), X‐rays, γ‐irradiation, multiple DNA toxins, among other cellular stressors 8. These environmental stressors and DNA damaging agents endanger the integrity of the HSC genome. GADD45A is specifically involved in DNA repair, and thus, delays the cell cycle when DNA damage is detected. Thereby Gadd45a is a tumor suppressor under the transcriptional control of p53 and the homologs p63 and p73 15. However, p53‐dependent and ‐independent pathways are involved that regulate Gadd45a induction by genotoxic stress; the later may use BRCA1‐related or MAPK‐mediated signals 16, 17.
The role of GADD45A in inducing a cell cycle arrest as a direct consequence of DNA damage is well‐established. GADD45A expression arrests the cell cycle progression at G2/M phase in normal and cancer cells by the interaction with the protein kinase cell division cycle 2, cyclin B1, and p53‐inducing proteins such as PCNA and p21 18, 19, 20. Also cellular senescence in human fibroblasts is associated with a p53‐dependent induction of GADD45A 21. Along the same line, mouse embryonal fibroblasts, lymphocytes, and bone marrow (BM) myeloid cells from Gadd45a −/− mice showed an increased proliferation with an accelerated mitotic index upon methylmethane sulfonate or UV‐induced stress 13.
GADD45A plays a dual function in cell survival and death control in a cell type‐specific manner. Ectopic expression of GADD45A in leukemia cells and solid cancer cells triggers apoptosis via the Transforming growth factor, beta (TGFb)/MAP3K4/p38/c‐Jun N‐terminal kinase (JNK) pathway 20, 22. Conversely, GADD45A acts antiapoptotic and increases hematopoietic cell survival under UV‐irradiation or treatment with chemotherapeutic drugs 23. While thymocytes from Gadd45a −/− mice had no change in gamma irradiation (γ‐IR)‐ or UV‐induced apoptosis 13, GADD45A‐deficient BM hematopoietic cells were more sensitive to UV‐radiation and chemotherapy‐induced apoptosis compared to wild‐type cells 23, 24, 25. GADD45A induces the Nuclear factor of kappa light polypeptide gene enhancer in B‐cells (NFκB)‐mediated prosurvival pathway via p38 in myeloid cells 24. In contrast, HSCs from Gadd45a −/− mice were more resistant to γ‐IR than their wild‐type counterparts, suggesting a specific proapoptotic function of GADD45A in HSCs but not in myeloid‐committed progenitors 14.
These controversies on the functional consequences of GADD45A expression motivated us to elucidate the changes in cell fate decision and function of long‐term repopulating HSCs (LT‐HSCs) after GADD45A expression at single cell resolution. GADD45A is higher expressed in LT‐HSCs than in more differentiated progenitor populations in steady‐state hematopoiesis 14, albeit at low levels in general, and is strongly induced by genotoxic noxins. We hypothesized that the induction of GADD45A in LT‐HSCs would cause alterations in cell survival and a delay in cell cycle progression in murine LT‐HSCs. Surprisingly, ectopic GADD45A expression did not change cell expansion, cell cycle progression or survival in murine LT‐HSCs, neither in vitro nor in vivo. GADD45A expression caused an immediate and accelerated differentiation of LT‐HSCs allowing all cell lineages to emerge. The differentiation induction is mediated by the activation of the MAP3K4‐p38 axis via GADD45A, and may provide a mechanism to circumvent the long‐term existence of damaged HSCs to avoid fatal transformation and leukemogenesis.
Materials and Methods
Mice
C57BL/6J and B6.SJL‐Ptprca Pepcb/BoyJ were used at 10–14 weeks of age. The mice were bred and maintained under specific pathogen‐free conditions. Animal experiments were performed in accordance with the German animal welfare legislation and were approved by the relevant authorities (Regierungspräsidium Darmstadt).
Ex Vivo Differentiation
One hundred LT‐HSCs per well (96‐well format) were lentivirally transduced (multiplicity of infection (MOI) 100) and cultured in serum‐free expansion medium (SFEM) (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) supplemented with 100 ng/ml SCF and Thrombopoietin (TPO) (both PeproTech, Rocky Hill, NJ, http://www.peprotech.com). Cells were analyzed by fluorescence activated cell sorting (FACS) (antibodies against CD48, CD117, CD16/32, CD11b). For Tetracycline (TET)‐inducible GADD45A expression studies, 300 LT‐HSCs were seeded per well in SFEM supplemented with stem cell factor (SCF), TPO, and Doxycycline at the indicated concentrations. The cells were lentivirally transduced and analyzed after 8 days of culture as described above.
For analyzing the LT‐HSC fate upon DNA‐damage inducing conditions, 1,000 LT‐HSCs per well (96‐well format) were seeded in SFEM supplemented with SCF and TPO. After 24 hours, the cells were stimulated with γ‐IR (2 Gy) and were analyzed for their marker expression after 5 days of culture as described above.
For inhibitor studies 20 ng/ml Interleukin 3 (IL3) (PeproTech) was additionally supplemented. Inhibitors were used as follows: VX‐702 (1 μM), LY2228820 (0.1 μM, all from Selleck, http://www.selleckchem.com). FACS antibodies are listed in Supporting Information Table S1.
FACS Analysis and Sorting of Stem and Progenitor Cells
BM cells from femurs, tibias, coxae, and sternum were either crushed or flushed (without sternum) followed by a lineage depletion of lineage positive cells (EasySepTM Biotin Selection Kit, Stem Cell Technologies) using biotin‐labeled antibodies (CD3, CD45R, CD19, CD11b, CD41, Ter119, Gr1). Hematopoietic stem and progenitor cell (HSPC) populations were sorted with a FACS Aria I or III (BD, http://www.bd.com) or analyzed using a LSR Fortessa (BD). Following surface marker combinations were used to identify and sort the different populations: CD117, Sca1, CD150, CD34, CD16/32, Streptavidin, and CD48 (Supporting Information Table S1 and Fig. S1A). Viable sorted cells were counted with trypan blue exclusion.
Western Blot
20,000 KSL (CD117+ Sca1+ Lin− cells) were lentivirally transduced with either control or GADD45A (MOI 100) and cultured in SFEM supplemented with SCF, TPO (both 100 ng/ml), IL3, and IL6 (both 20 ng/ml) for 9 days. An aliquot of the cells was used to determine the transduction efficiency via flow cytometry. Cells were lysed for 30 minutes on ice in lysis buffer (50 mM Tris pH 7, 4, 150 mM NaCl, 2 mM EDTA, 1% Triton X‐100, protease inhibitors (Roche, Basel, Switzerland, http://www.roche-applied-science.com)), and the supernatant was collected after centrifugation. 30 µg of protein was applied to SDS‐PAGE and transferred to a Polyvinylidene difluoride (PVDF) membrane using the Trans‐Blot Turbo‐Transfer System (Bio‐Rad, Hercules, CA, http://www.bio-rad.com). The bound primary antibodies (GADD45A (D17E8) rabbit mAb and HSP90 rabbit mAb; both Cell Signaling, Beverly, MA, http://www.cellsignal.com) were detected by the use of horseradish peroxidase‐conjugated secondary antibody and the ECL Prime detection system (GE Health). The band density was semiquantified using ImageJ software.
