Summary
Experimental data show that Arabidopsis thaliana is able to decode different calcium signatures to produce specific gene expression responses. It is also known that calmodulin‐binding transcription activators (CAMTAs) have calmodulin (CaM)‐binding domains. Therefore, the gene expression responses regulated by CAMTAs respond to calcium signals. However, little is known about how different calcium signatures are decoded by CAMTAs to produce specific gene expression responses.
A dynamic model of Ca2+–CaM–CAMTA binding and gene expression responses is developed following thermodynamic and kinetic principles. The model is parameterized using experimental data. Then it is used to analyse how different calcium signatures are decoded by CAMTAs to produce specific gene expression responses.
Modelling analysis reveals that: calcium signals in the form of cytosolic calcium concentration elevations are nonlinearly amplified by binding of Ca2+, CaM and CAMTAs; amplification of Ca2+ signals enables calcium signatures to be decoded to give specific CAMTA‐regulated gene expression responses; gene expression responses to a calcium signature depend upon its history and accumulate all the information during the lifetime of the calcium signature.
Information flow from calcium signatures to CAMTA‐regulated gene expression responses has been established by combining experimental data with mathematical modelling.
Keywords: Arabidopsis, calcium signatures, calmodulin (CaM), calmodulin‐binding transcription activators (CAMTAs), gene expression, mathematical modelling
Introduction
Plants are sessile organisms and therefore they must adapt their metabolism, growth and architecture to a changing environment. The majority of their defence against stress is realized by changes in gene expression in order to produce proteins required to combat the conditions they encounter. It is thus vital that the correct proteins are produced in response to different environmental conditions; that is, different genes need to be switched on in response to different stimuli. Calcium is a ubiquitous second messenger in eukaryotes and it is a ubiquitous intermediate between stimulus perception and responses in plants. It has been observed that different stimuli produce calcium signatures with different characteristics in plants (McAinsh et al., 1995; Allen et al., 2001; Love et al., 2004; Miwa et al., 2006; McAinsh & Pittman, 2009; Dodd et al., 2010; Kudla et al., 2010; Short et al., 2012). Given that calcium is an intermediate between stimulus perception and gene expression (Whalley et al., 2011), it is possible that the specific characteristics of the calcium signatures produced by different stresses encode stimulus‐specific information. Recent experimental data demonstrate that Arabidopsis thaliana is able to decode specific calcium signatures and interpret them, leading to distinct gene expression profiles (Whalley et al., 2011; Whalley & Knight, 2013).
Calmodulin‐binding transcription activators (CAMTAs) are well‐characterized Ca2+/calmodulin (CaM)‐regulated transcription factors (Kim et al., 2009; Galon et al., 2010; Reddy et al., 2011; Bickerton & Pittman, 2012; Poovaiah et al., 2013), and they have CaM‐binding domains (Finkler et al., 2007). Therefore, gene expression responses regulated by CAMTAs respond to calcium signals (Whalley et al., 2011; Whalley & Knight, 2013).
Although experimental data (Whalley et al., 2011; Whalley & Knight, 2013) show that A. thaliana is able to decode different calcium signatures to produce specific gene expression responses, little is known about how complex calcium signatures are decoded to generate gene expression responses. In this work, we establish the principles of information flow from calcium signatures to CAMTA‐regulated gene expression responses by combining experimental data with mathematical modelling.
Materials and Methods
A dynamic model describing the information flow from calcium signatures to CAMTA‐regulated gene expression
The information flow from calcium signatures to CAMTA‐regulated gene expression in Arabidopsis thaliana is described in Fig. 1. The left pane of Fig. 1 describes the binding of Ca2+, CaM and CAMTAs. CaM has two pairs of Ca2+‐binding EF‐hand domains located at the N and C termini, respectively (Finn & Forsen, 1995; Valeyev et al., 2008). Ca2+‐binding kinetics for the pairs of EF‐hands at the N and C termini are significantly different and their Ca2+‐binding kinetics display cooperativity (Linse et al., 1991). The cooperative binding between Ca2+ and the four binding sites of CaM has been previously subjected to both experimental and modelling studies (Fajmut et al., 2005; Shifman et al., 2006; Pepke et al., 2010) and the kinetic parameters have been determined (Shifman et al., 2006; Pepke et al., 2010). Table 1 summarizes those parameters (Shifman et al., 2006; Pepke et al., 2010). In addition, experimental data show that the CAMTA proteins consist of multiple functional domains associated with binding of CaM and CaM‐like proteins (Bouche et al., 2002; Finkler et al., 2007). Mapping of a Ca2+‐dependent CaM‐binding domain in A. thaliana AtCAMTA1 revealed a single high‐affinity binding site (the binding affinity K d = 1.2 × 10−3 μM) (Bouche et al., 2002; Finkler et al., 2007) and a similar binding site exists in rice (Oryza sativa) (Choi et al., 2005). As binding of the Ca2+–CaM complex to CAMTAs is tighter than binding of free CaM to CAMTAs (Bouche et al., 2002; Finkler et al., 2007), the K d for R15 in Fig. 1 is always larger than 1.2 × 10−3 μM. These parameters are also included in Table 1.
Figure 1.
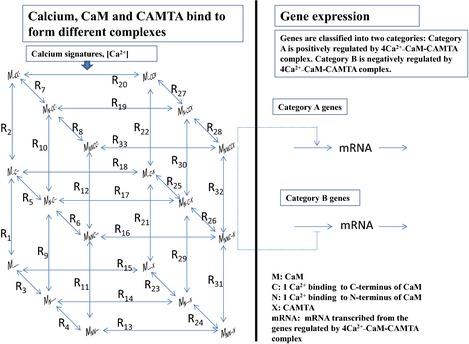
A dynamic model that describes the information flow from calcium signatures to calmodulin‐binding transcription activator (CAMTA)‐regulated gene expression in Arabidopsis thaliana. Left panel: Ca2+, calmodulin (CaM) and CAMTA bind to form different complexes. When [Ca2+] changes, this binding process responds following thermodynamic principles. Right panel: gene expression is regulated by the active complex 4Ca2+–CaM–CAMTA (MNNCCX) using the two simplest gene expression mechanisms. This figure illustrates a generic model for studying the information flow from calcium signatures to CAMTA‐regulated gene expression.
Table 1.
Parameters for the model described in Fig. 1
| 1. Parameters derived using experimental data for the binding of Ca2+, CaM and CAMTA (left pane of Fig. 1) | |||
| Reaction | Reaction description | Equilibrium constant (K d) | Kinetic constants (k on; k off) |
| R1, R9, R11 | Binding of first Ca2+ to CaM C terminus | 10 μM (Linse et al., 1991; Shifman et al., 2006; Kubota et al., 2007; Pepke et al., 2010) | k on = 4 μM−1 s−1; k off = 40 s−1 (Martin et al., 1992; Persechini et al., 1996; Gaertner et al., 2004; Pepke et al., 2010) |
| R2, R10, R12 | Binding of second Ca2+ to CaM C terminus | 0.925 μM (Linse et al., 1991; Shifman et al., 2006; Kubota et al., 2007; Pepke et al., 2010) | k on = 10 μM−1 s−1; k off = 9.25 s−1 (Gaertner et al., 2004; Pepke et al., 2010) |
| R3, R5, R7 | Binding of first Ca2+ to CaM N terminus | 25 μM (Linse et al., 1991; Shifman et al., 2006; Kubota et al., 2007; Pepke et al., 2010) | k on = 100 μM−1 s−1; k off = 2500 s−1 (Brown et al., 1997; Peersen et al., 1997; Gaertner et al., 2004; Pepke et al., 2010) |
| R4, R6, R8 | Binding of second Ca2+ to CaM N terminus | 5 μM (Linse et al., 1991; Shifman et al., 2006; Kubota et al., 2007; Pepke et al., 2010) | k on = 150 μM−1 s−1; k off = 750 s−1 (Brown et al., 1997; Peersen et al., 1997; Gaertner et al., 2004; Pepke et al., 2010) |
| R14 | Binding of Ca2+–CaM complex to CAMTA | 1.2 × 10−3 μM (Bouche et al., 2002; Finkler et al., 2007) |
k
on = 1 μM−1 s−1; k
off = 1.2 × 10−3 s−1
Notes: k on is an adjustable parameter in this work. k off = K d k on |
| R15 | Binding of free CaM to CAMTA |
K
d(R14)/P = 1.2 × 10−3 μM/P. P = 0.1, which is always smaller than 1, is an adjustable parameter, indicating that binding of Ca2+–CaM complex to CAMTA is tighter than binding of free CaM to CAMTA (Bouche et al., 2002; Finkler et al., 2007) |
k
on = k
on(R14)/Q. Q = 1.0 is an adjustable parameter and it describes the cooperative binding between CaM and CAMTA in the presence of Ca2+ due to on binding rate. k off = K d k on = (K d(R14) k on(R14))/(PQ) = k off(R14)/(PQ) |
| 2. Parameters derived based on the detailed balance conditions following thermodynamic principles and the assumption that the affinity for the binding of any Ca2+–CaM complex to CAMTA is always the same (left pane of Fig. 1) | |||
|
K
d(R13) = K
d(R14) = k
d(R16) = k
d(R17) = K
d(R18) = K
d(R19) = K
d(R20) = K
d(R33), K
d(R2) = K
d(R22), K
d(R4) = K
d(R24), K
d(R5) = K
d(R25), K
d(R6) = K
d(R26), K
d(R7) = K
d(R27), K
d(R8) = K
d(R28), K
d(R9) = K
d(R29), K
d(R10) = K
d(R30), K
d(R11) = K
d(R31), K
d(R12) = K
d(R32)
As long as the binding affinities (K d) for two reactions are the same, we consider their respective k on and k off are also the same. | |||
| 3. Parameters for gene expression (right pane of Fig. 1) | |||
| k 1 = 5.0 × 10−6 μM s−1, k 2 = 5.0 × 10−2 μM s−1, n = 2, k 3 = 3.75 × 10−4 s−1, k 4 = 1.1 × 10−2 μM | |||
CaM, calmodulin; CAMTA, calmodulin‐binding transcription activator.
