Abstract
BACKGROUND
Molecular stratification of prostate cancer (PCa) based on genetic aberrations including ETS or RAF gene‐rearrangements, PTEN deletion, and SPINK1 over‐expression show clear prognostic and diagnostic utility. Gene rearrangements involving ETS transcription factors are frequent pathogenetic somatic events observed in PCa. Incidence of ETS rearrangements in Caucasian PCa patients has been reported, however, occurrence in Indian population is largely unknown. The aim of this study was to determine the prevalence of the ETS and RAF kinase gene rearrangements, SPINK1 over‐expression, and PTEN deletion in this cohort.
METHODS
In this multi‐center study, formalin‐fixed paraffin embedded (FFPE) PCa specimens (n = 121) were procured from four major medical institutions in India. The tissues were sectioned and molecular profiling was done using immunohistochemistry (IHC), RNA in situ hybridization (RNA‐ISH) and/or fluorescence in situ hybridization (FISH).
RESULTS
ERG over‐expression was detected in 48.9% (46/94) PCa specimens by IHC, which was confirmed in a subset of cases by FISH. Among other ETS family members, while ETV1 transcript was detected in one case by RNA‐ISH, no alteration in ETV4 was observed. SPINK1 over‐expression was observed in 12.5% (12/96) and PTEN deletion in 21.52% (17/79) of the total PCa cases. Interestingly, PTEN deletion was found in 30% of the ERG‐positive cases (P = 0.017) but in only one case with SPINK1 over‐expression (P = 0.67). BRAF and RAF1 gene rearrangements were detected in ∼1% and ∼4.5% of the PCa cases, respectively.
CONCLUSIONS
This is the first report on comprehensive molecular profiling of the major spectrum of the causal aberrations in Indian men with PCa. Our findings suggest that ETS gene rearrangement and SPINK1 over‐expression patterns in North Indian population largely resembled those observed in Caucasian population but differed from Japanese and Chinese PCa patients. The molecular profiling data presented in this study could help in clinical decision‐making for the pursuit of surgery, diagnosis, and in selection of therapeutic intervention. Prostate 75:1051–1062, 2015. © 2015 The Authors. The Prostate, published by Wiley Periodicals, Inc.
Keywords: genetic rearrangements, TMPRSS2‐ERG, SPINK1, RAF kinase, PTEN
INTRODUCTION
Prostate cancer (PCa) is one of the most common malignancies after lung cancer and the second most common cause of cancer‐related death among men worldwide. The incidence of PCa in India has been steadily increasing concomitant with an increase in life expectancy. According to the National Cancer Registry program by the Indian Council of Medical Research, New Delhi, PCa is estimated to increase by 140% in the next few years 1, 2. The challenges posed in diagnosing and treating prostate cancer is attributed to the clinical and molecular heterogeneity associated with this disease. The seminal discovery of the recurrent (>50%) genetic rearrangements involving the androgen‐regulated gene transmembrane protease, serine 2 (TMPRSS2), with v‐ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) in PCa prompted the molecular categorization of PCa into distinct molecular subtypes 3, similar to hematologic malignancies, for identifying patients with aggressive subtypes and distinct therapeutic targets. Subsequently, discovery of genetic rearrangements involving TMPRSS2 with other 3' erythroblastosis virus E26 transformation‐specific (ETS) transcription factor family members such as ETV1 (∼5%), ETV4 (∼5%), or ETV5 (∼1%) led to further classification of PCa.
In a separate study using Cancer Outlier Profile Analysis (COPA), SPINK1 (serine peptidase inhibitor, Kazal type 1) was shown to be over‐expressed in ∼10–15% of the total PCa cases 4. Interestingly, SPINK1 and ETS over‐expression demonstrated mutually exclusive pattern across multiple independent PCa cohorts, and SPINK1 outlier expression was associated with an aggressive subset of prostate cancers 4, 5, 6.
Another important molecular event in PCa is the inactivation of the phosphatase and tensin homolog (PTEN) gene by genomic deletion or rearrangement, including intragenic breakage and translocation. PTEN deletion or rearrangement has been reported in 20–30% of PCa and is also associated with aggressive cancers 7, 8, 9. Interestingly, mouse model of PCa demonstrated that PTEN loss and ERG genetic rearrangement might cooperate in the development of prostate adenocarcinoma 10, 11. Many independent studies have shown that PTEN deletion and ERG genetic alterations frequently coexist, and incidences of PTEN deletions are more frequent in ETS fusion‐positive PCa than in fusion‐negative cancers 7, 8, 12, 13.
Using paired‐end next‐generation sequencing approach, the druggable RAF kinase genetic rearrangements, SLC45A3‐BRAF (solute carrier family 45, member 3–v‐raf murine sarcoma viral oncogene homolog B1) and ESRP1‐RAF1 (epithelial splicing regulatory protein‐1–v‐raf‐1murine leukemia viral oncogene homolog‐1) were discovered in ∼2% of the PCa patients 14. Most importantly, the RAF genetic rearrangements demonstrated sensitivity to sorafenib, a US Food and Drug Administration (FDA) approved RAF/MEK inhibitor suggesting that RAF genetic rearrangements, although rare, may benefit patients positive for RAF rearrangements 14.