Quantitative Reverse Transcriptase Polymerase Chain Reaction of GADD45A Expression upon DNA‐Damage Stimulus
Freshly FACS‐sorted KSL were subjected to γ‐IR (2 Gy), UV (1200 J/m2), or Cisplatin (10 µg/ml) in SFEM supplemented with 100 ng/ml SCF. Cells were harvested 4 hours post‐IR and UV or after 5 hours Cisplatin stimulation. The RNA was isolated and analyzed using TaqMan Gene Expression Cells‐to‐CT Kit (Life Technologies, Rockville, MD, http://www.lifetech.com). Cells were lysed and RNA was transcribed into cDNA according to the manufacturer's protocol. The cDNA was preamplified for 14 cycles using the TaqMan PreAmp Master Mix (Life Technologies) according to the manufacturer's protocol. The TaqMan Gene expression MasterMix (Life Technologies) was used for the quantitative polymerase chain reaction (qPCR) performed on a StepOne instrument (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com). The expression of GADD45A (Mm00432802_m1) was normalized to Hypoxanthine guanine phosphoribosyl transferase (HPRT) (Mm00446968_m1) using the ΔCT method.
Quantitative RT‐PCR of GADD45B and GADD45G Expression in GADD45A‐Transduced LT‐HSCs
3,000 LT‐HSCs per well (96‐well format) were lentivirally transduced with control or GADD45A (MOI 100) and cultured in SFEM supplemented with 100 ng/ml SCF and TPO (all PeproTech). After 3 days VENUS+, CD48− cells were directly sorted into the lysis buffer of the TaqMan Gene Expression Cells‐to‐CT Kit (Life Technologies). The whole lysate was used for the RT‐reaction according to the manufacturer's protocol. The cDNA was preamplified for 14 cycles using the TaqMan PreAmp Master Mix (Life Technologies) according to the manufacturer's protocol. The expression of GADD45B (Mm00435123_m1) and GADD45G (Mm00442225_m1) was normalized to B2m (Mm00437762_m1) using the ΔCT method.
Colony Formation Assay
100 LT‐HSCs per plate and 300 MEPs (megakaryocyte‐erythroid progenitor cell) per plate were lentivirally transduced (MOI 100), seeded 24 hours later in M3434 medium (Stem Cell Technologies), and scored microscopically after 9–12 (LT‐HSCs) and 6–8 days (MEPs) days, respectively, for transduction and colony formation (CellObserver, Zeiss, http://www.zeiss.com).
Time‐Lapse Imaging
FACS sorted LT‐HSCs were seeded in 24‐well plates (SFEM medium, 100 ng/ml SCF and TPO) equipped with silicon culture inserts (IBIDI, Martinsried, Germany http://www.ibidi.com) and immediately transduced with lentiviral particles (MOI 100). Allophycocyanin (APC)‐conjugated anti‐CD16/32 was added to the medium at 50 ng/ml. After preincubation at 5% CO2/37 °C for 19 hours plates were gas‐tight sealed with adhesive tape. Microscopy was performed using a CellObserver (Zeiss) at 37 °C. Phase contrast images were acquired every 2–3 minutes using a × 10 phase contrast objective (Zeiss), and an AxioCamHRm camera (at 13 1,388 × 1,040 pixel resolution) with a self‐written VBA module remote controlling Zeiss AxioVision 4.8 software. Fluorescence was detected every 2 hours with HXP illumination (Osram) and the filter sets for YFP (F46‐003) and APC (HC628/40, ET66LP XR, ET700/75, AHF Analysetechnik).
Cell Tracking
Cell tracking was performed using a self‐written computer program (TTT) as described 10, 26, 27, until the fate of all progeny in the third cell generation was determined. The generation time of an individual cell was defined as the time span from cytokinesis of its mother cell division to its own division. The first entry into mitosis of the purified LT‐HSCs was defined as time of first division. Dead cells are easily depicted by their shrunk, nonrefracting appearance with immobility. All cell tracking was done by scientists; the current analysis does not rely on data generated by an unsupervised computer algorithm for automated tracking.
Competitive Repopulation Assay
350 FACS‐sorted LT‐HSCs from C57.BL/6J mice (CD45.2), which were lentivirally transduced (MOI 100) 24 hours prior transplantation were tail vein injected into lethally irradiated B6.SJL‐Ptprca Pepcb/BoyJ (CD45.1) recipients together with 2 × 105 BM competitor recipient cells from B6.SJL‐Ptprca Pepcb/BoyJ. Transduction efficiency of the transplanted LT‐HSCs was determined with a remaining cell aliquot after three additional days in culture by FACS. Multilineage reconstitution was determined every 2–18 weeks post‐transplantation in peripheral blood (PB). Shortly, red blood cells were lysed with PharmLysis Buffer (BD), and cells were stained with antibodies against CD45.1, CD45.2, CD3, B220, Ter119, CD11b/Gr1, and a dead/live cell exclusion (Fixable Viability Dye, eBioscience). Lentivirally transduced hematopoietic cells were analyzed for VENUS expression by FACS (FACS Canto II or LSR Fortessa).
Short‐Term Transplantation
A total of 100,000 KSL cells from B6.SJL‐Ptprca Pepcb/BoyJ (CD45.1) were lentivirally transduced (MOI 100) and after 24 hours equally distributed and intravenously transplanted into seven lethally irradiated C57.BL/6J (CD45.2) recipient mice per group together with 5 × 105 BM mononuclear recipient bystander cells (CD45.2). Mice were sacrificed 21 days after injection and analyzed for donor reconstitution in BM and spleen. For cell proliferation analyses 1.5 mg Bromodeoxyuridine (BrdU) (BD) were injected intraperitoneally 4 hours before sacrificing the mice. Dead cells were determined by fixable Viability Dye staining (eBioscience). BrdU incorporation and DNA content 7‐Amino Actinomycin D (7AAD) in living cells were measured according to the manufacturer's instructions (BD) with an APC‐labeled anti‐BrdU antibody (BD) by FACS (Fortessa, BD).
Vector Construction
The open reading frame (ORF) of the fluorescent reporter protein VENUS‐hImportin subunit α1 (AA2‐67) was cloned into the third generation self‐inactivating lentiviral vector pRRL.PPT.SFFV.IRES.eGFP.wPRE by replacing the ORF of green fluorescent protein 28. A multiple cloning site (MCS) was inserted after the SFFV promoter. The ORF of murine Gadd45a was amplified by RT‐PCR (forward 5′‐TTGGCCGGCCGAGGGACTCGCACTTGCAATATG‐3′, reverse 5′‐TTACTAGTGAACTCGGCCCCTTGACAT‐3′) from murine cDNA and cloned into the MCS.
The ORF of GFP in the TET‐responsive vector pRRL.PPT.TetT11‐GFP‐PGK‐M2‐pre 29 was replaced by the cassette containing Gadd45a‐IRES‐VENUS‐hImportin subunit α1 (AA2‐67) of the above described plasmid pRRL.PPT.SFFV.Gadd45a‐IRES‐VENUS‐hImportin.wPRE via BsrGI and SalI.