The unknown equilibrium constants and on/off rates in the left pane of Fig. 1 are derived using the detailed balance condition following thermodynamic principles. For example, following the detailed balance condition, Eqn (1) is always valid.
| (1) |
| (2) |
As binding of the Ca2+–CaM complex to CAMTAs is tighter than binding of free CaM to CAMTAs (Bouche et al., 2002; Finkler et al., 2007), P is always < 1 (Table 1). P describes the cooperative binding between CaM and CAMTAs in the presence of Ca2+. As P has not been experimentally determined, it is an adjustable parameter in this work. Moreover, as K d is K off/K on, the difference between K d(R3) and K d(R23) or the difference between K d(R14) and K d(R15) could be caused by the difference in k on, k off or both. In order to examine the effects of changes in k on, k off or both on modelling results, we define
| (3) |
where Q is an adjustable parameter. If Q = 1.0, this implies that cooperativity is realized solely by the changes in k off. Similarly, Q = 1/P implies that cooperativity is realized solely by the changes in k on. Other values of Q imply that cooperativity is realized by the changes in both k on and k off. After applying the detailed balance condition following thermodynamic principles to all other loops in Fig. 1, the equilibrium constants (K d) are linked.
Based on experimental data, binding of the Ca2+–CaM complex to CAMTAs is tighter than binding of free CaM to CAMTAs (Bouche et al., 2002; Finkler et al., 2007). However, experimental measurements are not able to identify the binding affinity of each different Ca2+–CaM complex (i.e. with different numbers of calcium ions at different positions) to CAMTAs. Here, we assume that the affinity for the binding of any Ca2+–CaM complex to CAMTAs is always the same, regardless of how many Ca2+ bind to CaM. The advantage of this assumption is to greatly reduce adjustable parameters. However, for the sake of completeness, we have randomly tested the effects of different binding affinities for the binding between some Ca2+–CaM complexes and CAMTAs. Under the conditions that binding of the Ca2+–CaM complex to CAMTAs is tighter than binding of free CaM to CAMTAs (Bouche et al., 2002; Finkler et al., 2007), we have tested the effects of changes in binding affinity of up to 2 orders with reference to a value of 1.2 × 10−3 μM for some Ca2+–CaM complexes (e.g. R20 and R33). The qualitative conclusions we will draw in this work do not change if these binding affinities change, as shown in Supporting Information Figs S1–S4. After introducing the detailed balance condition following thermodynamic principles and based on the assumption that the affinity for the binding of any Ca2+–CaM complex to CAMTAs is always the same, we are able to derive all other unknown equilibrium constants and on/off rates, as summarized in Table 1.
After using the parameters determined experimentally and introducing thermodynamic constraints, there are only five adjustable parameters left for the left pane of Fig. 1, as summarized as follows. P describes the cooperative binding between CaM and CAMTAs in the presence of Ca2+; k on(R14) is the on rate for the binding of the Ca2+–CaM complex to CAMTAs; Q describes how the cooperative binding between CaM and CAMTAs in the presence of Ca2+ is realized by k on, k off or both. CaM_t describes the total concentration of CaM, which is the summation of free CaM and all CaM complexes. X_t describes the total concentration of CAMTAs, which is the summation of free CAMTAs and all CAMTA complexes.
The mass balance of each complex in the left pane of Fig. 1 is described using a differential equation (Notes S1). By coupling these differential equations together, we are able to calculate the concentration of any complex for any calcium signature at any time. It is known that 4Ca2+–CaM is the active CaM–Ca2+ binding complex (Pifl et al., 1984). Therefore, this work assumes that the complex 4Ca2+–CaM–CAMTA (MNNCCX in Fig. 1) is the active complex for the gene expression response.
The right pane of Fig. 1 describes CAMTA‐regulated gene expression. CAMTAs can be either activators or suppressors of gene expression, as evidenced by the experiments using CAMTA mutants (Galon et al., 2008; Doherty et al., 2009). In addition, the process of gene expression may have multiple entry points of Ca2+ signal as a consequence of the interactions between Ca2+ signal and CAMTAs (Miller et al., 2013; Zhang et al., 2014). Clearly, different genes may be regulated by different expression mechanisms. Even if they are regulated by the same mechanism, the parameter values controlling their expression may be different. Moreover, for most genes, gene expression mechanisms have not been experimentally determined. In this work, our focus is to investigate how different calcium signatures are decoded by CAMTAs to produce specific gene expression responses. Our primary interest is to establish the information flow from calcium signatures to gene expression rather than the gene expression mechanisms themselves. Therefore, here we use as simple as possible generic gene expression mechanisms. Our method can be generally extended to include any gene expression mechanism by replacing the right pane of Fig. 1, if the specific expression mechanism of that specific gene is known.
The simplest gene expression process includes: (1) gene transcription is activated or supressed by a transcription factor; and (2) the mRNA is decayed or consumed. The right pane of Fig. 1 describes these simplest mechanisms. For category A genes in Fig. 1, the differential equation for describing gene expression is as follows.
| (4) |
(k 1, the base rate for gene transcription; k 2, the maximal rate for CAMTA‐regulated gene transcription; k 3, the decay rate constant for the mRNA; k 4, the binding affinity between the 4Ca2+–CaM–CAMTA complex and DNA; n, the Hill coefficient.)
Similarly, for category B genes in Fig. 1, the differential equation for describing gene expression is as follows.
| (5) |
The parameters for gene transcription are included in Table 1. In the Results section, we will also test and discuss how the parameters relating to gene transcription affect the information flow from calcium signatures to gene expression.
Time delay
It is evident that the information flow from calcium signatures to changes in gene expression will generally be subjected to a time delay. When a calcium signal emerges, a change in gene expression cannot occur instantly, as the transcriptional pre‐initiation complex (containing specific transcription factors, e.g. CAMTAs, general transcription factors, mediators and RNA polymerase) needs to be recruited and assembled and an elongation complex needs to form to allow transcription of the coding region (Lee & Young, 2000). In this work, we consider that there is a time delay, τ, between the calcium signal and the gene expression response. After all the concentrations in the left pane of Fig. 1 are calculated, the complex 4Ca2+–CaM–CAMTA induces gene expression after a time delay (τ). The effects of τ on modelling results will be examined.
Numerical method
The model is implemented using the simulator Berkeley Madonna (www.berkeleymadonna.com). The Rosenbrock (Stiff) method is used with a tolerance of 1.0 × 10−5. Much smaller tolerances have also been tested and the numerical results show that further reduction of tolerances does not improve the accuracy of numerical results. Before a calcium signature is introduced, the system of ordinary differential equations is settled at a steady state using the average Ca2+ concentration of the control experiment as an input. When a calcium signature is introduced, the response of the system of ordinary differential equations is calculated using the time‐dependent Ca2+ concentration as an input.
Results
Ca2+ signals are nonlinearly amplified as a result of the Ca2+–CaM–CAMTA interaction
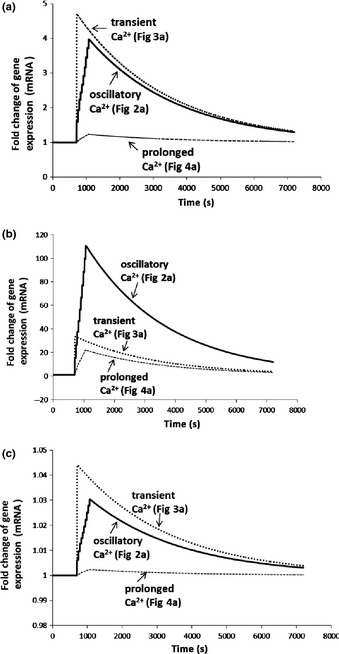
As shown in Fig. 1, under thermodynamic constraints, CaM binds to Ca2+, forming complexes with different numbers of calcium ions at different positions. It is known that 4Ca2+–CaM is the active CaM–Ca2+ binding complex (Pifl et al., 1984). Figs 2, 3 and 4 summarize the amplification of Ca2+ signals for three different Ca2+ signatures, which were experimentally generated (Whalley et al., 2011). As a result of the interaction of Ca2+–CaM–CAMTA, Ca2+ signals are nonlinearly amplified into the signals of the active functioning complex 4Ca2+–CaM–CAMTA.
