Summary
The sugar nucleotide dTDP‐L‐rhamnose is critical for the biosynthesis of the Group A Carbohydrate, the molecular signature and virulence determinant of the human pathogen Group A S treptococcus (GAS). The final step of the four‐step dTDP‐L‐rhamnose biosynthesis pathway is catalyzed by dTDP‐4‐dehydrorhamnose reductases (RmlD). RmlD from the Gram‐negative bacterium S almonella is the only structurally characterized family member and requires metal‐dependent homo‐dimerization for enzymatic activity. Using a biochemical and structural biology approach, we demonstrate that the only RmlD homologue from GAS, previously renamed GacA, functions in a novel monomeric manner. Sequence analysis of 213 Gram‐negative and Gram‐positive RmlD homologues predicts that enzymes from all Gram‐positive species lack a dimerization motif and function as monomers. The enzymatic function of GacA was confirmed through heterologous expression of gac A in a S. mutans rml D knockout, which restored attenuated growth and aberrant cell division. Finally, analysis of a saturated mutant GAS library using Tn‐sequencing and generation of a conditional‐expression mutant identified gac A as an essential gene for GAS. In conclusion, GacA is an essential monomeric enzyme in GAS and representative of monomeric RmlD enzymes in Gram‐positive bacteria and a subset of Gram‐negative bacteria. These results will help future screens for novel inhibitors of dTDP‐L‐rhamnose biosynthesis.
Introduction
The cell wall of Gram‐positive bacteria is an intricate network of peptidoglycan, proteins and secondary cell wall polymers (SCWPs) that are covalently linked to peptidoglycan. Teichoic or teichuronic acids are typical and well‐studied SCWP in Gram‐positive bacteria and play an important role in normal cell function and infection (Weidenmaier and Peschel, 2008). Many β‐hemolytic streptococcal species appear to lack expression of typical teichoic or teichuronic acid structures (Sutcliffe et al., 2008; Caliot et al., 2012) and instead express a rhamnose‐rich polymer, which comprises approximately half of the cell wall mass (McCarty, 1952). Historically, expression of these evolutionary conserved glycans underlies classification of β‐hemolytic streptococci in Lancefield groups (A, B, C, G …) (Lancefield, 1933), a feature that is still applied in contemporary rapid test kits to diagnose streptococcal infections.
Streptococcus pyogenes, also referred to as Group A Streptococcus (GAS), is a β‐hemolytic human‐restricted pathogen and ranks in the top 10 of infection‐related causes of mortality worldwide (Carapetis et al., 2005). GAS is the causative agent of a wide spectrum of clinical disease, including common localized infections (∼ 700 million cases per year worldwide) and approximately 1.8 million cases of severe disease (Carapetis et al., 2005), including necrotizing fasciitis, streptococcal toxic shock syndrome and post‐infectious streptococcal sequelae, i.e. acute rheumatic fever and rheumatic heart disease. A better understanding of GAS pathogenesis and development of new drugs and protective vaccines is crucial. GAS expresses a characteristic SCWP known as Lancefield Group A Antigen or Group A Carbohydrate (GAC) (Lancefield, 1933). The GAC structure consists of a polyrhamnose core decorated with alternating immunodominant N‐acetylglucosamine (GlcNAc) side‐chains (Coligan et al., 1978). Recently, van Sorge et al. (2014) identified the gene cluster responsible for GAC biosynthesis and demonstrated that the GAC GlcNAc side‐chain contributes to GAS virulence.