Virus Production
Vesicular Stomatitis Virus‐G‐pseudotyped lentiviral particles were produced in a split genome approach by calcium‐phosphate‐mediated transient transfection of human embryonic kidney HEK293T producer cells as recently described 10, 30. After 48 hours, supernatant was collected, filtered (45 µm), and enriched by ultracentrifugation (50,000g, 2 hours). Viral titers were determined by transduction of NIH3T3 cells with different concentrations of virus supernatant and FACS.
Phosphoflow Cytometry
FACS‐sorted LT‐HSCs were transduced with indicated lentiviral particles (MOI 100) and cultured for 4 days. After staining cells with Fixable Viability Dye (eBioscience) for live/dead cell exclusion, the cells were fixed with Fix Buffer I (BD) at 37 °C for 10 minutes and permeabilized with ice‐cold Perm Buffer III (BD) for 30 minutes on ice. Permeabilized cells were subsequently stained with antibodies against P‐p38‐Phycoerythrin (PE) (BD) to analyze the phosphorylation status of the cells by FACS. Four‐day cultured LT‐HSCs or MPPs were stimulated with 10 µg/ml Anisomycin (Sigma) for 30 minutes prior to fixation as positive control.
Statistics
Statistical analysis was performed with GraphPadPrism‐6.0 software. Statistical significance was determined by a Student's t test (two‐tailed, unpaired, equal variances) if not otherwise mentioned. The significance level for all tests was set to α = 5%.
Results
GADD45A Neither Inhibits Cell Cycle Progression Nor the Survival in LT‐HSCs and their Progeny
In order to assess the influence of GADD45A expression on the expansion of LT‐HSCs and their progeny, we lentivirally transduced FACS‐sorted LT‐HSCs (CD150+ CD48− CD34low CD117+ Sca1+ Lin−) with GADD45A or a control vector (Fig. 1A; Supporting Information Fig. S1A, S1B). The ectopic GADD45A expression enhanced GADD45A protein levels about twofold in lentivirally transduced cells (Fig. 1A). The number of transduced cells, that coexpressed a fluorescent VENUS reporter protein, was assessed at 5, 8, and 10 days in culture. There was no influence on the expansion of the cells by the expression of GADD45A (Fig. 1B). This unexpected result indicated that there might be no gross changes in proliferation and cell death caused by GADD45A. However, to confirm that these fate decisions were not skewed we investigated individual LT‐HSCs during differentiation via time‐lapse‐microscopy‐based cell tracking (Fig. 1C). This technology allows following individual LT‐HSCs and their progeny during their differentiation process over weeks in culture without losing cell identity. FACS‐sorted LT‐HSCs were subjected to time‐lapse microscopy while lentivirally transduced with GADD45A. The induction of transgene expression (VENUS+) could be followed in real time at a single cell level. We compared the time of first LT‐HSC division and the cell cycle lengths of subsequent progeny of LT‐HSCs transduced with GADD45A or control for several generations (Fig. 1D, 1E; Supporting Information Fig. S2). Neither did we determine any delay of the first entry into LT‐HSC division nor any increase in the cell cycle length in GADD45A‐expressing cells as originally expected by the expression of a protein normally involved in growth arrest. To the contrary, in later generations, the cell cycle was even shorter in GADD45A‐expressing cells, which might be a consequence of a further advanced differentiation stage rather than a direct influence of GADD45A on the cell cycle (Fig. 1E). Next, we assessed the cell cycle of hematopoietic cells after GADD45A expression in vivo (Fig. 1F). Since we sought to determine the influence of GADD45A on HSPC proliferation shortly after transplantation, a sufficient number of donor cells were required. Therefore, we transplanted KSL cells transduced with GADD45A or control and assessed the cell cycle status of GADD45A‐expressing progeny in the recipient BM 3 weeks after transplantation via a short BrdU pulse of 4 hours (Fig. 1F; Supporting Information Fig. S1C). When we analyzed the cell cycle phases of all cells, lineage− cKIT+ progenitor cells or immature HSPCs (KSL cells), neither of these subpopulations showed any signs of alterations upon GADD45A expression determined via FACS. The cell cycle distribution was unchanged in the presence of GADD45A, indicating that GADD45A does not alter the proliferation of HSPCs (Fig. 1G).
Figure 1.
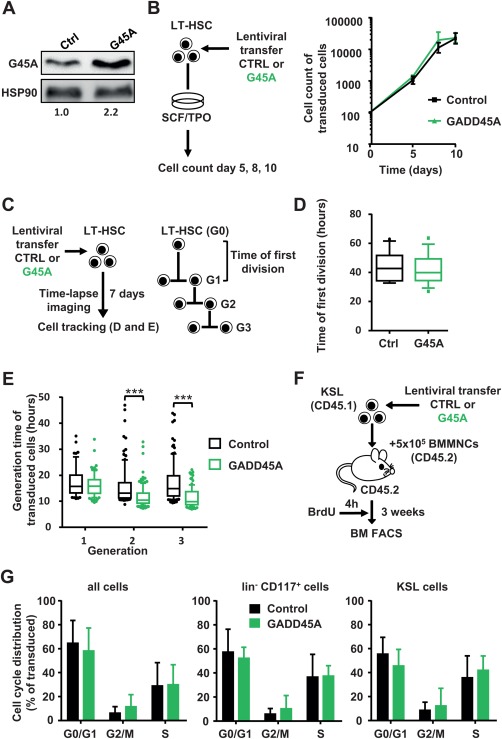
GADD45A expression does not lead to a cell cycle arrest in HSCs and their progeny (A): Relative quantification of the GADD45A protein expression via immunoblot of KSL cells lentivirally transduced with GADD45A or control. The transduction efficiency was 91% and 86% for control and GADD45A‐transduced cells, respectively. (B): Experimental scheme and quantification of in vitro expansion of GADD45A expressing compared to control‐transduced LT‐HSCs. N = 3 individual experiments. Data represented as mean ± SD. (C): Experimental scheme of video‐microscopy based cell tracking of GADD45A‐ or control‐expressing LT‐HSCs and their progeny. Tracking results are shown as boxplots (box represents 25%–75% and the median with whiskers indicating 10%–90%, outliers are shown as dots) in (D) and (E). (D): The time of first division of 15 control and 14 GADD45A‐expressing LT‐HSCs. (E): Cell cycle length (generation time) in three subsequent generations. Data were generated using two independent experiments analyzing N = 265 cells for control and N = 313 cells for GADD45A. Only transduced cells (VENUS+) were included. Statistical significance was determined using the Mann‐Whitney test. (F): Experimental scheme of BrdU incorporation analysis after in vivo transplantation. (G): Cell cycle phases of all cells, lineage− CD117+ cells, and KSL cells in the bone marrow 3 weeks after transplantation are displayed. Only transduced (VENUS+) cells were included. N = 6 mice per group. Data represented as mean + SD. ***, p < .001. Abbreviations: BM, bone marrow; BMMNCs, bone marrow mononuclear cells, BrdU, bromodeoxyuridine; Ctrl, control; FACS, fluorescence activated cell sorting; G45A, GADD45A; G0‐3, generation 0‐3; LT‐HSC, long‐term repopulating hematopoietic stem cells; SCF, stem cell factor; TPO, thrombopoietin.