Figure 2.
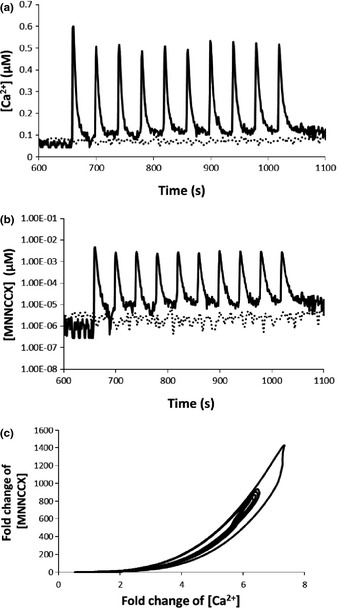
Oscillatory calcium signature induced in experiments that use controlled electrical stimulations (Whalley et al., 2011) and amplification of Ca2+ signal as a result of 4Ca2+–CaM–CAMTA binding in Arabidopsis thaliana. (a) Solid line: experimental Ca2+ elevation. Dashed line: control. (b) Computational results for the response of the active complex 4Ca2+–CaM–CAMTA to the calcium signature (solid line) and to the control experiment (dashed line), respectively. (c). Fold‐change analysis shows that Ca2+ signals are nonlinearly amplified by 4Ca2+–CaM–CAMTA binding. CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
Figure 3.
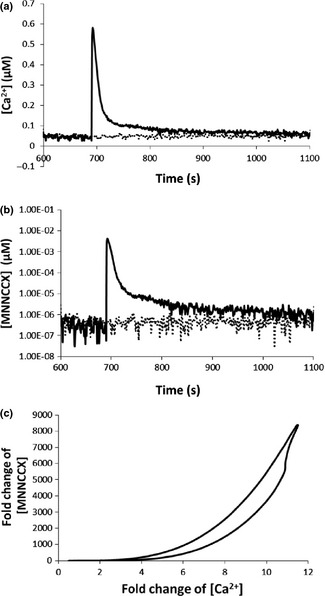
Transient calcium signature induced in experiments that use controlled electrical stimulations (Whalley et al., 2011) and amplification of Ca2+ signal as a result of 4Ca2+–CaM–CAMTA binding in Arabidopsis thaliana. (a) Solid line: experimental Ca2+ elevation. Dashed line: control. (b) Computational results for response of the active complex 4Ca2+–CaM–CAMTA to the calcium signature (solid line) and to the control experiment (dashed line), respectively. (c) Fold‐change analysis shows that Ca2+ signals are nonlinearly amplified by 4Ca2+–CaM–CAMTA binding. CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
Figure 4.
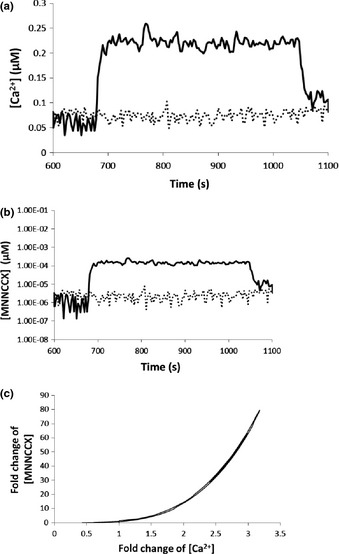
Prolonged calcium signature induced in experiments that use controlled electrical stimulations (Whalley et al., 2011) and amplification of Ca2+ signal as a result of 4Ca2+–CaM–CAMTA binding in Arabidopsis thaliana. (a) Solid line: experimental Ca2+ elevation. Dashed line: control. (b) Computational results for response of the active complex 4Ca2+–CaM–CAMTA to the calcium signature (solid line) and to the control experiment (dashed line), respectively. (c) Fold‐change analysis shows that Ca2+ signals are nonlinearly amplified by 4Ca2+–CaM–CAMTA binding. CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
The three Ca2+ signatures (Figs 2a, 3a, 4a) are the inputs for the interaction of Ca2+–CaM–CAMTA (Fig. 1), and the respective concentrations of the active functioning complex 4Ca2+–CaM–CAMTA for the three Ca2+ signatures are shown in Figs 2(b), 3(b), and 4(b). Fig. 2(c) shows that, for the oscillatory Ca2+ signature (Fig. 2a), a c. 7‐fold change in Ca2+ concentration (relative to the experimental measurement of average Ca2+ concentration in control experiments) is amplified to a c. 1400‐fold change in the concentration of the 4Ca2+–CaM–CAMTA complex (relative to the average computed concentration using Ca2+ concentration in control experiments). Fig. 3(c) shows that, for the transient Ca2+ signature (Fig. 3a), a c. 12‐fold change in Ca2+ concentration is amplified to a c. 8000‐fold change in the concentration of the 4Ca2+–CaM–CAMTA complex. Similarly, Fig. 4(c) shows that, for the prolonged Ca2+ signature (Fig. 4a), a c. 3.2 ‐fold change in Ca2+ concentration is amplified to a c. 80‐fold change in the concentration of the 4Ca2+–CaM–CAMTA complex.
Combination of Figs 2, 3 and 4 reveals that the amplification of Ca2+ signals by the interaction of Ca2+–CaM–CAMTA is nonlinear. For example, with reference to the steady‐state value of the Ca2+ concentration and the corresponding concentration of the 4Ca2+–CaM–CAMTA complex, a c. 2‐, 4‐ or 10‐fold increase in Ca2+ concentration leads to a c. 10‐, 200‐ or 5000‐fold increase in 4Ca2+–CaM–CAMTA, respectively. Therefore, although the fold changes of Ca2+ concentration in three Ca2+ signatures are relatively small, the resulting fold changes of the active complex (4Ca2+–CaM–CAMTA) are large and different for each individual signature.
We have further examined how the five adjustable parameters relating to the left pane of Fig. 1 affect the modelling results shown in Figs 2, 3, 4. The nonlinear fold‐change relationship between Ca2+ signals and the corresponding active complex (4Ca2+–CaM–CAMTA) always exists across a wide range of values for these five adjustable parameters, as shown in Figs 5, S5 and S6.
Figure 5.
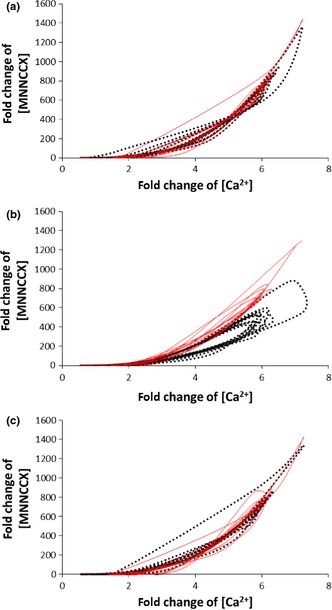
Evaluating the effects of the adjustable parameters on the amplification of Ca2+ signals. (a) Effects of altering the on rate for the binding between the Ca2+–calmodulin (CaM) complex and calmodulin‐binding transcription activators (CAMTAs) (k on(R14)) on the amplification of Ca2+ signals. Solid line: k on(R14) = 100 μM−1 s−1. Dashed line: k on(R14) = 0.01 μM−1 s−1. The reference value is k on(R14) = 1 μM−1 s−1 (Fig. 2). (b) Effects of altering the cooperative binding between CaM and CAMTA in the presence of Ca2+ by altering the binding rate (Q in Eqn (3)) on the amplification of Ca2+ signals. Solid line: Q = 0.01 μM−1 s−1. Dashed line: Q = 100 μM−1 s−1. The reference value is Q = 1 μM−1 s−1 (Fig. 2). (c) Effects of altering the total CAMTA concentration on the amplification of Ca2+ signals. Solid line: X_t = 1000 μM. Dashed line: X_t = 0.1 μM. The reference value is 10 μM (Fig. 2).