In contrast to detailed insights into the biosynthesis of classical SCWP like teichoic acids, information regarding the biosynthesis of rhamnose‐rich polysaccharides GAC is limited. The production of dTDP‐L‐rhamnose is critical for GAC biosynthesis but also more broadly for the viability or virulence of other medically important bacteria including Mycobacterium spp. (Ma et al., 2002), Pseudomonas spp. (Engels et al., 1985) and Enterococcus faecalis (Teng et al., 2005). dTDP‐L‐rhamnose is synthesized from α‐glucose‐1‐phosphate through a four‐step enzymatic process catalyzed by the enzymes RmlA‐D (Kornfeld and Glaser, 1961; Pazur and Shuey, 1961). Structural analysis of RmlA‐D from Pseudomonas aeruginosa (RmlA, Blankenfeldt et al., 2000), Streptococcus suis (RmlB, Beis et al., 2003 and RlmC, Dong et al., 2003) and Salmonella enterica (Se) (RmlD, Blankenfeldt et al., 2002), have provided valuable insights into the mechanism of action for these enzymes, including requirement for Mg2+‐dependent dimerization in the case of RmlD. In general, RmlD enzymes are members of the large short‐chain dehydro‐genases/reductases (SDR) superfamily, which act on a wide family of substrates and commonly form homo‐dimeric or multimeric complexes (Kavanagh et al., 2008).
In GAS, homologues of RmlA, B and C that catalyze the first steps of the dTDP‐rhamnose biosynthesis pathway can be identified through bioinformatics and are clustered on the genome. The GAS RmlD homologue appears to be encoded by the gacA gene, which is annotated as a dTDP‐4‐dehydrorhamnose reductase, but experimental data supporting this function is currently lacking. The goal of this study was to identify the function and structure of the gacA gene product through biochemistry, structural biology and bacterial genetics. We show that gacA is an essential gene of GAS that encodes a metal‐independent dTDP‐4‐dehydrorhamnose reductase representative of a new class of monomeric RmlD enzymes.
Results and discussion
GacA encodes a functional metal‐independent dTDP‐4‐dehydrorhamnose reductase (RmlD)
Bioinformatics analysis suggests that gacA encodes a dTDP‐4‐dehydrorhamnose reductase, an enzyme that catalyzes the final step in the production of dTDP‐L‐rhamnose (Giraud and Naismith, 2000). In contrast to the rmlD genes in other species like Shigella flexneri (Macpherson et al., 1994) and Streptococcus pneumoniae serotype 19F (Morona et al., 1997), the only GAS rmlD homologue gacA is not part of an rmlABCD rhamnose biosynthesis operon. Instead, gacA is located at the beginning of the recently identified GAC gene cluster and hence named gacA (van Sorge et al., 2014). A similar split genomic architecture of the rhamnose biosynthesis genes rmlA‐C and rmlD was previously observed in Streptococcus mutans (S. mutans), a cariogenic Gram‐positive bacterium (Tsukioka et al., 1997).
We set out to investigate the potential function of GacA as a dTDP‐4‐dehydrorhamnose reductase. We cloned and expressed full‐length GacA fused to a cleavable GST‐His6 tag. The GAS GacA protein sequence is 36% identical to the Salmonella enterica serovar Typhimurium RmlD protein (SeRmlD) (Fig. 1), the only reported RmlD structure (Blankenfeldt et al., 2002). To confirm the enzymatic activity of GacA in vitro, we also cloned, expressed and purified the putative RmlB and RmlC GAS homologues, as the RmlD substrate dTDP‐4‐dehydrorhamnose is not commercially available. In this biochemical assay, oxidation of NADPH is a read‐out for GacA activity. No activity was observed with any combination of two enzymes (Fig. 2A). However, we observed significant oxidation of NADPH when all three enzymes were present, suggesting that GacA is indeed acting as a dTDP‐4‐dehydrorhamnose reductase (Fig. 2A). As mentioned above, the enzyme activity of SeRmlD was shown to be metal dependent (Blankenfeldt et al., 2002). Similarly, RmlD from Mycobacterium tuberculosis (MtbRmlD) was assayed in presence of 10 mM MgCl2, despite the lack of biochemical evidence that the enzyme requires a divalent cation for enzymatic activity (Wang et al., 2011). To investigate the metal‐dependent activity of GacA, we performed the assay in presence and absence of 10 mM MgCl2 and/or 10 mM EDTA. No significant changes in enzymatic activity were observed for GacA (Fig. 2A), demonstrating that unlike SeRmlD (Blankenfeldt et al., 2002), the proper positioning of the cofactor is not dependent on a metal‐ion (Graninger et al., 1999). The GacA enzyme activity assay was subsequently performed in absence of MgCl2.