GADD45A has been implicated in the decision of cells to either survive or to go into apoptosis upon cell damage. Both scenarios have been reported as a consequence of the absence of GADD45A in hematopoietic cells 14, 23, 24. We measured the rate of dying cells at a single cell level by observing GADD45A‐expressing LT‐HSCs and their progeny by time‐lapse‐microscopy‐based cell tracking (Fig. 2A). There was no increase in apoptosis in HSPCs upon GADD45A expression for several cell generations (Fig. 2A). Also in vivo, 3 weeks after transplanting GADD45A‐expressing KSL, there was no increase in apoptosis in BM cells originating from GADD45A‐expressing KSL cells (Fig. 2B).
Figure 2.
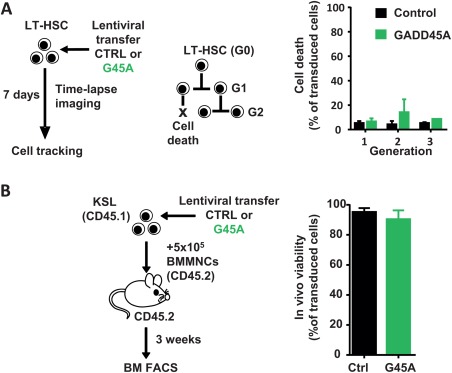
GADD45A does not induce cell death in HSCs and their progeny. (A): Occurrence of cell death events upon ectopic expression of GADD45A in LT‐HSCs was assessed by video‐microscopy and subsequent single cell tracking. Data were generated by tracking cells from two independent experiments. Only transduced cells (VENUS+) were included. (B): Experimental scheme and quantification of viability of donor‐derived cells in recipient bone marrow 3 weeks after transplantation determined via FACS. N = 6 mice per group. All data are represented as mean + SD. Abbreviations: BM, bone marrow; BMMNCs, bone marrow mononuclear cells; CTRL, control; FACS, fluorescence activated cell sorting; G45A, GADD45A; LT‐HSCs, long‐term repopulating hematopoietic stem cells.
GADD45A Expression in LT‐HSCs Induces and Accelerates Differentiation
In order to investigate consequences of GADD45A on HSC differentiation and self‐renewal, we assessed the differentiation kinetics of LT‐HSCs that were lentivirally transduced with GADD45A by the analyses of surface marker expression via FACS (Fig. 3A). Already 5 days after transduction, almost 40% of the GADD45A‐expressing LT‐HSCs have differentiated into granulocyte‐macrophage progenitor (GMP)‐like cells, whereas only 4% of the control‐transduced LT‐HSCs have developed into this stage and the majority of cells were still considered immature (Fig. 3A, 3B). Along the same line, after 8 days of differentiation, 66% of GADD45A expressing cells developed into mature cells of the granulocyte/monocyte (GM) lineage. Only 12% of control‐transduced cells reached this stage of differentiation after 8 days (Fig. 3B). Since the level of GADD45A expression by a lentiviral vector may influence the true physiological cell fate decision of LT‐HSCs upon induced endogenous GADD45A expression, we additionally used an inducible and scalable TET‐regulated lentiviral system (Supporting Information Fig. S3A). The expression of Gadd45a‐IRES‐VENUS was under the control of a TET‐inducible promoter, and limiting concentrations of Doxycycline induced a linear range of GADD45A expression (Fig. 3C). Even at maximal induction, the GADD45A expression level was only 22% of the expression level of the SFFV promoter‐driven lentiviral vector and could be scaled down to 6% (Fig. 3C). Only 22% of the previously ectopically expressed GADD45A was required to efficiently induce and accelerate the differentiation in LT‐HSCs to the same extend as shown with the SFFV‐driven GADD45A expression (Fig. 3D; Supporting Information Fig. S3B). The stepwise reduction of ectopic GADD45A expression below 22% of the original level decreased the impact on differentiation in a linear way. Strikingly, even as little as 6% was sufficient to induce differentiation in LT‐HSCs, although not to the same extent (Fig. 3D; Supporting Information Fig. S3B). This result clearly showed that even low levels of GADD45A, once expressed in LT‐HSCs, initiate the differentiation program in these cells and accelerate the differentiation. To test whether LT‐HSCs would start to differentiate after GADD45A expression, or whether they are able to remain self‐renewing in their in vivo niche, we transplanted LT‐HSCs after transduction of GADD45A or control in lethally irradiated recipients (Fig. 3E). We assessed donor blood cell reconstitution in PB up to 18 weeks after transplantation. After 2 weeks, GADD45A‐expressing LT‐HSCs cells initially contributed almost equally to donor blood cell production, suggesting a successful homing to the recipient BM. Thereafter, the donor cell chimerism did not increase and completely disappeared after 8 weeks, and there was no contribution of GADD45A‐expressing cells at 18 weeks after transplantation, whereas a robust reconstitution was detectable using control‐transduced cells (Fig. 3E). As a consequence, the serial transplantation of GADD45A‐expressing cells into secondary recipients was obsolete. When we analyzed the BM and spleen of GADD45A‐expressing KSL recipient mice, it became apparent that already 3 weeks after transplantation, the donor blood cells were markedly reduced (Supporting Information Fig. S3C, S3D). These in vivo transplantations demonstrated that LT‐HSC differentiate upon GADD45A expression and were unable to maintain their self‐renewal program even in their natural niche environment.
Figure 3.
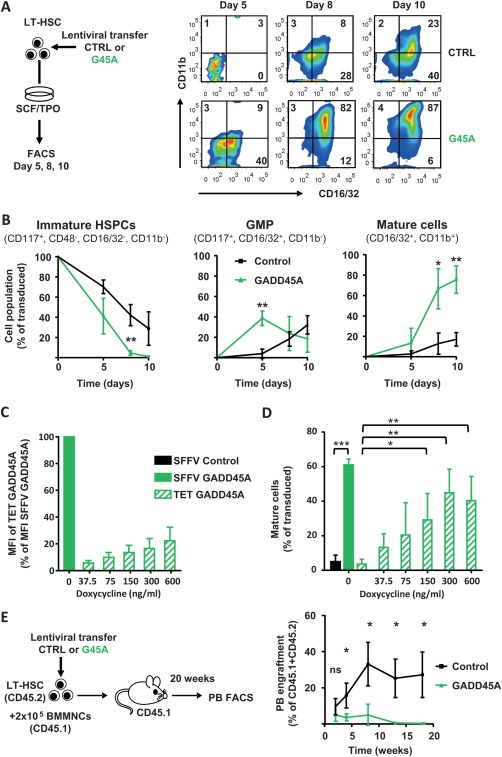
GADD45A expression in LT‐HSCs induces and accelerates differentiation. (A): Experimental scheme and exemplary FACS plots showing the vitro differentiation upon lentiviral expression of GADD45A compared to control over time. (B): Quantification of the differentiation kinetic after 5, 8, and 10 days in culture. Transduced cells (VENUS+) were gated for immature HSPCs, GMP‐like, and mature GM cells using the indicated surface marker expression profile. N = 4 individual experiments. Data represented as mean ± SD. (C, D): Quantification of the differentiation of LT‐HSCs transduced with a lentiviral Tet‐inducible Gadd45a‐IRES‐VENUS expression system after 8 days in culture via FACS. (C): Ectopic expression levels using increasing concentrations of Doxycycline determined by VENUS fluorescence. (D): Transduced cells (VENUS+) were gated for mature GM cells using the indicated surface marker expression profile. N = 3–4 individual experiments. Data represented as mean + SD. (E): In vivo blood reconstitution ability of GADD45A‐expressing LT‐HSCs compared to control in recipient mice. N = 3 for GADD45A, N = 6 mice for control group. Similar transduction efficiencies > 70%. Statistical significance was determined using the Mann‐Whitney test. *, p < .05; **, p < .01; ***, p < .001. Abbreviations: CTRL, control; G45A, GADD45A; GMP, granulocyte‐macrophage progenitor; HSPC, hematopoietic stem and progenitor cells; LT‐HSC, long‐term repopulating hematopoietic stem cells; MFI, mean fluorescence intensity; PB, peripheral blood; SCF, stem cell factor; SFFV, spleen focus‐forming virus; TET, tetracycline; TPO, thrombopoietin.