Although TMPRSS2‐ERG gene fusion is one of the highly recurrent (∼50%) oncogenic drivers in PCa, it has been a challenging therapeutic target. A recent study demonstrated a potential strategy to therapeutically target ERG via its interacting partner protein, poly‐ADP‐ribose polymerase (PARP) for the treatment of ERG fusion‐positive PCa 15. Interestingly, lower incidence of ERG genetic rearrangement has been reported in African Americans (∼28%) in comparison to ∼50–60% recurrence in Caucasian Americans. Moreover, incidence of PTEN deletion is low (7%) in African American men compared to Caucasian men and the prevalence of SPINK1 over‐expression is high (∼23%) among African American men compared to their Caucasian counterpart 16. Much lower recurrence of ERG alteration (∼10–20%) has been reported among the Japanese 17 and Chinese PCa cohort 18 compared to the Western population. Moreover, Filipinos with PCa are at increased risk of progressing to advanced stages of PCa and have lower survival rates compared to other Asians. Prevalence of TMPRSS2‐ERG rearrangement is 23% among Filipino PCa patients with an increased rate (∼33%) observed in the advanced PCa cohort 19. Comprehensive landscape of genetic alterations in PCa patients from the Indian sub‐continent is currently lacking. Recently, a report on PCa patients from India reported low recurrence (∼27%) of TMPRSS2‐ERG genetic rearrangement 20. However, inherent limitations of this study include the small cohort size (n = 30) and all samples analyzed were from a single institution, thus extrapolation of these results to broader population is difficult. Here, to study the genetic heterogeneity of PCa in the Indian population, we performed comprehensive characterization of patient samples to determine the prevalence of genetic rearrangements involving ETS gene family members (ERG, ETV1, and ETV4); non‐ETS genetic rearrangements such as RAF kinase family fusions (BRAF or RAF1), SPINK1 over‐expression and the status of the tumor suppressor gene PTEN. Our data revealed that the ETS gene rearrangement and SPINK1 over‐expression patterns in the North Indian population largely resembled those observed in the Caucasian population but differed from Japanese and Chinese PCa patients.
MATERIALS AND METHODS
Prostate Specimens
A total of 121 PCa specimens (including five benign prostate specimens) were procured from All India Institute of Medical Sciences, New Delhi; King George's Medical University, Lucknow; Digdarshika Pathology Laboratory, Lucknow; and GSVM Medical College, Kanpur. A total of 22 PCa specimens were excluded from the study based on low tumor content and integrity of the cancer tissues. All PCa specimens were collected from patients with written informed consent and upon Institutional Review Board approval. The study PCa cohort included specimens from men who underwent prostatectomy (n = 40), needle core biopsies (n = 48), and transurethral resection of the prostate (TURP) (n = 33) to relieve obstructive symptoms from locally advanced disease at the participating institutions between year 2010 and 2014. None of the patients received preoperative radiation or androgen deprivation therapy. All patients included in this study were of Indian descent residing in northern India. All specimen included in the study were de‐identified.
ERG and SPINK1 Immunohistochemistry
Immunohistochemistry was performed on unstained formalin‐fixed, paraffin‐embedded (FFPE) tissue sections obtained from the prostatectomy, needle core biopsies, or TURP specimen blocks. Primary monoclonal antibody against ERG, clone EPR 3864 (Epitomics, Burlingame, CA) or monoclonal antibody against SPINK1 (H00006690‐M01‐Abnova, Taiwan) was used on the automated Ventana Benchmark staining platform (Ventana Medical Systems‐A Roche group, Tucson, AZ). Staining of blood vessels was used as a positive control and slides displaying no staining of vessels were excluded from further analysis. All IHC slides were evaluated by a board certified pathologist (LPK).
Fluorescence In Situ Hybridization (FISH)
FISH assays were performed on unstained FFPE sections obtained from the PCa specimen blocks. BAC clones (ERG 5'‐RP11‐830D9; ERG 3'‐RP11‐143L14; PTEN locus probe‐RP11‐165M8; chr10 control probe RP11‐351D16; ETV4 5'‐RP11‐147C10; ETV4 3'‐CTD3215I16; BRAF 5'‐RP11‐767F15; BRAF 3'‐RP11‐248P7; RAF1 5'‐RP11‐767C1; RAF1 3'‐RP11‐586C12) were selected from the UCSC genome browser and purchased through BACPAC resources (Children's Hospital, Oakland, CA). Following colony purification DNA was prepared using QiagenTips‐100 (Qiagen, Valencia, CA). DNA was labeled by nick translation method with biotin‐16‐dUTP and digoxigenin‐11‐dUTP for 3' and 5' probes and PTEN locus and CEN10 control probes respectively (Roche, USA). Probe DNA was precipitated and dissolved in hybridization mixture containing 50% formamide, 2X SSC, 10% dextran sulphate, and 1% Denhardt's solution. Approximately two hundred nanogram of labeled probe was hybridized to normal human chromosomes to confirm the map position of each BAC clone. FISH signals were obtained using anti‐digoxigenin‐fluorescein and AlexaFluor‐594 conjugate to obtain green and red colors, respectively. Fluorescence images were captured using a high resolution CCD camera controlled by ISIS image processing software (Metasystems, Germany).
Deletion of PTEN was defined as fewer than two copies of the gene specific probe in the presence of two reference signals in >20% of the tumor nuclei. For both ERG rearrangement and PTEN deletion, at least one hundred tumor nuclei per case were evaluated under a fluorescence microscope (Carl Zeiss & Metasystems, Germany).