Fig. 5(a) shows the effects of varying the on rate for the binding between the Ca2+–CaM complex and CAMTAs (k on(R14)) on the amplification of Ca2+ signals. For a 4‐order change in k on(R14), the amplification of Ca2+ signals is qualitatively similar. If k on(R14) is further increased from 100 μM−1 s−1, the amplification of Ca2+ signals is similar to the solid line in Fig. 5(a). However, if k on(R14) is further decreased from 0.01 μM−1 s−1, the amplification of Ca2+ signals will be markedly smaller. Fig. 5(b) shows that effects of varying the cooperative binding between CaM and CAMTAs in the presence of Ca2+ by altering the binding rate (Q in Eqn (3)). For a 4‐order change, the qualitative trend for the amplification of Ca2+ signals is always similar. In addition, we have tested the effects of varying the cooperative binding between CaM and CAMTAs in the presence of Ca2+ (P in Eqn (2)) in a 3‐order range, as P can only be increased to 1.0. Fig. S5 shows that the qualitative trend for the amplification of Ca2+ signals is always similar. Fig. 5(c) shows the effects of varying the total CAMTA concentration on the amplification of Ca2+ signals. Change of the total CAMTA concentration in a 4‐order range gives rise to qualitatively similar amplification of Ca2+ signals. In a similar manner, Fig. S6 shows that the qualitative trend for the amplification of Ca2+ signals is always similar for a 4‐order change of the total CaM concentration. In addition, we have also examined the effects of simultaneous variations of all five adjustable parameters. When all parameters are varied, there are a large number of possible combinations. In this work, therefore, we are only able to test certain combinations. As shown in Figs S7 and S8, the qualitative trend for the amplification of Ca2+ signatures is similar when all parameters vary.
Therefore, as a result of the interaction of Ca2+–CaM–CAMTA, Ca2+ signals are always nonlinearly amplified (Figs 2c, 3c, 4c). Moreover, for three different calcium signatures (Figs 2a, 3a, 4a), as a result of the differences in the amplitude of Ca2+ signatures, the maximum amplification fold change of the three calcium signatures is significantly different (Figs 2, 3, 4).
Amplification of Ca2+ signals enables calcium signatures to be decoded to give specific CAMTA‐regulated gene expression responses
Experimental data for fold change in CAMTA‐regulated gene expression level for three calcium signatures (Figs 2a, 3a, 4a) are included in Table 2. We defined CAMTA‐regulated genes as the 20 genes that were induced by any calcium signature described in Whalley et al. (2011) which contained the CAMTA‐binding motif 5′‐ACGCGT‐3′ within 500 bp of their promoters (Whalley et al., 2011). As shown in Table 2, both oscillatory (Fig. 2a) and transient (Fig. 3a) calcium signatures are able to induce > 1.5‐fold expression change in CAMTA‐regulated genes, while the prolonged (Fig. 4a) calcium signature cannot induce > 1.5‐fold change in any CAMTA‐regulated gene (Whalley et al., 2011). As the elevated Ca2+ increases CAMTA‐regulated gene expression for oscillatory (Fig. 2a) and transient (Fig. 3a) calcium signatures, we consider that, under our experimental conditions (Whalley et al., 2011), Ca2+ only activates (but does not decrease) CAMTA‐regulated gene expression (Table 2). Thus, we use Eqn (4) to calculate the effects of different calcium signatures on CAMTA‐regulated gene expression.
Table 2.
Experimental results for the fold change of calmodulin‐binding transcription activator (CAMTA)‐regulated gene expression at 1 h in Arabidopsis thaliana for the three calcium signatures that were induced using controlled electrical stimulations (Whalley et al., 2011)
| Arabidopsis Genome Initiative (AGI) code | Fold change for oscillatory calcium signature; Fig. 2a | Fold change for transient calcium signature; Fig. 3a | Fold change for prolonged calcium signature; Fig. 4a |
|---|---|---|---|
| AT2G20630 | 3.13 | 2.36 | Not induced |
| AT3G10300 | Not induced | 2.14 | Not induced |
| AT3G18420 | 1.71 | 2.06 | Not induced |
| AT1G19180 | 1.54 | 2.26 | Not induced |
| AT5G15650 | 1.80 | 2.27 | Not induced |
| AT3G05500 | 3.14 | 3.90 | Not induced |
| AT1G07890 | 1.58 | 2.12 | Not induced |
| AT1G18610 | Not induced | 4.56 | Not induced |
| AT1G19380 | 1.89 | 2.29 | Not induced |
| AT1G63750 | 3.08 | No data | Not induced |
| AT3G03020 | 1.82 | 2.11 | Not induced |
| AT3G19150 | 2.20 | 1.85 | Not induced |
| AT3G43680 | Not induced | 5.49 | Not induced |
| AT3G45970 | Not induced | 2.02 | Not induced |
| AT4G19200 | 2.18 | 2.02 | Not induced |
| AT4G22610 | 1.62 | 1.99 | Not induced |
| AT4G29670 | 2.26 | 1.89 | Not induced |
| AT4G30210 | 1.74 | 2.53 | Not induced |
| AT5G24810 | 2.11 | 2.06 | Not induced |
| AT5G45350 | 2.40 | 3.24 | Not induced |
‘Not induced’ refers to a < 1.5‐fold change (Whalley et al., 2011).
The capability of the Ca2+–CaM–CAMTA interaction to nonlinearly amplify Ca2+ signals allows different calcium signatures to be differentially decoded to generate specific gene expression responses. If Ca2+ signals were not amplified by the Ca2+–CaM–CAMTA interaction, the fold changes in the three calcium signatures (Figs 2a, 3a, 4a) would be small (from c. 3.5‐fold (for a prolonged calcium signature; Fig. 4a) to c. 11‐fold (for a transient calcium signature; Fig. 3a). Such differences in Ca2+ signals would, on their own, be too small to be distinguished and to allow different gene expression responses if they were not amplified. Thus, the role of the Ca2+–CaM–CAMTA interaction in amplifying Ca2+ signals is important for inducing specific gene expression responses. Fig. 6 shows how different calcium signatures are decoded to generate specific gene expression responses by virtue of the Ca2+–CaM–CAMTA interaction.
Figure 6.
Fold change in gene expression induced by three different calcium signatures with three binding affinities between the active complex 4Ca2+–CaM–CAMTA and DNA in Arabidopsis thaliana. The delay time between calcium signature and gene expression is 600 s for all three calcium signatures. (a) Binding affinity (K d) is 1.1 × 10−2 μM. Both oscillatory and transient calcium signatures induce c. 2‐fold gene expression increase at 1 h, while the prolonged calcium signature induces c. 1.05‐fold gene expression increase at 1 h. (b) Binding affinity (K d) is 1.1 × 10−3 μM. Oscillatory, transient and prolonged calcium signatures induce c. 43‐, 12‐ and 9‐fold gene expression increases at 1 h, respectively. (c) Binding affinity (K d) is 1.1 × 10−1 μM. Oscillatory, transient and prolonged calcium signatures all induce < 1.02‐fold gene expression increases at 1 h. CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
As shown in Fig. 6(a), as a result of the large fold amplification of Ca2+ signals in oscillatory and transient calcium signatures (Figs 2a, 3a), large fold changes in gene expression level are induced. Similarly, as a result of the small fold amplification of Ca2+ signals in prolonged calcium signatures (Fig. 4a), only a small fold change in gene expression level is induced. Thus, at 1 h, the fold change of gene expression level is generally larger than 1.5‐fold (Fig. 6a) for oscillatory and transient calcium signatures (Figs 2a, 3a), while it is < 1.5‐fold (Fig. 6a) for the prolonged calcium signature (Fig. 4a). Figs S9, S10 and S11 show that fold change for all three calcium signatures at a specific time (e.g. 1 h) depends on the delay time, which is the time when gene expression starts to responds to Ca2+ signals. In Fig. 6(a), we assume that the delay time is always 600 s for three calcium signatures. If we assume that the delay time is different for different genes and/or different calcium signatures, the fold change of gene expression for oscillatory and transient calcium signatures at 1 h will change (Figs S9, S10). However, the fold change of gene expression for a prolonged calcium signature is always < 1.5‐fold, independently of the delay time (Fig. S11). Thus, Fig. 6(a) explains the experimental observations in Table 2 (Whalley et al., 2011), and it shows that calcium signatures are differentially decoded to give specific CAMTA‐regulated gene expression responses.
Modelling analysis further reveals that the binding affinity between the 4Ca2+–CaM–CAMTA complex and DNA is an important parameter for inducing gene expression by calcium signatures. To our knowledge, this parameter has not been experimentally determined. When the binding affinity is reduced to 1.1 × 10−3 μM from 1.1 × 10−2 μM, all three calcium signatures in Figs 2(a), 3(a) and 4(a) are able to induce different large fold changes in gene expression (Fig. 6b). At 1 h, oscillatory (Fig. 2a), transient (Fig. 3a) and prolonged (Fig. 4a) calcium signatures induce c. 43‐, 12‐ and 9‐fold changes in gene expression, respectively. Thus, Fig. 6(b) shows that even a relatively small fold amplification of calcium signals (e.g. a prolonged calcium signature; Fig. 4a) is able to induce a relatively large fold induction of gene expression, with an oscillatory calcium signature inducing the largest fold change in gene expression . By contrast, the largest fold change in gene expression is induced by a transient calcium signature (Fig. 3a) in Fig. 6(a). Comparison of Fig. 6(a,b) shows that the binding affinity between the 4Ca2+–CaM–CAMTA complex and DNA can change how CAMTA‐regulated gene expression responds to different calcium signatures. In addition, when the binding affinity is decreased to 1.1 × 10−1 μM from 1.1 × 10−2 μM, all three calcium signatures in Figs 2(a), 3(a) and 4(a) are unable to induce fold changes larger than 1.05 in gene expression (Fig. 6c). Therefore, gene expression induced by different calcium signatures is quantitatively dependent on the binding affinity between the 4Ca2+–CaM–CAMTA complex and DNA.