In order to measure the kinetics of differentiation induction in individual LT‐HSCs in real time, we investigated the onset of the differentiation marker CD16/32 expression that indicates GM‐committed cells, in the presence or absence of GADD45A. We transduced LT‐HSCs with GADD45A and control and subjected the cells to time‐lapse microscopy. The expression of the surface marker CD16/32 was determined by the binding of a fluorescently labeled antibody during differentiation in culture. In average, the time gap between the first GADD45A expression and the onset of CD16/32 was only 29 hours (±19 SD). However, almost no cells expressed detectable CD16/32 after 6 days of cell observation and tracking under control conditions.
DNA‐Damage Causing Conditions Upregulate GADD45A and Result in an Enhanced Differentiation of LT‐HSCs
So far we only provided evidence that ectopic GADD45A rapidly induces and accelerates differentiation in LT‐HSCs. Next we assessed whether conditions that cause DNA‐damage in cells, such as γ‐IR, UV, or cisplatin treatment induce endogenous GADD45A expression in stem and progenitor cells. We therefore FACS‐sorted KSL cells and treated them with IR, UV, or cisplatin and compared the levels of Gadd45a mRNA to untreated control KSL. As previously shown 14, 24, all these regimens triggered the expression of GADD45A (Fig. 4A). To test whether these DNA‐damage causing conditions also lead to an induction and acceleration of differentiation in LT‐HSCs, as seen with the ectopic expression (Fig. 3), we cultured γ‐IR‐treated LT‐HSCs for 5 days and determined the level of differentiation by FACS. Indeed, a significant lower proportion of immature HSPCs remained in the population of γ‐IR‐treated LT‐HSCs in comparison to untreated LT‐HSCs, and the reduction was similar to the effects caused by ectopic GADD45A expression (Fig. 4B). The additional ectopic expression of GADD45A in γ‐IR‐treated LT‐HSCs further enhanced their differentiation, probably by further increasing the levels of GADD45A (Fig. 4B). These results suggest that DNA‐damage causing conditions can indeed trigger the differentiation in LT‐HSCs, most likely caused by the upregulation of GADD45A.
Figure 4.
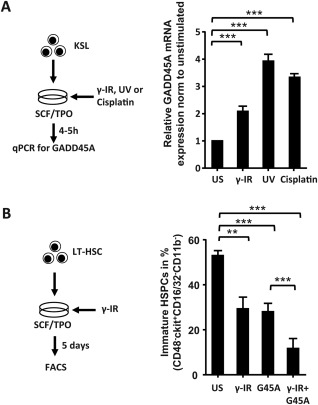
DNA damage‐causing conditions induce the expression of GADD45A and mediate the differentiation in LT‐HSCs. (A): GADD45A expression after DNA damage‐causing regimens in KSL cells via quantitative real‐time polymerase chain reaction. Cells were harvested 4 hours post γ‐IR (2 Gy), or UV (1,200 J/m2), or after 5 hours Cisplatin (10 µg/ml) stimulation. The GADD45A expression was normalized to HPRT and relative to the unstimulated control (N = 3). (B): LT‐HSCs were stimulated with γ‐IR (2 Gy) and analyzed for their differentiation status via FACS on day 5 (N = 3). All data represented as mean + SD. **, p < .01; ***, p < .001. Abbreviations: Cis, cisplatin; G45A, GADD45A; HSPCs, hematopoietic stem and progenitor cells; IR, irradiation; LT‐HSC, long‐term repopulating hematopoietic stem cells; qPCR, quantitative polymerase chain reaction; SCF, stem cell factor; TPO, thrombopoietin; US, unstimulated; UV, ultraviolet.
GADD45A Expression Allows the Generation of all Blood Cell Lineages
To elaborate whether GADD45A expression influences the lineage choice of LT‐HSCs and their progeny, we lentivirally transduced LT‐HSCs with either GADD45A or control and analyzed their colony‐forming ability in methylcellulose‐based medium (Fig. 5A). Mature cells of all myeloid lineages arose from LT‐HSCs transduced with GADD45A without major shifts in the lineage distribution in comparison to control‐transduced LT‐HSCs (Fig. 5A). Neither the colony size nor the lineage contribution was changed upon GADD45A expression. The video‐microscopy‐based tracking of GADD45A‐transduced LT‐HSCs elucidated their differentiation into megakaryocytes in culture (Supporting Information Fig. S4A; Video 1). Also in in vivo transplantations, PB B, T, and myeloid cell lineages were generated from LT‐HSCs transduced with GADD45A (Fig. 5B). The increased ratio of long‐lived lymphoid cells versus short lived myeloid cells upon GADD45A expression further suggest an advanced differentiation of immature HSPCs 8 weeks after transplantation. Later on, almost no GADD45A‐expressing cells were detectable for multilineage analyses. Furthermore, progenitor cells for GMPs and MEPs were unchanged in the BM 3 weeks after transplantation indicating that GADD45A‐expressing KSL give rise to all myeloid lineages (Supporting Information Fig. S4B). Since the other family member GADD45G induced a lineage selection against megakaryocyte‐erythroid cell fate, and MEPs could not handle the GADD45G‐induced program and died 10, we wanted to confirm that the program of GADD45A is not inhibiting cells of this lineage. In contrast to GADD45G, however, GADD45A‐transduced MEPs (CD150+ CD117+ Sca1− Lin−, Supporting Information Fig. S1A, FACS‐sorting according to 31, 32) were largely unaffected in their ability to form mature erythroid/megakaryocytic colonies (Fig. 5C). Taken together, GADD45A expression did not lead to a skewing of the generated lineages as GADD45G expression does by selecting against megakaryocyte‐erythroid lineage choice 10. Therefore, we assessed the expression of endogenous GADD45 Beta and Gamma upon ectopic GADD45A expression in LT‐HSCs (Fig. 5D). While the GADD45B mRNA levels remained constant, the ectopic expression of GADD45A by lentiviral transduction of LT‐HSCs caused a sharp downregulation of endogenous GADD45G, thereby allowing the development of megakaryocytes and erythrocytes (Fig. 5D).
Figure 5.