RNA In Situ Hybridization and Evaluation Criteria
RNA in situ hybridization (RNA‐ISH) was performed as described previously using RNAscope FFPE Reagent Kit 2.0 (Advanced Cell Diagnostics, Hayward, CA) 21. Briefly, FFPE sections were baked at 60 °C for 1 hr. ETV1 RNA probe (ETV1 Accession Number NM_004956.4, region 998‐2031) and POLR2A (as a positive control) were designed by Advanced Cell Diagnostics (Hayward, CA). Tissues were deparaffinized in xylene twice for 15 min each with periodic agitation, slides were then immersed in 100% ethanol twice for 3 min each with periodic agitation and air‐dried for 5 min. Tissues were circled using a pap pen (Vector, H‐4000), allowed to dry and treated with pretreatment 1 buffer for 10 min. The slides were processed using previously established protocol 21 and mounted in Cytoseal XYL (Thermo Scientific, #8312‐4) for viewing under bright‐field microscope. Positive controls were performed for all runs using a POLR2A gene‐specific RNA probe.
RNA‐ISH expression intensity scoring guidelines were established to classify tumor foci as ETV1 positive or ETV1 negative 22. ETV1 expression by RNA‐ISH appeared as distinct cytoplasmic punctate dots. All tumor foci were evaluated and scanned at 20× magnification. Scoring for an entire tumor focus was based on the highest ETV1 intensity using criteria previously reported 21. All ETV1 RNA‐ISH slides were reviewed by study pathologist (LPK).
Statistical Analysis
Fisher's exact tests were used to evaluate association between categorical variables. For analyzing the association between PTEN deletion status (response, normal vs. deletion) with co‐occurrence of ERG or SPINK1 positive status logistic regression has been applied. For assessing any association between ERG positive, SPINK1 positive, or SPINK1 negative statuses and Gleason score (using numeric Gleason score as a response) linear regression model was used. For all statistical tests, a P < 0.05 was considered statistically significant.
RESULTS
Prevalence of ERG, ETV1, and ETV4 Genetic Rearrangements
ERG oncoprotein expression was evaluated by IHC on 94 PCa biopsies and five benign prostate specimens. A total of 46 out of 94 PCa (48.9%) were found to be positive for ERG expression (Fig. 1A) and all five benign PCa specimens were negative for ERG over‐expression. We next confirmed ERG gene rearrangement in a subset of patients (n = 17) that were positive for ERG staining by FISH. As anticipated, all cases showed ERG rearrangement either by ERG split signal or 5' deletion of ERG (Fig. 1A, bottom panel). Results from the FISH assay of ERG rearrangement‐positive cases revealed that ERG gene fusion occurred in neoplastic cells but not in adjacent benign nuclei or stromal cells. The majority of the cases included in this study had index tumor Gleason scores of 7 or higher (82/94, 87%) and all available clinical information associated with the patients' specimens are listed in Supplementary Table 1. For the remaining 43 ERG‐negative cases, ETV1, and ETV4 rearrangement detection was performed by RNA‐ISH and FISH, respectively. None of the 43 (0/43) PCa specimens tested by ETV4 break‐apart FISH were positive and only one PCa specimen was detected as ETV1 positive as evaluated by RNA‐ISH (Fig. 1B). Of note, the ETV1‐positive case was negative for ERG and SPINK1 over‐expression by IHC. None of the benign specimens (n = 5) were positive for ERG, ETV1, or ETV4 genetic rearrangements.
Figure 1.
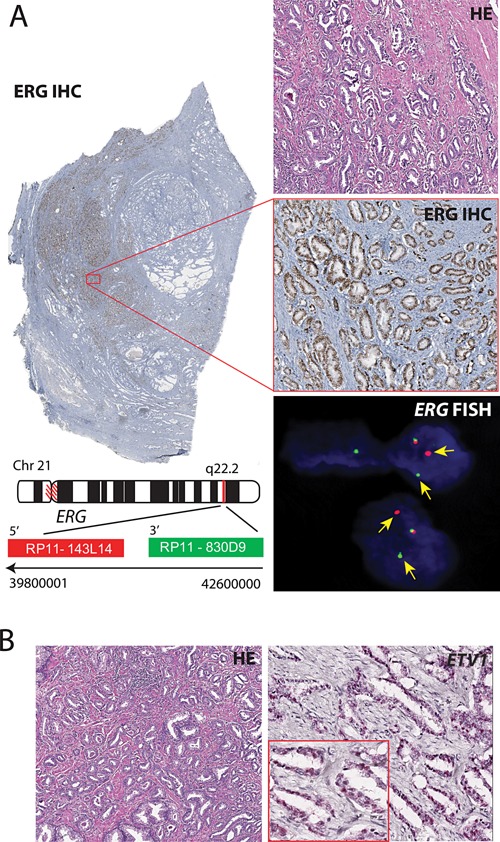
IHC staining and FISH for ERG rearrangement and RNA‐ISH for ETV1. Panel A, whole section view of a prostate carcinoma tissue (Gleason score 3 + 3) showing ERG expression by IHC staining (panel on left). Right panel showing IHC staining for ERG over‐expression (×20 magnification) with the corresponding H&E stained sections (top). Bottom panel shows FISH for ERG genetic rearrangement using ERG break‐apart probes. Yellow arrow shows rearranged ERG, and co‐localizing red‐green signal depicts intact ERG locus. Panel B shows ETV1 expression by RNA‐ISH with a maximum intensity score 4 (×20 magnification).