In addition, the effects of other parameters relating to gene expression were also examined. Increasing or decreasing k 1 (the base rate for gene transcription) or k 2 (the maximal rate for 4Ca2+–CaM–CAMTA complex‐regulated gene transcription) by 2‐fold does not qualitatively change the modelling results (Figs S12, S13). The Hill coefficient (n) taking a value of 1, 2 or 3 also qualitatively leads to similar results (Fig. S14). However, the decay constant of mRNA (k 3) is an important parameter that affects the shape of the curve for gene expression (Fig. 6). If k 3 is very small, gene expression continues to increases for the computational time we have tested (2 h). If k 3 is very large, gene expression approaches the original steady state very quickly. An approximately 2‐fold increase or decrease of k 3 from its reference value (3.75 × 10−4 s−1) generally maintains the shape of the curve for gene expression, as shown in Fig. 6 (Fig. S15).
Gene expression response to a calcium signature depends on its history during its lifetime
Modelling analysis reveals that the gene expression response to a specific calcium signature depends on its history during its lifetime. Fig. 7 shows how this occurs for the oscillatory calcium signature (Fig. 2a) using the three binding affinities between the 4Ca2+–CaM–CAMTA complex and DNA, which are used in Fig. 6.
Figure 7.
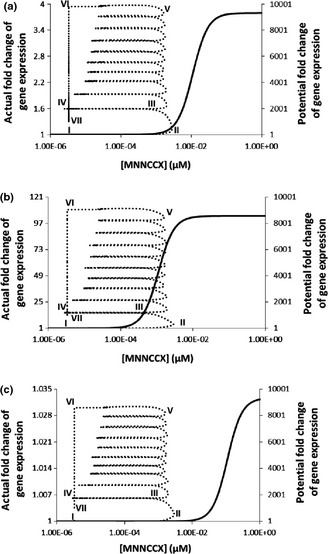
Gene expression accumulates all information during the lifetime of the oscillatory calcium signature (Fig. 2a) for three binding affinities between the active complex 4Ca2+–CaM–CAMTA and DNA in Arabidopsis thaliana. Solid line (right y‐axis): potential fold change of gene expression if the concentration of 4Ca2+–CaM–CAMTA stays at each concentration sufficiently long that a steady‐state is established at each concentration. Dashed line (left y‐axis): actual fold change of gene expression for 10 cycles of Ca2+ oscillation (Fig. 2a). Binding affinity (K d) between the active complex 4Ca2+–CaM–CAMTA and DNA: (a) 1.1 × 10−2 μM; (b) 1.1 × 10−3 μM; (c) 1.1 × 10−1 μM. CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
The oscillatory calcium signature (Fig. 2a) is nonlinearly amplified into a functional signal (4Ca2+–CaM–CAMTA complex) (Fig. 2c). In order to understand why the calcium signature induces gene expression in the specific ways described for Fig. 6, we calculate the potential steady‐state gene expression fold change by varying the 4Ca2+–CaM–CAMTA complex concentration (solid curve, Fig. 7) for three binding affinities between the 4Ca2+–CaM–CAMTA complex and DNA. The steady‐state gene expression fold change represents the maximum possible fold change in gene expression if time is sufficiently long so that steady‐state gene expression can become established for each 4Ca2+–CaM–CAMTA complex concentration. However, when a calcium signature emerges, the 4Ca2+–CaM–CAMTA complex concentration is a transient signal corresponding to the calcium signature and it does not establish a steady state (Figs 2c, 3c, 4c). Thus, actual gene expression follows the potential, but does not reach the potential, as explained in Fig. 7. At point I in Fig. 7(a), the oscillatory calcium signature has not emerged yet, and gene expression is at a steady state. When Ca2+ concentration elevates (Fig. 2a), the 4Ca2+–CaM–CAMTA complex concentration increases. At point II, gene expression has the potential to increase c. 800‐fold. However, the 4Ca2+–CaM–CAMTA complex concentration does not stay at point II and it starts to decrease from point II as a result of the decrease in Ca2+ concentration. At point III, gene expression has the potential to increase only c. 10‐fold. From point III to point IV, the 4Ca2+–CaM–CAMTA complex concentration continues to decrease following the calcium signature (Fig. 2a–c), and the potential fold increase of gene expression diminishes. At point IV, gene expression has no potential to increase at all, as the 4Ca2+–CaM–CAMTA complex concentration at point IV is the same as the original steady‐state concentration. From point I to point IV, gene expression continuously accumulates all the information from the calcium signature. During the first cycle of the calcium signature, gene expression increases to 1.6‐fold (Fig. 7a), although at point II it has the potential to increase c. 800‐fold and at point IV it has the potential to recover to the original steady‐state gene expression level, which is the level for which a calcium signature has not emerged. At point IV, gene expression memorizes the 1.6‐fold gene expression level and uses it as a starting point to read out the second cycle of the calcium signature (Fig. 2a). Gene expression response to the second cycle of the calcium signature follows the same principle as that for the first cycle. However, as gene expression memorizes the 1.6‐fold increase at point IV, at the end of the second cycle, it establishes a c. 1.9‐fold gene expression level (Fig. 7a). Again, the gene expression response memorizes this 1.9‐fold increase and continues to read out the third cycle of the calcium signature. After 10 cycles, gene expression increases to c. 4‐fold (point VI). At point VI, the calcium signature (Fig. 2a) ends and Ca2+ concentration recovers its original steady‐state level. Correspondingly, the 4Ca2+–CaM–CAMTA complex concentration also recovers its steady‐state level. Thus, at point VI, gene expression also starts to approach its original steady‐state level through point VII to point I. From point I to point VII, gene expression has continuously accumulated all the information during the lifetime of this calcium signature. Therefore, Fig. 7(a) reveals that the gene expression response depends on the history of the oscillatory calcium signature (Fig. 2a) and accumulates all information during the lifetime of this calcium signature.
Binding affinity between the 4Ca2+–CaM–CAMTA complex and DNA affects the dependence of the gene expression response on the history of a calcium signature. Fig. 7(b,c) show that, when the binding affinity is reduced or increased, the curve for the potential fold change of gene expression moves to the left or right, respectively. Thus, for the same oscillatory calcium signature (Fig. 2a), the reduced or increased binding affinity leads to larger (Fig. 7b) or smaller (Fig. 7c) gene expression fold changes, respectively. For example, when binding affinity is reduced (Fig. 7b), point II corresponds to a c. 8000‐fold potential gene expression change. However, when binding affinity is increased (Fig. 7c), point II corresponds to a c. 2‐fold potential gene expression change. This leads to a c. 16‐fold (Fig. 7b) and 1.006‐fold (Fig. 7c) actual gene expression change after the first cycle of the calcium signature, respectively. After 10 cycles of the calcium signature, a c. 115‐fold (Fig. 7b) or 1.03‐fold (Fig. 7c) actual gene expression change has been reached, respectively.
For both the transient calcium signature (Fig. 3a) and the prolonged calcium signature (Fig. 4a), how gene expression depends on the history of a calcium signature can also be analysed using the method summarized in Fig. 7. Specifically, gene expression accumulates information from both these calcium signatures in a similar manner to the first cycle of Fig. 7 (points I–IV), as shown in Figs S16 and S17. Therefore, for these two calcium signatures, gene expression also accumulates all information during their lifetimes. In summary, for all three types of calcium signatures (i.e. oscillatory (Fig. 2a), transient (Fig. 3a) and prolonged (Fig. 4a) calcium signatures), the gene expression response always depends on the history of the individual calcium signature and accumulates all information from the individual calcium signature, as shown in Figs 7, S16, S17. This explains phenomena such as why two signatures with equal areas under the curve (e.g. prolonged and transient) can give different gene expression responses (Whalley et al., 2011).