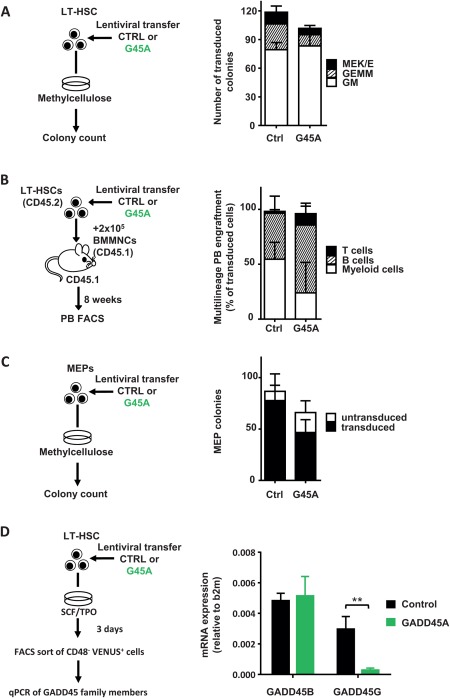
GADD45A expression allows HSCs to form all mature blood cell lineages. (A): Colony formation of 200 LT‐HSCs transduced with GADD45A or control vector. N = 3 independent experiments. (B): Multilineage PB engraftment monitored after 8 weeks upon transplantation of LT‐HSC as shown in the experimental scheme. N = 3 for GADD45A, N = 6 mice for control group. (C): Colony formation assay with sorted MEPs. N = 3 experiments. (D): Experimental scheme to analyze the mRNA expression of GADD45B and GADD45G in LT‐HSCs ectopically expressing control or GADD45A. After 3 days VENUS+ CD48− cells were directly sorted into the lysis buffer and RNA was extracted. The expression level of GADD45B and GADD45G was normalized to B2m. N = 5. All data represented as mean + SD. **, p < .01. Abbreviations: BMMNCs, bone marrow mononuclear cells; b2m, Beta‐2 microglobulin; Ctrl, control; FACS, fluorescence activated cell sorting; G45A, GADD45A; GEMM, granulocyte‐erythrocyte‐monocyte/macrophage‐megakaryocyte; LT‐HSC, long‐term repopulating hematopoietic stem cells; MEK/E, megakaryocyte‐erythrocyte; GM, granulocyte/monocyte; MEP, megakaryocyte‐erythrocyte progenitor; PB, peripheral blood.
GADD45A Activates the p38 MAPK for Differentiation Induction in HSCs
To enlighten the molecular mechanism how GADD45A activates the differentiation program in LT‐HSCs, we focused on the recently unraveled mechanism of GADD45G‐mediated differentiation induction in LT‐HSCs 10. GADD45 proteins bind to mitogen‐activated protein kinase kinase kinase 4 (MAP3K4) and release its autoinhibitory domain, leading to its activation and phosphorylation of downstream MAPK kinases (such as MKK3 and MKK6). Thereby, the binding of GADD45A to MAP3K4 initiates a MAPK cascade resulting in p38 and JNK activation 33. We showed that GADD45G specifically leads to p38 phosphorylation, and that the inhibition of p38, but not of JNK, resulted in a block in differentiation initiation of GADD45G 10. In order to see whether GADD45A also uses the same pathway we tested the induction of p38 phosphorylation by GADD45A via phosphoflow cytometry (Fig. 5A). As hypothesized, GADD45A‐expressing LT‐HSCs showed a stronger p38 phosphorylation than control‐transduced LT‐HSCs, indicating that the differentiation induction pathway is shared between GADD45G and GADD45A (Fig. 6A). To further corroborate that activated p38 is necessary for the GADD45A‐induced differentiation, we tested the differentiation kinetics of LT‐HSCs in the presence of p38 inhibitors (Fig. 6B). We lentivirally transduced LT‐HSCs with GADD45A or control and cultured them for 5 days in the presence or absence of two p38 inhibitors Vx702 and Ly2228820. In the presence of any p38 inhibitor, the differentiation induction and acceleration mediated by GADD45A was blocked, and the cells behaved like the control transduced cells (Fig. 6B). This clearly indicates that GADD45A induces its differentiation program by activation of the MAP3K4‐MKK6‐ p38 pathway.
Figure 6.
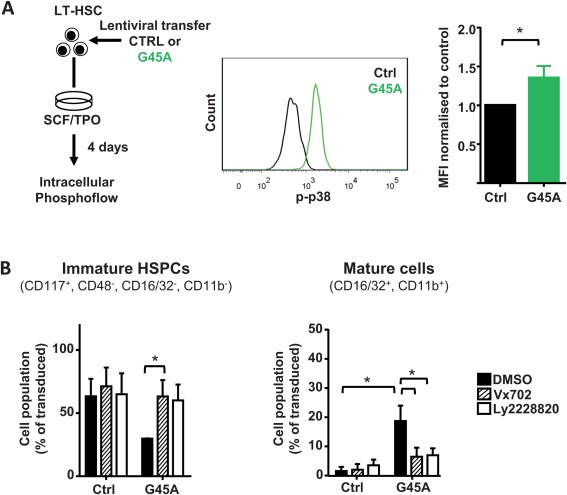
GADD45A induces differentiation in HSCs via p38 activation. (A): Intracellular Phosphoflow analysis of 4 day cultured LT‐HSCs after lentiviral transduction of GADD45A. Representative histograms of GADD45A and control vector expressing LT‐HSCs and quantification of the MFI normalized to control are shown. N = 3 individual experiments. (B): Specific inhibition of the GADD45A induced differentiation using two p38 inhibitors (Vx702 and Ly2228820) in LT‐HSCs cultured for 5 days. Transduced cells (VENUS+) were gated for immature HSPCs, GMP‐like, and mature GM cells using the indicated surface marker expression. N = 3 individual experiments. All data represented as mean + SD. *, p < .05. Abbreviations: CTRL, control; DMSO, dimethyl sulfoxide; G45A, GADD45A; HSPCs, hematopoietic stem and progenitor cells; MFI, mean fluorescence intensity; p‐p38, phosphorylated p38; SCF, stem cell factor; TPO, thrombopoietin.
Discussion
The function of GADD45A in hematopoiesis long remained controversial. Here we revisited the functional consequences of GADD45A on HSC fate decisions at single cell resolution. Constitutive GADD45A expression did not inhibit the cell cycle progression or forced HSCs into apoptosis but rapidly induced and accelerated multilineage differentiation by activating the p38 MAPK pathway. The absence of Gadd45a in HSCs has little effect on steady state hematopoiesis 14. In contrast, once genotoxic stress was applied to LT‐HSCs by γ‐IR, the absence of GADD45A provided increased radioprotection and survival in HSCs at the expense of DNA integrity that resulted in a higher incidence of B cell leukemia 14. Additionally, ATM−/− Gadd45a −/− mice showed an increased incidence of hematopoietic malignancies as well as an increased rate of metastasis compared with ATM−/− mice 34. Gadd45a deletion aggravated the DNA damage accumulation, which subsequently resulted in a further impaired self‐renewal capacity and an increased malignant transformation in ATM−/− HSCs 34. Since GADD45A is obviously required for the genomic stability in HSPCs by allowing proper DNA repair, we expected to observe a cell cycle arrest upon GADD45A expression in LT‐HSCs. Surprisingly, GADD45A expression did not slow the cell cycle of HSCs and their progeny, neither in vitro nor in vivo, but rather shortened the cell cycle lengths in subsequent generations. Since GADD45A strongly accelerates the differentiation in HSCs, this might be a consequence of further differentiated progenitor stages with shorter generation times rather than a directly permitted effect by GADD45A on cell cycle progression. We cannot exclude that the absence of GADD45A may have an influence on the cell cycle progression 14, or that there are potential effects of GADD45A expression on the cell cycle of largely quiescent LT‐HSCs in steady‐state hematopoiesis. However, despite the fact that the GADD45 family members acquired their name as growth arrest genes it seems unlikely that cell cycle inhibition is of high relevance in the hematopoietic system.