Prevalence of SPINK1 Over‐Expression
SPINK1 over‐expression was evaluated by performing IHC on all the ERG‐negative cases and the cases where the ERG‐positive immunostaining was heterogeneous (7 cases). A total of 66 PCa specimens were immunostained for the SPINK1 over‐expression, 12 cases were confirmed as SPINK1‐positive representing 12.76% of the 94 total PCa specimens (Fig. 2A–D). Importantly, none of the SPINK1‐positive cases were ERG rearrangement‐positive, confirming mutual exclusivity of both genetic alterations. Moreover, all seven ERG‐positive heterogeneous cases were found to be negative for SPINK1. Of note, SPINK1 prevalence in our cohort is similar to the SPINK1 incidence (∼10–15%) reported in the Caucasian cohorts 4, 5. As anticipated none of the benign specimen (n = 5) were positive for SPINK1 over‐expression. No significant association between ERG positive status and Gleason score was found when adjusted for SPINK1 status (P = 0.42) using a linear model (numeric Gleason scores) and an ordinal proportional odds regression with fine ordinal categorization of Gleason scoring (<6, 3 + 4, 4 + 3, 8–10). Although SPINK1 negative cases showed a trend for increased Gleason score (0.58, SE = 0.29, P = 0.053) using a linear regression model. The effect reached significance in the ordinal model with the cumulative logOR of 1.26 (SE = 0.64, P = 0.049).
Figure 2.
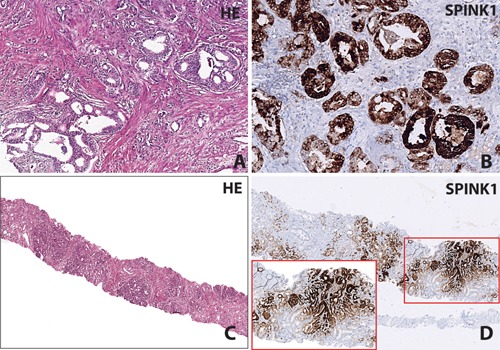
IHC staining for SPINK1. Panel A and B show a prostatic adenocarcinoma with SPINK1 positive immunostaining with the corresponding H&E stained section; Gleason score 3 + 4 (×20 magnification). Panel C and D show a needle core biopsy which demonstrates SPINK1 positive immunostaining with the corresponding H&E stained section; Gleason score 3 + 3 (×4 and ×10 magnification in inset).
Concomitant Presence of ERG Rearrangement, SPINK1 Over‐Expression, and PTEN Deletion
To assess PTEN genomic deletion, a two color interphase FISH approach was used as PTEN deletions are interstitial and are usually limited to small regions of chromosome 10. The PTEN FISH was performed on 92 PCa specimens, of those 13 specimens were excluded from the analysis due to no signal or no hybridization on the tissue sections. Homozygous deletion of PTEN was observed in 11.39% (9 out of 79) of the PCa cases whereas hemizygous deletion was observed in 10.13% (8 out of 79) of the PCa cases (Fig. 3A–C). We also observed aneuploidy (>2 copies of PTEN) in 17.72% (14 out of 79) of the PCa cases (Fig. 3A–B). Interestingly, 30% (14 out of 46) of the ERG‐positive cases showed PTEN deletion compared to only one SPINK1‐positive case (1 out of 12) with PTEN deletion. Logistic regression was applied to model the association of the PTEN deletion status (response, normal vs. deletion) with ERG and SPINK1 positive status. ERG positivity leads to an increase in the chances to develop a PTEN deletion of 1.65 on the logit scale (SE = 0.69, P = 0.017). Conversely, the association of SPINK1 positive status with PTEN deletion was non‐significant (P = 0.67), and point estimate of the effect was much smaller (0.64 on logit scale, SE = 1.47). Thus, our findings corroborate with the previous studies demonstrating a significant overlap between ERG genetic rearrangements and PTEN deletions 12, 23. Moreover, the incidence of ERG positivity in patients with PTEN deletions was 0.82, exact 95% Confidence Interval (CI) (0.59, 0.94). Likewise the incidence of SPINK1 positive status with PTEN deletion was much smaller, 0.33, exact 95% CI (0.008, 0.91), although the sample size for the latter is too small to be interpreted.
Figure 3.
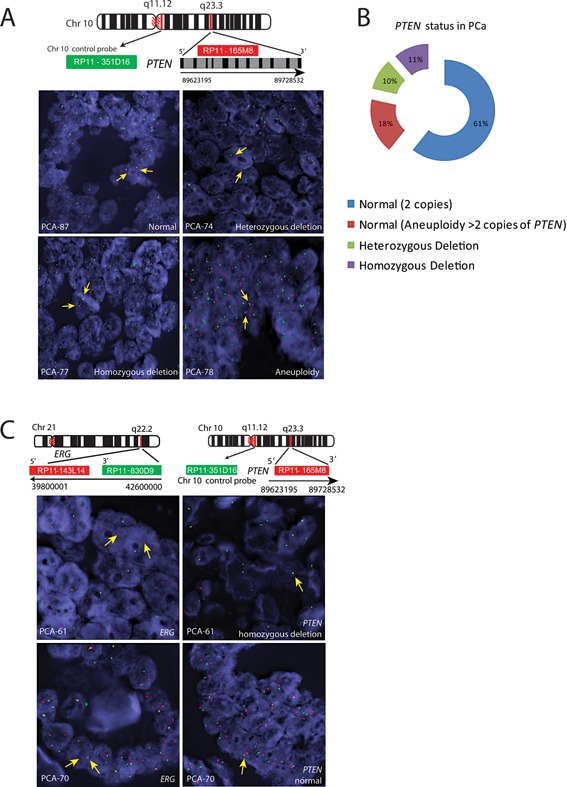
FISH for PTEN aberrations. Panel A, schematic diagram showing the genomic organization of PTEN gene on Chr10 q23.3 and chromosome 10 control probe (q11.12). Green and red bars indicate the chromosome 10 control probe and PTEN BAC clones, respectively. FISH images of the PCa specimens, PCA‐87 shows normal PTEN copy number with two red and two green signals corresponding to PTEN and Chr10 control probe, respectively. PCA‐74 shows heterozygous deletion of PTEN showing loss of one red signal, but two green centromere signals. PCA‐77 depicts homozygous deletion of PTEN (loss of red signals), but two green centromere signals. PCA‐78 shows aneuploidy with 3‐4 copies of chromosome 10 and PTEN. Panel B, percent distribution of the PTEN aberrations. Panel C shows ERG genetic rearrangement using ERG break‐apart probes and homozygous deletion of PTEN in the same patient (PCA‐61). Likewise, PCA‐70 shows ERG genetic rearrangement and normal PTEN status.