The three calcium signatures (i.e. oscillatory (Fig. 2a), transient (Fig. 3a) and prolonged (Fig. 4a)) examined above have distinctive kinetics. We further investigate how the parameters in oscillations are linked to gene expression. To do so, we reconstruct piecewise calcium signatures using the oscillatory calcium signature (Fig. 2a). For a piecewise calcium signature, the following relationship is always valid.
| (6) |
(A, the average calcium concentration of the calcium signature; t max and t min, the times for calcium concentration to be [Ca2+]max and [Ca2+]min, respectively; T, the period.) For the oscillatory calcium signature (Fig. 2a) A = 0.16 μM and T = 40 s. The average maximum and minimum calcium concentrations of the 10 calcium spikes in Fig. 2(a) are [Ca2+]max = 0.52 μM and [Ca2+]min = 0.10 μM, respectively. Thus, using Eqn (6), a piecewise calcium signature is constructed (Fig. 8a). This piecewise calcium signature has the same key parameters (average, maximum and minimum calcium concentrations, periods and durations) as those in Fig. 2(a), but it has a piecewise shape (Fig. 8a). Similarly, we can use Eqn (6) to construct other piecewise calcium signatures with the same key parameters (average, maximum and minimum calcium concentrations and durations) as those in Fig. 2(a), but with different periods (Figs S18, S19). As a consequence of the difference in oscillatory period, for a duration of 400 s, a piecewise oscillatory calcium signature with a period of 8 s (Fig. S18), 40 s (Fig. 8a) and 200 s (Fig. S19) will contain 50, 10 and two spikes, respectively. Using the reconstructed three oscillatory calcium signatures, we have investigated how gene expression depends on both the shape and period of calcium signatures (Fig. 8b). First, Fig. 8(b) reveals that a piecewise calcium signature induces a larger fold change in gene expression than the oscillatory calcium signature in Fig. 2(a) (the curve for the gene expression induced by the reconstructed piecewise calcium signature with a period of 40 s in Fig. 8(b) is compared with the curve for the gene expression induced by an experimental calcium signature with a period of 40s in Fig. 6a). Second, gene expression fold change depends on the period of oscillatory piecewise calcium signatures. Specifically, at 1 h, a piecewise oscillatory calcium signature with a period of 8, 40 and 200 s induces a 4.8‐, 6.0‐ and 6.6‐fold change in gene expression, respectively. Thus, for a fixed duration, increasing the number of calcium spikes by decreasing the oscillatory period decreases the fold change of gene expression.
Figure 8.
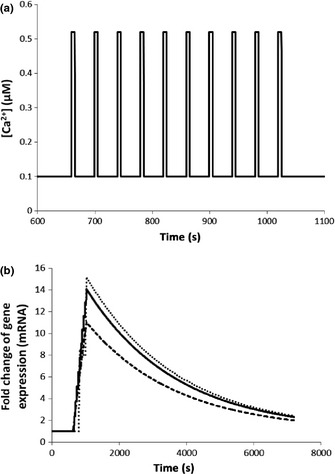
Fold change in gene expression induced by three piecewise calcium signatures that are reconstructed using the oscillatory calcium signature (Fig. 2a). Binding affinity (K d) between the active complex 4Ca2+–CaM–CAMTA and DNAis 1.1 × 10−2 μM. (a) The reconstructed piecewise calcium signature with A = 0.16 μM, [Ca2+]max = 0.52 μM and [Ca2+]min = 0.10 μM, T = 40 s. (b) Fold change in gene expression induced by three piecewise calcium signatures: bottom: T = 8 s (Fig. S18); middle: T = 40 s (a); top: T = 200 s (Fig. S19). CAMTA, calmodulin‐binding transcription activator; CaM, calmodulin.
An alternative way to vary the number of calcium spikes is to alter the duration of a calcium signature while its oscillatory period is fixed. Fig. S20 shows that increasing the number of calcium spikes by increasing the duration of a calcium signature increases the fold change of gene expression. At 1 h, a piecewise calcium signature with 5, 40 and 75 spikes induces a 1.3‐, 4.0‐ and 6.9‐fold change in gene expression, respectively. Experimentally, it has been shown that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell (Miwa et al., 2006). Combination of Figs 8(b) and S20 shows that calcium spike number is an important parameter regulating gene expression in our study. Moreover, both the oscillatory period and the duration of an oscillatory calcium signature also play their roles in gene expression (Figs 8b, S20). We note that our modelling results (Figs 8b, S20) are only applicable to CAMTA‐regulated gene expression and the gene expression mechanism we have used to calculate Figs 8(b) and S20 is the simple mechanism shown in Fig. 1. In general, gene expression may be regulated by other transcription factors and its expression mechanism may be different. In addition, Fig. 8(b) further indicates that the gene expression response represents an accumulation of all information on oscillatory periods in the three piecewise oscillatory calcium signatures during their lifetimes, as the only difference between the three calcium signatures (Figs 8a, S18, S19) is period.
Discussion
Experimental data show that A. thaliana is able to decode different calcium signatures to produce specific gene expression responses (Whalley et al., 2011; Whalley & Knight, 2013). Some of these calcium‐dependent genes are targets for CAMTA. It is also known that CAMTAs have calmodulin‐binding domains (Finkler et al., 2007). Therefore, gene expression responses regulated by CAMTAs respond to calcium signals. In this work, we develop a modelling methodology that establishes the information flow from calcium signatures to CAMTA‐regulated gene expression. Specifically, the 4Ca2+–CaM–CAMTA interaction nonlinearly amplifies different calcium signatures. Then, amplification of Ca2+ signals allows the calcium signatures to be differentially decoded to give specific CAMTA‐regulated gene expression responses. Finally, mathematical modelling reveals that the gene expression response depends on the history of a calcium signature and accumulates all information during the lifetime of this calcium signature. This could account for why oscillations of different frequencies can activate different downstream calcium decoders, for example, CaM kinase II, rather than the amplitude and duration of spikes being responsible for the activation, as previously suggested (De Koninck & Schulman, 1998).
For plants to survive stress, it is vital that their responses are specific and appropriate to the particular stimulus. This means that the identity of the primary stimulus must be encoded in a ‘language’ the cell can understand. Most stimuli lead to transient elevations in cellular calcium concentrations. Importantly, different stimuli produce calcium elevations with different characteristics: a unique ‘calcium signature’. Consequently, the specific properties of different calcium signatures have been proposed to encode information on the identity of the stimulus (McAinsh et al., 1995; Allen et al., 2001; Love et al., 2004; Miwa et al., 2006; McAinsh & Pittman, 2009; Dodd et al., 2010; Short et al., 2012). For example, temperature stress responses are associated with specific calcium signatures (Knight & Knight, 2012). In plants, there are different mechanisms of Ca2+‐regulated gene expression (Kim et al., 2009; Galon et al., 2010; Bickerton & Pittman, 2012). One of the possible mechanisms is through the binding of Ca2+, CaM and transcription factors. Using the transcription factor CAMTA as an example, this work has developed a general methodology to establish the links between calcium signatures and gene expression. First, Ca2+ binds to its target proteins following the principles of thermodynamics. This process nonlinearly amplifies the Ca2+ signal. As the binding of Ca2+ to its target proteins may follow different binding mechanisms (Kim et al., 2009; Galon et al., 2010; Bickerton & Pittman, 2012), how different binding processes for Ca2+ and its target proteins amplify Ca2+ signals should be investigated for each type of transcription factor. As demonstrated in this work, a relatively small fold amplification of signal is able to induce a relatively large fold change in gene expression (Fig. 6b). If the binding affinity between the active complex 4Ca2+–CaM–CAMTA and DNA is further reduced from that in Fig. 6(b) (1.1 × 10−3 μM), any small fold amplification of signal is able to induce a relatively large fold change in gene expression. This demonstrates that any calcium signature, even a modest one, is able to induce gene expression. This explains how even very modest increases in cytosolic free calcium, for example, in response to ozone, can lead to increases in gene expression (Clayton et al., 1999). Moreover, different calcium signatures are thus capable of inducing specific gene expression patterns (Fig. 6). Second, which Ca2+ and protein binding complex is active for DNA binding should be experimentally explored. Based on experimental observation, the 4Ca2+–CaM complex is the active complex for Ca2+–CaM binding (Pifl et al., 1984). Moreover, the binding affinity between active complex and DNA should be measured, as modelling analysis reveals that it is a key parameter for specific gene expression responses to calcium signatures. Third, the mechanisms of gene expression should be investigated for all relevant genes. In particular, Ca2+ signals may affect several processes relating to gene expression. For example, it has been proposed that the expression of the genes downstream of Enhanced Disease Susceptibility 1 protein may be simultaneously positively and negatively regulated by calcium signals (Zhang et al., 2014). In addition, during symbiosis signalling, it has been shown that calcium/calmodulin‐dependent protein kinase is negatively and positively regulated by calcium (Miller et al., 2013). For these cases, although the gene expression mechanisms include multiple interaction points with Ca2+ signals, how Ca2+ signals affect gene expression can also be analysed using the methodology developed in this work. This can be done by introducing a more complex gene expression mechanism in the right pane of our Fig. 1. In the work presented here, as the gene expression mechanism is generally unknown, we use the simplest gene expression mechanisms (Fig. 1) to establish the links between calcium signatures and gene expression, demonstrating how different calcium signatures are decoded to produce specific CAMTA‐regulated gene expression responses. As actual gene expression mechanisms may be more complicated than those we used in this work, our results shown in Fig. 6 can only be qualitatively (not quantitatively) compared with our Table 2. The quantitative fold change of a specific gene should be further investigated if its expression mechanism and the related parameters are determined in the future. Finally, as the gene expression response accumulates all information during the lifetime of a calcium signature, it is important to accurately record the kinetics of a calcium signature during its lifetime. This work reveals that the information flow from calcium signatures to gene expression is an integrative dynamical system (Fig. 1) following thermodynamic principles. A combined experimental and modelling approach is able to establish this information flow. Based on the experimental data in Table 2, we assume that expression of all genes we have studied in this work is positively regulated by calcium signatures (no down‐regulated genes were empirically observed). Therefore, we use Eqn (4) to analyse gene expression. However, if expression of other (non‐CAMTA‐regulated) genes is negatively regulated by calcium signals, Eqn (5) should be used to analyse gene expression following the methodology established in this work. The same methodology can also be extended to analyse gene expression regulated by other Ca2+‐dependent transcription factors.