In contrast to a report that GADD45A expression resulted in increased apoptosis in LT‐HSCs 14 we did not observe increased cell death upon GADD45A expression, neither determined at the single cell level by continuous cell tracking nor after transplantation in vivo. This is in line with reports that propose a prosurvival function of GADD45A in hematopoietic cells 23, 24. Since ectopic GADD45A expression did not alter cell survival under normal conditions, it needs to be tested whether GADD45A expression increases HSC resistance to various noxins. We rather observed that LT‐HSCs were rapidly differentiating upon GADD45A expression with a loss of their self‐renewal ability. The differentiation‐promoting function of GADD45A is supported by the notion that GADD45A‐deficient LT‐HSCs exhibited increased self‐renewal 14 and colony‐replating abilities 25 albeit with a higher incidence of mutations. Similar to what we have previously reported for GADD45G, a GADD45 member that is under differentiation‐promoting cytokine control 10, GADD45A works as a toggle switch in inducing differentiation in LT‐HSCs. The trigger for GADD45A expression is mediated by DNA‐damaging events. We showed that GADD45G selectively activates the p38 pathway, most likely by the activation of MAP3K4 and MKK6, but not of JNK 10. Again, here we could reveal that GADD45A, albeit not induced by hematopoietic cytokines, uses a common pathway shared with GADD45G to induce p38‐mediated differentiation. However, the lineage selection against megakaryocyte‐erythroid fate by GADD45G expression 10 is not mirrored by GADD45A. As a possible explanation for this difference, we showed that GADD45A expression caused a sharp downregulation of GADD45G, thereby allowing all lineages to develop. Nevertheless, the exact molecular machinery that is initiated by different GADD45 family members to control lineage choice in HSPCs needs further clarification. It will be intriguing to unravel the downstream effectors that might be common or unique for individual GADD45 family members in HSC differentiation induction.
The role of GADD45A in preserving the genomic integrity of HSCs is vital, but its mode of action as tumor suppressor for hematologic malignancies, especially the cell cycle inhibition and proapoptotic function, has been unclear so far. We showed that the differentiation induction might be the major antileukemia effect of GADD45A in the hematopoietic system, besides providing DNA repair and genomic stability. Importantly, we provide strong evidence that various DNA‐damaging conditions such as γ‐IR and UV upregulate GADD45A expression in stem and progenitor cells and that these conditions ultimately lead to a differentiation induction and acceleration in LT‐HSCs. Damaged cells are forced to terminally differentiate and thereby cleared from the system. The hypothesis of differentiation check‐points for DNA‐damage response has been previously postulated 35, 36, 37. As one prominent example, the induction of the transcription factor BATF upon hematopoietic stress instructs terminal lymphoid differentiation 36. Our data supports the notion that also GADD45A exerts its important function in response to DNA damage at least partly in this respect. GADD45A and GADD45G are epigenetically silenced in many leukemias and solid cancers 9. Furthermore, GADD45A is frequently deleted in breast cancer 38. The reactivation of GADD45A by, for example, the histone deacetylase inhibitor Trichostatin promoted terminal differentiation and death of human carcinoma cells 39, 40.
Enhanced p38 activation induces cell survival in many cancer cell types but obviously promotes a strong differentiation signal in HSCs, maybe by the phosphorylation of GATA3 41. Our data suggest that anticancer strategies using inhibiting agents against p38 MAPK should be carefully evaluated in blood cell malignancies, and reactivation of GADD45 might be of benefit for leukemia treatment.
Conclusion
The long‐disputed functional consequences of genotoxic stress‐induced GADD45A expression in hematopoietic cells were revisited in this study, elucidating the role of GADD45A expression in murine LT‐HSCs at single cell resolution. We showed here that neither cell cycle arrest nor cell death was induced by GADD45A, but a strong differentiation program via the initiation of the p38 MAPK signaling pathway. Forcing terminal differentiation of damaged HSPCs might be a prominent function of GADD45A in the hematopoietic system, besides providing DNA repair and genomic stability.
Author Contributions
S.W.: conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; F.B.T., N.H., and M.R.: collection and/or assembly of data and data analysis and interpretation; T.S.: developed the tracking software and comments on the manuscript; M.A.R.: conception and design, data analysis and interpretation, financial support, manuscript writing, and final approval.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article
Supporting Information Fig. S1
Supporting Information Fig. S2
Supporting Information Fig. S3
Supporting Information Fig. S4
Supporting Information Table S1
Supporting Information 1
Supporting Information 2
Acknowledgments
We thank C. Jourdan, S. Bothur, and M. Engel for excellent technical assistance. This work was supported by the LOEWE Center for Cell and Gene Therapy (HMWK III L 4‐518/17.004 (2013)) and the German Research Foundation (SFB 834).