Incidence of BRAF and RAF1 Rearrangements
BRAF and RAF1 rearrangements were detected by FISH using break‐apart probes on 88 and 63 PCa specimens, respectively. Interestingly, one out of 88 (∼1%) PCa cases harbored BRAF genetic rearrangement and two cases displayed focal amplification with 5–7 copies of BRAF (Fig. 4A). These results are consistent with our previous finding that reported ∼2% incidence of BRAF rearrangement in prostate cancer 14. RAF1 rearrangement was observed in ∼4.5% (3 out of 66) of PCa cases; PCA‐23 harbored RAF1 rearrangement, second case (PCA‐56) displayed 3' deletion and third case (PCA‐40) displayed 3' deletion in one tumor foci and RAF amplification and 5' deletion in another tumor foci (Fig. 4B, top panel). We also observed a single case (PCA‐16) with 4 copies of RAF1 resulting from aneuploidy (Fig. 4B, bottom panel). Notably, all the cases that were positive for rearrangements or amplification of BRAF or RAF1 had features of advanced PCa including high Gleason score (four cases with Gleason score 9 and two cases with Gleason score 7). Intriguingly, all the RAF rearrangement‐positive cases were also positive for ERG rearrangement, but not the sample harboring BRAF amplification.
Figure 4.
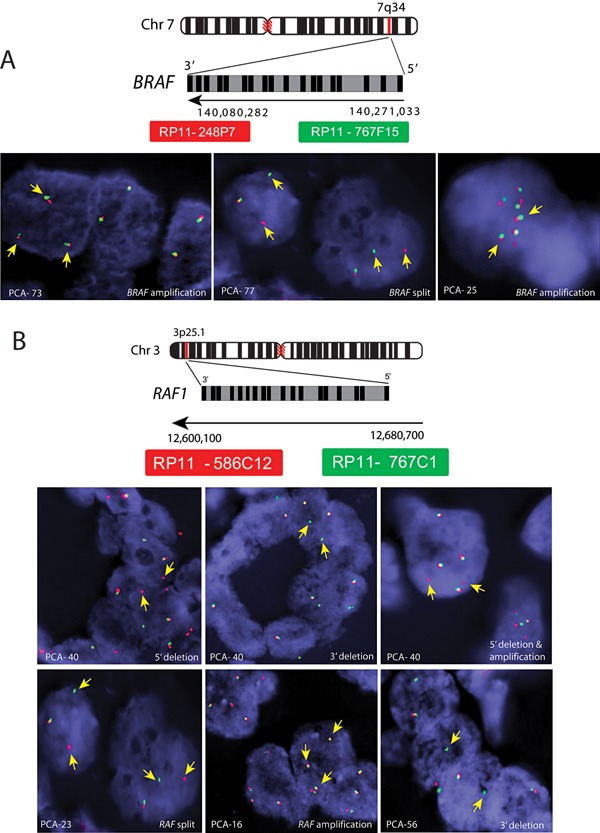
FISH for RAF genetic rearrangements. Panel A, schematic diagram showing the genomic organization of BRAF gene on chr7q34. Green and red bars indicate the 5' and 3' BAC clones, respectively. FISH images of the PCa specimens (PCA‐73 & 25) show BRAF amplification (left) and BRAF rearrangement (middle) in PCA‐77 (right). Panel B, schematic diagram showing the genomic organization of the RAF1 gene on chr3p25. Green and red bars indicate 5' and 3' BAC clones, respectively. FISH images of the PCa specimens (PCA‐40) show RAF1 rearrangement with 5' deletion (green signal missing), 3' deletion (red signaling missing), and 5' deletion with 3 copies of normal RAF1 in the top panel. Likewise, PCa specimens PCA‐23, PCA‐16, and PCA‐56 show RAF rearrangement, amplification, and 3' deletion, respectively (bottom panel).
DISCUSSION
In this study, we stratified Indian PCa cohort based on the molecular alterations known to be prevalent among different PCa populations in the world 3, 4, 24, 25, 26, 27. To our knowledge, this is the first study to comprehensively evaluate the entire spectrum of driving aberrations in Indian PCa samples that include ERG, ETV1, ETV4, RAF kinase genetic rearrangements, SPINK1 over‐expression, and PTEN deletion. Our findings indicate a higher incidence of ERG rearrangement (49%) in the Indian PCa cohort compared to a recent report by Rawal and colleagues (27%) 20. This discrepancy could arise from the smaller cohort size (total 30 PCa cases) and the samples collected from a single medical institution. Additionally, they characterized only the ERG gene rearrangement using a different ERG antibody (9FY clone, Biocare Medical Inc.) 20 for IHC analysis as well as variation in evaluation criteria, and possibly therapeutic interventions (anti‐androgen therapy). Most importantly, Park et al., demonstrated that ERG immunostaining using same monoclonal antibody that we have used against ERG had an overall 95.7% sensitivity and 96.5% specificity for ERG rearrangements, suggesting that ERG expression by IHC has high concordance with FISH 28.