Parameterization of kinetic models is generally a challenging task (Liu et al., 2010; Almquist et al., 2014). In this work, we use the following process to parameterize the kinetic model (Fig. 1): (1) parameters that have been experimentally determined are used; (2) the relationship of parameters is constrained following thermodynamic principles; (3) the model sensitivity is evaluated by varying each of the adjustable parameters; (4) the model sensitivity is tested for certain parameter combinations by simultaneously varying all adjustable parameters. Our analysis shows that the modelling results presented in this work are robust to variations in the parameter values across a wide range. In addition, while our model has integrated a wide range of knowledge about Ca2+–CaM–CAMTA binding, many other aspects relating to Ca2+–CaM–CAMTA binding and activity have not been included in the current model. For instance, different CAMTA isoforms are expressed in different cell types (Mitsuda et al., 2003) and different CAMTA isoforms have been suggested to be involved in responses to different primary signals (Kim et al., 2013; Pandey et al., 2013; Benn et al., 2014). It is also not known whether CAMTAs are subject to posttranslational modifications, so this feature is also not included in our model. Therefore, we consider the current model to be a starting point for establishing the relationship between calcium signatures and gene expression responses.
Mathematical modelling is an important tool with which to establish the link between stimulus, calcium signatures and gene expression. Currently, modelling analysis concentrates on different aspects of this link. For example, this work establishes the link between calcium signatures and gene expression for CAMTA‐regulated genes in A. thaliana cells. For other cells such as hepatocytes, various modelling efforts have also been made in an attempt to understand the decoding of calcium signals (Larsen et al., 2004; Schuster et al., 2005; Dupont et al., 2011). Information transfer in Ca2+ signalling pathways was also studied by combining experimental data and mathematical modelling (Pahle et al., 2008). In plant cells, other modelling work includes how different calcium signatures are generated from different stimuli. Specifically, a simple model for the cytosolic pool was used to explain the generation of calcium signatures by assuming that calcium‐permeable channels depend solely on the cooling rate and that calcium pumps are dependent on the absolute temperature (Plieth, 1999). A model of action potential in cells of vascular plants for the cytosolic pool was developed by incorporating K+, Cl− and Ca2+ channels; H+ and Ca2+ ATPases; the 2H+/Cl− symporter; and the H+/K+ antiporter. The model supports a hypothesis that H+ ATPase participates in AP generation (Vladimir & Vladimir, 2009). Recently, an integrative model that incorporates the interactions of Ca2+, H+, K+, Cl− and ATP in both cytosolic and vacuolar pools reveals how multiple ions in both the cytosol and the vacuole interplay to shape low‐temperature calcium signatures in plant cells (Liu et al., 2012). In addition, noisy time series of calcium oscillations (Granqvist et al., 2011) and generation of calcium signatures in other subcellular compartments such as the nucleus (Granqvist et al., 2012) have been studied. All of these studies and other modelling work in plant cells made efforts to establish links between stimuli and calcium signatures. Thus, it is plausible that, by integrating the links between stimuli and calcium signatures in the literature with the links between calcium signatures and gene expression response as described in this work, future research will be able to establish the relationship of stimuli, calcium signatures and gene expression responses. Thus, an integrative view of calcium signalling in plant cells can be formulated by integrating modelling and experimental studies.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The effects of varying the K d of R33, K d(R33), by changing k on.
Fig. S2 The effects of varying the K d of R33, K d(R33), by changing k off.
Fig. S3 The effects of varying the K d of R20, K d(R20), by changing k on.
Fig. S4 The effects of varying the K d of R20, K d(R20), by changing k off.
Fig. S5 The effects of varying the cooperative binding between CaM and CAMTA in the presence of Ca2+.
Fig. S6 The effects of varying the total CaM concentration on the amplification of Ca2+ signals.
Fig. S7 The effects of simultaneously varying all five adjustable parameters (example 1).
Fig. S8 The effects of simultaneously varying all five adjustable parameters (example 2).
Fig. S9 Dependence of fold change in gene expression induced by an oscillatory calcium signature (Fig. 2a) on the delay time.
Fig. S10 Dependence of fold change in gene expression induced by a transient calcium signature (Fig. 3a) on the delay time.
Fig. S11 Dependence of fold change in gene expression induced by a prolonged calcium signature (Fig. 4a) on the delay time.
Fig. S12 The effects of varying the base rate for gene transcription (k 1) on the fold change of gene expression.
Fig. S13 The effects of varying the maximal rate for 4Ca2+–CaM–CAMTA complex‐regulated gene transcription (k 2) on fold change of gene expression.
Fig. S14 The effects of varying the Hill coefficient (n) on fold change of gene expression.
Fig. S15 The effects of varying the decay constant of mRNA (k 3) on fold change of gene expression.
Fig. S16 Gene expression accumulates all information during the lifetime of the transient calcium signature (Fig. 3a).
Fig. S17 Gene expression accumulates all information during the lifetime of the prolonged calcium signature (Fig. 4a).
Fig. S18 The reconstructed piecewise calcium signature with T = 8 s.
Fig. S19 The reconstructed piecewise calcium signature with T = 200 s.
Fig. S20 Effects of the number of calcium spikes on fold change in gene expression.
Notes S1 Modelling equations.
Acknowledgements
The authors gratefully acknowledge Research Councils UK and the Biotechnology & Biological Sciences Research Council for funding in support of this study.
References
- Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffmann T, Tang YY, Grill E, Schroeder JI. 2001. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Almquist J, Cvijovic M, Hatzimanikatis V, Nielsen J, Jirstrand M. 2014. Kinetic models in industrial biotechnology‐improving cell factory performance. Metabolic Engineering 24: 38–60. [DOI] [PubMed] [Google Scholar]
- Benn G, Wang CQ, Hicks DR, Stein J, Guthrie C, Dehesh K. 2014. A key general stress response motif is regulated non‐uniformly by CAMTA transcription factors. Plant Journal 80: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton PD, Pittman JK. 2012. Calcium signalling in plants, eLS. Chichester, UK: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0023722 [Google Scholar]
- Bouche N, Scharlat A, Snedden W, Bouchez D, Fromm H. 2002. A novel family of calmodulin‐binding transcription activators in multicellular organisms. Journal of Biological Chemistry 277: 21851–21861. [DOI] [PubMed] [Google Scholar]
- Brown SE, Martin SR, Bayley PM. 1997. Kinetic control of the dissociation pathway of calmodulin‐peptide complexes. Journal of Biological Chemistry 272: 3389–3397. [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim MC, Yoo JH, Moon BC, Koo SC, Park BO, Lee JH, Koo YD, Han HJ, Lee SY et al 2005. Isolation of a calmodulin binding transcription factor from rice (Oryza sativa L.). Journal of Biological Chemistry 280: 40820–40831. [DOI] [PubMed] [Google Scholar]
- Clayton H, Knight MR, Knight H, McAinsh MR, Hetherington AM. 1999. Dissection of the ozone‐induced calcium signature. Plant Journal 17: 575–579. [DOI] [PubMed] [Google Scholar]
- De Koninck P, Schulman H. 1998. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279: 227–230. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. 2010. The language of calcium signaling. In: Merchant S, Briggs WR, Ort D, eds.Annual Review of Plant Biology 61: 593–620. [DOI] [PubMed] [Google Scholar]
- Doherty DJ, Van Buskirk HA, Myers SJ, Thomashow MF. 2009. Roles for Arabidopsis CAMTA transcription factors in cold‐regulated gene expression and freezing tolerance. Plant Cell 21: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont G, Combettes L, Bird GS, Putney JW. 2011. Calcium oscillations. Cold Spring Harbor Perspectives in Biology 3: a004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajmut A, Brumen M, Schuster S. 2005. Theoretical model of the interactions between Ca2+, calmodulin and myosin light chain kinase. FEBS Letters 579: 4361–4366. [DOI] [PubMed] [Google Scholar]
- Finkler A, Ashery‐Padan R, Fromm H. 2007. CAMTAs: calmodulin‐binding transcription activators from plants to human. FEBS Letters 581: 3893–3898. [DOI] [PubMed] [Google Scholar]
- Finn BE, Forsen S. 1995. The evolving model of calmodulin structure, function and activation. Structure 3: 7–11. [DOI] [PubMed] [Google Scholar]
- Gaertner TR, Putkey JA, Waxham MN. 2004. RC3/Neurogranin and Ca2+/calmodulin dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. Journal of Biological Chemistry 279: 39374–39382. [DOI] [PubMed] [Google Scholar]