References
- 1. Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008;132:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rieger MA, Schroeder T. Hematopoiesis. Cold Spring Harbor Perspect Biol 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blanpain C, Mohrin M, Sotiropoulou PA et al. DNA‐damage response in tissue‐specific and cancer stem cells. Cell Stem Cell 2011;8:16–29. [DOI] [PubMed] [Google Scholar]
- 4. Walter D, Lier A, Geiselhart A et al. Exit from dormancy provokes DNA‐damage‐induced attrition in haematopoietic stem cells. Nature 2015;520:549‐552. [DOI] [PubMed] [Google Scholar]
- 5. Fornace AJ, Alamo I, Hollander MC. DNA damage‐inducible transcripts in mammalian cells. Proc Natl Acad Sci USA 1988;85:8800–8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdollahi A, Lord KA, Hoffman‐Liebermann B et al. Sequence and expression of a cDNA encoding MyD118: A novel myeloid differentiation primary response gene induced by multiple cytokines. Oncogene 1991;6:165–167. [PubMed] [Google Scholar]
- 7. Zhang W, Bae I, Krishnaraju K et al. CR6: A third member in the MyD118 and Gadd45 gene family which functions in negative growth control. Oncogene 1999;18:4899–4907 10.1038/sj.onc.1202885. [DOI] [PubMed] [Google Scholar]
- 8. Moskalev AA, Smit‐McBride Z, Shaposhnikov MV et al. Gadd45 proteins: Relevance to aging, longevity and age‐related pathologies. Ageing Res Rev 2012;11:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liebermann DA, Tront JS, Sha X et al. Gadd45 stress sensors in malignancy and leukemia. Crit Rev Oncog 2011;16:129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thalheimer FB, Wingert S, de Giacomo P et al. Cytokine‐regulated GADD45G induces differentiation and lineage selection in hematopoietic stem cells. Stem Cell Rep 2014;3:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith ML, Chen IT, Zhan Q et al. Interaction of the p53‐regulated protein Gadd45 with proliferating cell nuclear antigen. Science 1994;266:1376–1380. [DOI] [PubMed] [Google Scholar]
- 12. Smith ML, Ford JM, Hollander MC et al. p53‐mediated DNA repair responses to UV radiation: Studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol 2000;20:3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hollander MC, Sheikh MS, Bulavin DV et al. Genomic instability in Gadd45a‐deficient mice. Nat Genet 1999;23:176–184. [DOI] [PubMed] [Google Scholar]
- 14. Chen Y, Ma X, Zhang M et al. Gadd45a regulates hematopoietic stem cell stress responses in mice. Blood 2014;123:851–862. [DOI] [PubMed] [Google Scholar]
- 15. Levrero M, Laurenzi V de, Costanzo A et al. The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J Cell Sci 2000;113 (Pt 10):1661–1670. [DOI] [PubMed] [Google Scholar]
- 16. Harkin DP, Bean JM, Miklos D et al. Induction of GADD45 and JNK/SAPK‐dependent apoptosis following inducible expression of BRCA1. Cell 1999;97:575–586. [DOI] [PubMed] [Google Scholar]
- 17. Oh‐Hashi K, Maruyama W, Isobe K. Peroxynitrite induces GADD34, 45, and 153 VIA p38 MAPK in human neuroblastoma SH‐SY5Y cells. Free Radic Biol Med 2001;30:213–221. [DOI] [PubMed] [Google Scholar]
- 18. Liebermann DA, Hoffman B. Myeloid differentiation (MyD) primary response genes in hematopoiesis. Blood Cells Mol Dis 2003;31:213–228. [DOI] [PubMed] [Google Scholar]
- 19. Wang XW, Zhan Q, Coursen JD et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA 1999;96:3706–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang W, Hoffman B, Liebermann DA. Ectopic expression of MyD118/Gadd45/CR6 (Gadd45beta/alpha/gamma) sensitizes neoplastic cells to genotoxic stress‐induced apoptosis. Int J Oncol 2001;18:749–757. [PubMed] [Google Scholar]
- 21. Jackson JG, Pereira‐Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 2006;66:8356–8360. [DOI] [PubMed] [Google Scholar]
- 22. Selvakumaran M, Lin HK, Sjin RT et al. The novel primary response gene MyD118 and the proto‐oncogenes myb, myc, and bcl‐2 modulate transforming growth factor beta 1‐induced apoptosis of myeloid leukemia cells. Mol Cell Biol 1994;14:2352–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta M, Gupta SK, Balliet AG et al. Hematopoietic cells from Gadd45a‐ and Gadd45b‐deficient mice are sensitized to genotoxic‐stress‐induced apoptosis. Oncogene 2005;24:7170–7179. [DOI] [PubMed] [Google Scholar]
- 24. Gupta M, Gupta SK, Hoffman B et al. Gadd45a and Gadd45b protect hematopoietic cells from UV‐induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. J Biol Chem 2006;281:17552–17558. [DOI] [PubMed] [Google Scholar]
- 25. Gupta SK, Gupta M, Hoffman B et al. Hematopoietic cells from gadd45a‐deficient and gadd45b‐deficient mice exhibit impaired stress responses to acute stimulation with cytokines, myeloablation and inflammation. Oncogene 2006;25:5537–5546. [DOI] [PubMed] [Google Scholar]
- 26. Rieger MA, Hoppe PS, Smejkal BM et al. Hematopoietic cytokines can instruct lineage choice. Science 2009;325:217–218. [DOI] [PubMed] [Google Scholar]
- 27. Rengstl B, Newrzela S, Heinrich T et al. Incomplete cytokinesis and re‐fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed‐Sternberg cells. Proc Natl Acad Sci USA 2013;110:20729–20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schambach A, Mueller D, Galla M et al. Overcoming promoter competition in packaging cells improves production of self‐inactivating retroviral vectors. Gene Ther 2006;13:1524–1533. [DOI] [PubMed] [Google Scholar]
- 29. Heinz N, Schambach A, Galla M et al. Retroviral and transposon‐based tet‐regulated all‐in‐one vectors with reduced background expression and improved dynamic range. Human Gene Ther 2011;22:166–176. [DOI] [PubMed] [Google Scholar]
- 30. Naldini L. Lentiviruses as gene transfer agents for delivery to non‐dividing cells. Curr Opin Biotechnol 1998;9:457–463. [DOI] [PubMed] [Google Scholar]
- 31. Pronk, Cornelis J H, Rossi DJ, Månsson R et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007;1:428–442. [DOI] [PubMed] [Google Scholar]
- 32. Rieger MA, Smejkal BM, Schroeder T. Improved prospective identification of megakaryocyte‐erythrocyte progenitor cells. Br J Haematol 2009;144:448–451. [DOI] [PubMed] [Google Scholar]
- 33. Mita H, Tsutsui J, Takekawa et al. Regulation of MTK1/MEKK4 kinase activity by its N‐terminal autoinhibitory domain and GADD45 binding. Mol Cell Biol 2002;22:4544–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y, Yang R, Guo P et al. Gadd45a deletion aggravates hematopoietic stem cell dysfunction in ATM‐deficient mice. Protein Cell 2014;5:80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santos MA, Faryabi RB, Ergen AV et al. DNA‐damage‐induced differentiation of leukaemic cells as an anti‐cancer barrier. Nature 2014;514:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, Sun Q, Morita Y et al. A differentiation checkpoint limits hematopoietic stem cell self‐renewal in response to DNA damage. Cell 2012;148:1001–1014. [DOI] [PubMed] [Google Scholar]
- 37. Inomata K, Aoto T, Binh NT et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 2009;137:1088–1099. [DOI] [PubMed] [Google Scholar]
- 38. Hoggard N, Hey Y, Brintnell B et al. Identification and cloning in yeast artificial chromosomes of a region of elevated loss of heterozygosity on chromosome 1p31.1 in human breast cancer. Genomics 1995;30:233–243. [DOI] [PubMed] [Google Scholar]
- 39. Chen Z, Clark S, Birkeland M et al. Induction and superinduction of growth arrest and DNA damage gene 45 (GADD45) alpha and beta messenger RNAs by histone deacetylase inhibitors trichostatin A (TSA) and butyrate in SW620 human colon carcinoma cells. Cancer Lett 2002;188:127–140. [DOI] [PubMed] [Google Scholar]
- 40. Hirose T, Sowa Y, Takahashi S et al. p53‐independent induction of Gadd45 by histone deacetylase inhibitor: Coordinate regulation by transcription factors Oct‐1 and NF‐Y. Oncogene 2003;22:7762–7773. [DOI] [PubMed] [Google Scholar]
- 41. Frelin C, Herrington R, Janmohamed S et al. GATA‐3 regulates the self‐renewal of long‐term hematopoietic stem cells. Nat Immunol 2013;14:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article
Supporting Information Fig. S1
Supporting Information Fig. S2
Supporting Information Fig. S3
Supporting Information Fig. S4
Supporting Information Table S1
Supporting Information 1
Supporting Information 2