Although, ETS genetic rearrangement represents a highly prevalent genetic alteration (50–60%) among PCa patients of the Western world, therapeutic targeting of ETS transcription remains a challenge. Nevertheless, we have shown that ERG interacts with the enzyme poly (ADP‐ribose) polymerase 1 (PARP1) in a DNA‐independent manner and pharmacological inhibition of PARP1 inhibits ETS‐positive PCa xenograft 15. Our findings also showed that ETV1 genetic rearrangements are less recurrent (∼1%) in Indian PCa cohort than what has been reported in the Caucasian cohorts (∼5%) 24, 29, although studies with larger patient cohorts are needed to validate our finding. Interestingly, Baena et al., demonstrated that ERG and ETV1 regulates androgen receptor (AR) target genes inversely, ERG negatively regulates the AR transcriptional program whereas ETV1 enhances AR signaling and activation of the AR transcriptional program 30. Our data corroborates previous findings that ERG rearrangements are clonal in nature as all cells in a given cancer foci were positive for the ERG expression, while distinct cancer foci in a single prostate may have differing ERG rearrangement status 12, 31, 32, 33. However recently, existence of rare molecular subsets of PCa with dual gene rearrangements, such as ERG/SPINK1 34, ERG/ETV1, and ERG/ETV4 in different tumor focus of the same tumor has been noted 22.
Similar to the incidences reported in Caucasian PCa cohorts 4, 5, we also identified SPINK1 over‐expression in ∼12% of the PCa cases in Indian subcontinent. A recent report on African American men (n = 105) with PCa showed higher SPINK1 incidence (23.8%) compared to only ∼8% SPINK1‐positive cases in Caucasian PCa samples (n = 113) 16, highlighting differences at the molecular level in these two clinicopathologically matched PCa cohorts and racial disparities in prostate cancer 35, 36, 37. Several independent studies have confirmed the mutual exclusivity of SPINK1 and ERG rearrangements in PCa 4. SPINK1 over‐expression is associated with aggressive phenotype among ETS‐negative PCa cases, with higher risk of biochemical recurrence than SPINK1‐negative patients 4, 5. Previously, we showed that SPINK1 interacts with epidermal growth factor receptor (EGFR) and activates downstream signaling and EGFR dimerization 38. Moreover, monoclonal antibodies to either SPINK1 or EGFR (cetuximab) significantly slowed down tumor growth in SPINK1‐positive tumor xenografted mice 38. Of note, multiplex assay of SPINK1 and TMPRSS2‐ERG along with GOLPH2 and PCA3 transcript expression could be utilized as predictors of PCa, and most importantly these biomarkers could outperform serum PSA or PCA3 alone in detecting the disease 39.
Deletions or mutations in PTEN, which encodes a phosphoinositide 3‐phosphatase, have been found to be associated with higher Gleason score, metastasis, hormone resistance, and an overall poor prognosis 9, 26, 40. Furthermore, the clinical significance of molecular aberrations in PTEN, TMPRSS2‐ERG, and SPINK1 has been demonstrated in the development of castration‐resistant prostate cancer (CRPC) 23. Here, we found almost equal proportion of homozygous (11.39%) and heterozygous PTEN deletions (10.13%) in the Indian PCa cohort. A recent report demonstrated that heterozygous PTEN deletions are less frequent in African American PCa cohort (6.9%) than in Caucasian PCa cohort (19.8%) 16. Moreover, homozygous PTEN deletions are more prevalent in CRPC whereas PTEN heterozygous deletions occurred at higher frequency in localized PCa 6, 23. Importantly, loss of PTEN results in increased AKT and mTOR signaling, suggesting that alterations in the mTOR/AKT pathways could also be therapeutically targeted in patients harboring PTEN loss. Indeed, a recent study demonstrated promising therapeutic outcomes upon dual inhibition of AKT and mTORC1 in the preclinical genetically‐engineered mouse (GEM) model of CRPC 41.
In the present study, we found ∼4.5% (3 out of 66) RAF1 genetic rearrangements and ∼1% (1 out of 88) BRAF genetic rearrangement in the Indian PCa cohort. Likewise, lower incidence of the BRAF (2.5%) and RAF1 (1.5%) aberrations has been reported in the Chinese PCa cohort as well 42. Nevertheless, a high incidence of BRAF (29%) and RAF1 (15%) copy number gain was observed in the same cohort, suggesting activation of the RAS/RAF/MEK/ERK signaling pathway 42. In contrast to Chinese PCa cohort, we found only one case with BRAF focal amplification; however more studies with larger cohort size are needed to confirm our finding. One of the limitations of our study is the lack of patients' follow‐up information and the evaluation of associations with clinical outcome, as follow‐up of the cancer patients is poor in India due to inadequate health/medical awareness and socioeconomic status of the patients. Nevertheless, the current study provides an initial molecular stratification of the PCa in this patient population that could aid in clinical decision‐making for the pursuit of surgical, targeted therapy, hormonal and/or chemo, and radiation therapy. Moreover, in the current genomic and precision therapy era, the diagnosis and treatment of PCa is rapidly evolving 43, 44. Hence, comprehensive molecular characterization of PCa patients from Indian sub‐continent utilizing high‐throughput sequencing approaches will inform the pursuit of appropriate targeted therapies.