- Galon Y, Finkler A, Fromm H. 2010. Calcium‐regulated transcription in plants. Molecular Plant 3: 653–669. [DOI] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. 2008. Calmodulin‐binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Letters 582: 943–948. [DOI] [PubMed] [Google Scholar]
- Granqvist E, Oldroyd GED, Morris RJ. 2011. Automated Bayesian model development for frequency detection in biological time series. BMC Systems Biology 5: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granqvist E, Wysham D, Hazledine S, Kozlowski W, Sun J, Charpentier M, Martins TV, Haleux P, Tsaneva‐Atanasova K, Downie JA et al 2012. Buffering capacity explains signal variation in symbiotic calcium oscillations. Plant Physiology 160: 2300–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Chung WS, Yun DJ, Cho MJ. 2009. Calcium and calmodulin‐mediated regulation of gene expression in plants. Molecular Plant 2: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park S, Gilmour SJ, Thomashow MF. 2013. Roles of CAMTA transcription factors and salicylic acid in configuring the low‐temperature transcriptome and freezing tolerance of Arabidopsis. Plant Journal 75: 364–376. [DOI] [PubMed] [Google Scholar]
- Knight MR, Knight H. 2012. Low‐temperature perception leading to gene expression and cold tolerance in higher plants. New Phytologist 195: 737–751. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Putkey J, Waxham M. 2007. Neurogranin controls the spatiotemporal pattern of postsynaptic Ca2+/CaM signaling. Biophysical Journal 93: 3848–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. 2010. Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen AZ, Olsen LF, Kummer U. 2004. On the encoding and decoding of calcium signals in hepatocytes. Biophysical Chemistry 107: 83–99. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. 2000. Transcription of eukaryotic protein‐coding genes. Annual Review of Genetics 34: 77–137. [DOI] [PubMed] [Google Scholar]
- Linse S, Helmersson A, Forsen S. 1991. Calcium binding to calmodulin and its globular domains. Journal of Biological Chemistry 266: 8050–8054. [PubMed] [Google Scholar]
- Liu JL, Grieson CS, Webb AAR, Hussey PJ. 2010. Modelling dynamic plant cells. Current Opinion in Plant Biology 13: 744–749. [DOI] [PubMed] [Google Scholar]
- Liu JL, Knight H, Hurst CH, Knight MR. 2012. Modelling and experimental analysis of the role of interacting cytosolic and vacuolar pools in shaping low temperature calcium signatures in plant cells. Molecular BioSystems 8: 2205–2220. [DOI] [PubMed] [Google Scholar]
- Love J, Dodd AN, Webb AAR. 2004. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 16: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Maune J, Beckingham K, Bayley P. 1992. Stopped‐flow studies of calcium dissociation from calcium‐binding‐site mutants of Drosophila melanogaster calmodulin. European Journal of Biochemistry 205: 1107–1114. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. 2009. Shaping the calcium signature. New Phytologist 181: 275–294. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Webb AAR, Taylor JE, Hetherington AM. 1995. Stimulus‐induced oscillations in guard cell cytosolic‐free calcium. Plant Cell 7: 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JB, Pratap A, Miyahara A, Zhou L, Bornemann S, Morris RJ, Oldroyd GE. 2013. Calcium/calmodulin‐dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Isono T, Sato MH. 2003. Arabidopsis CAMTA family proteins enhance V‐PPase expression in pollen. Plant and Cell Physiology 44: 975–981. [DOI] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Downie JA. 2006. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant Journal 48: 883–894. [DOI] [PubMed] [Google Scholar]
- Pahle J, Green A, Dixon C, Kummer U. 2008. Information transfer in signaling pathways: a study using coupled simulated and experimental data. BMC Bioinformatics 9: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N, Ranjan A, Pant P, Tripathi RK, Ateek F, Pandey HP, Patre UV, Sawant SV. 2013. CAMTA 1 regulates drought responses in Arabidopsis thaliana . BMC Genomics 14: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen OB, Madsen TS, Falke JJ. 1997. Intermolecular tuning of calmodulin by target peptides and proteins: differential effects on Ca2+ binding and implications for kinase activation. Protein Science 6: 794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Kinzer‐Ursem T, Mihalas S, Kennedy MB. 2010. A dynamic model of interactions of Ca2+, calmodulin, and catalytic subunits of Ca2+/calmodulin‐dependent protein kinase II. PLoS Computational Biology 6: e1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persechini A, White HD, Gansz KJ. 1996. Different mechanisms for Ca2+ dissociation from complexes of calmodulin with nitric oxide synthase or myosin light chain kinase. Journal of Biological Chemistry 271: 62–67. [DOI] [PubMed] [Google Scholar]
- Pifl C, Plank B, Wyskovsky W, Bertel O, Hellmann G, Suko J. 1984. Calmodulin X (Ca2+)4 is the active calmodulin‐calcium species activating the calcium‐, calmodulin‐dependent protein kinase of cardiac sarcoplasmic reticulum in the regulation of the calcium pump. Biochimica et Biophysica Acta 773: 197–206. [DOI] [PubMed] [Google Scholar]
- Plieth C. 1999. Temperature sensing by plants: calcium‐permeable channels as primary sensors‐A model. The Journal of Membrane Biology 172: 121–127. [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Du L, Wang H, Yang T. 2013. Recent advances in calcium/calmodulin‐mediated signaling with an emphasis on plant‐microbe interactions. Plant Physiology 163: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Ali GS, Celesnik H, Day IS. 2011. Coping with stresses: roles of calcium‐ and calcium/calmodulin‐regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Knoke B, Marhl M. 2005. Differential regulation of proteins by bursting calcium oscillations‐a theoretical study. BioSystems 81: 49–63. [DOI] [PubMed] [Google Scholar]
- Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. 2006. Ca2+/Calmodulin dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proceedings of the National Academy of Sciences, USA 103: 13968–13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short EF, North KA, Roberts MR, Hetherington AM, Shirras AD, McAinsh MR. 2012. A stress‐specific calcium signature regulating an ozone‐responsive gene expression network in Arabidopsis. Plant Journal 71: 948–961. [DOI] [PubMed] [Google Scholar]
- Valeyev N, Bates D, Heslop‐Harrison P, Postlethwaite I, Kotov N. 2008. Elucidating the mechanisms of cooperative calcium‐calmodulin interactions: a structural systems biology approach. BMC Systems Biology 2: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimir S, Vladimir V. 2009. A mathematical model of action potential in cells of vascular plants. The Journal of Membrane Biology 232: 59–67. [DOI] [PubMed] [Google Scholar]
- Whalley HJ, Knight MR. 2013. Calcium signatures are decoded by plants to give specific gene responses. New Phytologist 197: 690–693. [DOI] [PubMed] [Google Scholar]
- Whalley HJ, Sargeant AW, Steele JFC, Lacoere T, Lamb R, Saunders NJ, Knight H, Knight MR. 2011. Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 23: 4079–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Du L, Shen C, Yang Y, Poovaiah BW. 2014. Regulation of plant immunity through ubiquitin‐mediated modulation of Ca2+‐calmodulin‐AtSR1/CAMTA3 signaling. Plant Journal 78: 269–281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The effects of varying the K d of R33, K d(R33), by changing k on.
Fig. S2 The effects of varying the K d of R33, K d(R33), by changing k off.
Fig. S3 The effects of varying the K d of R20, K d(R20), by changing k on.
Fig. S4 The effects of varying the K d of R20, K d(R20), by changing k off.
Fig. S5 The effects of varying the cooperative binding between CaM and CAMTA in the presence of Ca2+.
Fig. S6 The effects of varying the total CaM concentration on the amplification of Ca2+ signals.
Fig. S7 The effects of simultaneously varying all five adjustable parameters (example 1).
Fig. S8 The effects of simultaneously varying all five adjustable parameters (example 2).
Fig. S9 Dependence of fold change in gene expression induced by an oscillatory calcium signature (Fig. 2a) on the delay time.
Fig. S10 Dependence of fold change in gene expression induced by a transient calcium signature (Fig. 3a) on the delay time.
Fig. S11 Dependence of fold change in gene expression induced by a prolonged calcium signature (Fig. 4a) on the delay time.
Fig. S12 The effects of varying the base rate for gene transcription (k 1) on the fold change of gene expression.
Fig. S13 The effects of varying the maximal rate for 4Ca2+–CaM–CAMTA complex‐regulated gene transcription (k 2) on fold change of gene expression.
Fig. S14 The effects of varying the Hill coefficient (n) on fold change of gene expression.
Fig. S15 The effects of varying the decay constant of mRNA (k 3) on fold change of gene expression.
Fig. S16 Gene expression accumulates all information during the lifetime of the transient calcium signature (Fig. 3a).
Fig. S17 Gene expression accumulates all information during the lifetime of the prolonged calcium signature (Fig. 4a).
Fig. S18 The reconstructed piecewise calcium signature with T = 8 s.
Fig. S19 The reconstructed piecewise calcium signature with T = 200 s.
Fig. S20 Effects of the number of calcium spikes on fold change in gene expression.
Notes S1 Modelling equations.