In summary, this is the first comprehensive report demonstrating the prevalence of the ETS gene‐rearrangements, SPINK1 over‐expression, druggable RAF rearrangements and PTEN aberrations prevalent among Indian men with PCa. Taken together, Indian PCa population characterized in this study largely resembled the ETS gene rearrangement and SPINK1 over‐expression scenario observed in the Caucasian race, and differed from the prevalence reported in Japanese and Chinese patients; suggesting racial disparity and differences at the molecular level in prostate cancer. Most importantly, similar to ALK gene fusions (∼5% of the cases) in non‐small cell lung cancer patients, who benefit from ALK kinase inhibitors; the higher incidence of RAF rearrangements positive PCa patients (∼5% in Indian PCa) reported in this study may respond to the FDA‐approved RAF inhibitors or MEK inhibitors. Therefore, understanding the underlying common molecular driver alterations prevalent among Indian PCa patients will permit optimization of the screening methods and selection of the appropriate treatment regimen in this population.
DISCLOSURE/CONFLICT OF INTEREST
N.P. receives research funding and honoraria from the Ventana/Roche but this study was not supported by this funding A.M.C. serves on the advisory board of the Gen‐Probe; and is a co‐inventor on a patent filed by the University of Michigan covering the diagnostic and therapeutic field of use for ETS fusions in prostate cancer. The Ventana/ Roche and Gen‐Probe did not play a role in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article. The remaining authors declare no conflicts of interest.
Supporting information
Supplementary Table S1.
ACKNOWLEDGEMENT
We thank Saravana Mohan Dhanasekaran and Jyoti Athanikar for critically reading the manuscript. B.A. is an Intermediate Fellow of the Wellcome Trust/DBT India Alliance and a Young Investigator of the DST‐FAST Track scheme. This work is supported by the Wellcome Trust/ DBT India Alliance grant [IA/I(S)/12/2/500635 to B.A.]. A.M.C. is supported by the Howard Hughes Medical Institute, the Doris Duke Foundation, and the Prostate Cancer Foundation and is an American Cancer Research Professor and a Taubman Scholar. We thank Ventana Medical Systems/Roche for providing the Ventana Benchmark platform. We thank Indian Institute of Technology, Kanpur for providing infra‐structure support. We also thank Dr. Jonaki Sen for her assistance in the specimen procurement.
Bushra Ateeq and Nallasivam Palanisamy contributed equally to this work.
REFERENCES
- 1. Takiar R, Vijay CR. An alternative approach to study the changes in the cancer pattern of men in India (1988–2005). Asian Pac J Cancer Prev 2011; 12(4):875–878. [PubMed] [Google Scholar]
- 2. Swaminathan R, Shanta V, Ferlay J, Balasubramanian S, Bray F, Sankaranarayanan R. Trends in cancer incidence in Chennai city (1982–2006) and statewide predictions of future burden in Tamil Nadu (2007–16). Natl Med J India 2011; 24(2):72–77. [PubMed] [Google Scholar]
- 3. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Montie JE, Shah RB, Pienta KJ, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310(5748):644–648. [DOI] [PubMed] [Google Scholar]
- 4. Tomlins SA. The role of SPINK1 in ETS rearrangement‐negative prostate cancers. Cancer Cell 2008; 13(6):519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramaki OR, Visakorpi T. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine? Treated prostate cancer. Clin Cancer Res 2010; 16(10):2845–2851. [DOI] [PubMed] [Google Scholar]
- 6. Bismar TA, Yoshimoto M, Duan Q, Liu S, Sircar K, Squire JA. Interactions and relationships of PTEN, ERG, SPINK1, and AR in castration‐resistant prostate cancer. Histopathology 2012; 60(4):645–652. [DOI] [PubMed] [Google Scholar]
- 7. Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 2007; 97(5):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer 2008; 99(8):1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA. PTEN genomic deletion is associated with p‐Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol 2009; 218(4):505–513. [DOI] [PubMed] [Google Scholar]
- 10. King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL. Cooperativity of TMPRSS2‐ERG with PI3‐kinase pathway activation in prostate oncogenesis. Nat Genet 2009; 41(5):524–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon‐Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 2009; 41(5):619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 2009; 22(8):1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer‐Kornblum M, Kotschau D, Tennstedt P, Huang J, Gerhauser C, Mader M, Kurtz S, Sirma H, Saad F, Steuber T, Graefen M, Plass C, Sauter G, Simon R, Minner S, Schlomm T. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion‐positive and fusion‐negative prostate cancer. Am J Pathol 2012; 181(2):401–412. [DOI] [PubMed] [Google Scholar]
- 14. Palanisamy N, Ateeq B, Kalyana‐Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, Kumar‐Sinha C, Dhanasekaran SM, Chen YB, Esgueva R, Banerjee S, LaFargue CJ, Siddiqui J, Demichelis F, Moeller P, Bismar TA, Kuefer R, Fullen DR, Johnson TM, Greenson JK, Giordano TJ, Tan P, Tomlins SA, Varambally S, Rubin MA, Maher CA, Chinnaiyan AM. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer, and melanoma. Nat Med 2010; 16(7):793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, Palanisamy N, Siddiqui J, Yan W, Cao X, Mehra R, Sabolch A, Basrur V, Lonigro RJ, Yang J, Tomlins SA, Maher CA, Elenitoba‐Johnson KS, Hussain M, Navone NM, Pienta KJ, Varambally S, Feng FY, Chinnaiyan AM. Mechanistic rationale for inhibition of poly(ADP‐ribose) polymerase in ETS gene fusion‐positive prostate cancer. Cancer Cell 2011; 19(5):664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khani F, Mosquera JM, Park K, Blattner M, O'Reilly C, MacDonald TY, Chen Z, Srivastava A, Tewari AK, Barbieri CE, Rubin MA, Robinson BD. Evidence for molecular differences in prostate cancer between African American and Caucasian Men. Clin Cancer Res 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Furusato B, van Leenders GJ, Trapman J, Kimura T, Egawa S, Takahashi H, Furusato M, Visakorpi T, Hano H. Immunohistochemical ETS‐related gene detection in a Japanese prostate cancer cohort: Diagnostic use in Japanese prostate cancer patients. Pathol Int 2011; 61(7):409–414. [DOI] [PubMed] [Google Scholar]
- 18. Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Tian W, Wang J, Yang H, Sun Y. RNA‐seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer‐associated long noncoding RNAs and aberrant alternative splicings. Cell Res 2012; 22(5):806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymundo EM, Diwa MH, Lapitan MC, Plaza AB, Sevilleja JE, Srivastava S, Sesterhenn IA. Increased association of the ERG oncoprotein expression in advanced stages of prostate cancer in Filipinos. Prostate 2014; 74(11):1079–1085. [DOI] [PubMed] [Google Scholar]
- 20. Rawal S, Young D, Williams M, Colombo M, Krishnappa R, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. Low frequency of the ERG oncogene alterations in prostate cancer patients from India. J Cancer 2013; 4(6):468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warrick JI, Tomlins SA, Carskadon SL, Young AM, Siddiqui J, Wei JT, Chinnaiyan AM, Kunju LP, Palanisamy N. Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization‐a correlative study with urine PCA3 and TMPRSS2‐ERG . Mod Pathol 2014; 27(4):609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kunju LP, Carskadon S, Siddiqui J, Tomlins SA, Chinnaiyan AM, Palanisamy N. Novel RNA hybridization method for the in situ detection of ETV1, ETV4, and ETV5 gene fusions in prostate cancer. Appl Immunohistochem Mol Morphol 2014; 22(8):e32–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J, Squire JA. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int 2010; 107(3):477–485. [DOI] [PubMed] [Google Scholar]
- 24. Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007; 448(7153):595–599. [DOI] [PubMed] [Google Scholar]
- 25. Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2‐ERG gene fusion in prostate cancer. Neoplasia 2008; 10(2):177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, Saadati HR. Role of PTEN gene in progression of prostate cancer. Urol J 2007; 4(2):95–100. [PubMed] [Google Scholar]
- 27. Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2: ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res 2006; 66(7):3396–3400. [DOI] [PubMed] [Google Scholar]
- 28. Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody‐based detection of ERG rearrangement‐positive prostate cancer. Neoplasia 2010; 12(7):590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barros‐Silva JD, Paulo P, Bakken AC, Cerveira N, Lovf M, Henrique R, Jeronimo C, Lothe RA, Skotheim RI, Teixeira MR. Novel 5' fusion partners of ETV1 and ETV4 in prostate cancer. Neoplasia 2013; 15(7):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, Kim J, Nguyen M, Zhang X, Godinho FJ, Bronson RT, Mucci LA, Loda M, Yuan GC, Orkin SH, Li Z. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev 2013; 27(6):683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2‐ERG fusion heterogeneity in multifocal prostate cancer: Clinical and biologic implications. Urology 2007; 70(4):630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG, Dobi A, Srivastava S, Petrovics G, Sesterhenn IA. Mapping of TMPRSS2‐ERG fusions in the context of multi‐focal prostate cancer. Mod Pathol 2008; 21(2):67–75. [DOI] [PubMed] [Google Scholar]
- 33. Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: Molecular evidence for an independent group of diseases. Cancer Res 2007; 67(17):7991–7995. [DOI] [PubMed] [Google Scholar]
- 34. Bhalla R, Kunju LP, Tomlins SA, Christopherson K, Cortez C, Carskadon S, Siddiqui J, Park K, Mosquera JM, Pestano GA, Rubin MA, Chinnaiyan AM, Palanisamy N. Novel dual‐color immunohistochemical methods for detecting ERG‐PTEN and ERG‐SPINK1 status in prostate carcinoma. Mod Pathol 26(6):835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Bohm D, Perner S, Dobi A, McLeod DG, Sesterhenn IA, Srivastava S. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology 80(4):749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magi‐Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2‐ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African‐American and Japanese patients. Prostate 2010; 71(5):489–497. [DOI] [PubMed] [Google Scholar]
- 37. Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Bohm D, Perner S, Dobi A, McLeod DG, Sesterhenn IA, Srivastava S. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology 2012; 80(4):749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, Li Y, Wang X, Feng FY, Pienta KJ, Varambally S, Chinnaiyan AM. Therapeutic targeting of SPINK1‐positive prostate cancer. Sci Transl Med 2011; 3(72):17ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first‐generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008; 68(3):645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen MM, Abate‐Shen C. PTEN inactivation and the emergence of androgen‐independent prostate cancer. Cancer Res 2007; 67(14):6535–6538. [DOI] [PubMed] [Google Scholar]
- 41. Floc'h N, Abate‐Shen C. The promise of dual targeting AKT/mTOR signaling in lethal prostate cancer. Oncotarget 2012; 3(12):1483–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, Song R, Berney DM, Clark J, Cooper C, Lu YJ. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosomes Cancer 2012; 51(11):1014–1023. [DOI] [PubMed] [Google Scholar]
- 43. Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana‐Sundaram S, Sam L, Balbin OA, Quist MJ, Barrette T, Everett J, Siddiqui J, Kunju LP, Navone N, Araujo JC, Troncoso P, Logothetis CJ, Innis JW, Smith DC, Lao CD, Kim SY, Roberts JS, Gruber SB, Pienta KJ, Talpaz M, Chinnaiyan AM. Personalized oncology through integrative high‐throughput sequencing: A pilot study. Sci Transl Med 2011; 3(111):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roychowdhury S, Chinnaiyan AM. Advancing precision medicine for prostate cancer through genomics. J Clin Oncol 2013; 31(15):1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1.