Figure 1.
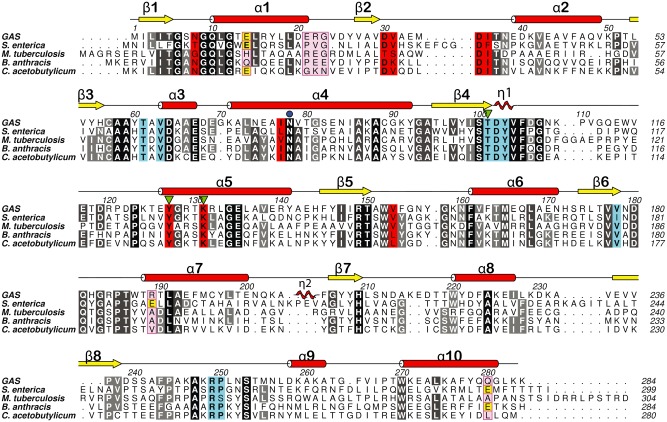
Sequence alignment for GacA and RmlD homologues. Sequence alignment of GAS GacA with RmlD from S almonella enterica serovar Typhimurium (Blankenfeldt et al., 2002), M ycobacterium tuberculosis (accession number WP_009938025), the hypothetical RmlD homologues from B acillus anthracis str. Ames (accession number NP_843703) and C lostridium acetobutylicum (accession number WP_010965612). Conserved residues are colored in black (> 80%) and gray (60–80%). Substrate binding site residues are colored in turquois and cofactor binding site residues in red. Secondary structure elements from GacA are indicated and labeled with red α‐helices and yellow β‐strands. Mg2+‐binding site residues from SeRmlD are colored in yellow (E1–E3) and the corresponding non‐conserved residues in the other species are colored in magenta (R1–R3). The terminal residues of the α1‐helix motif are highlighted with magenta boxes. N78 is indicated with a blue dot, and the catalytic triad is indicated with green triangles.
Figure 2.
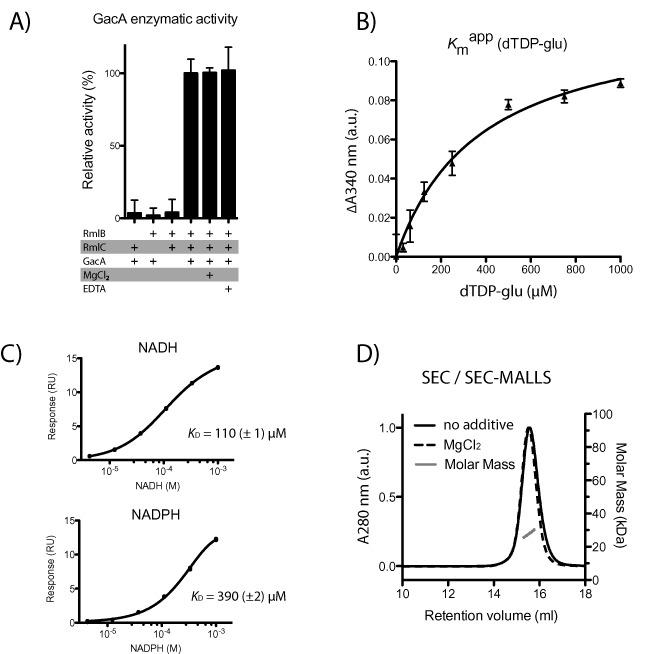
GacA is a functional metal‐independent monomeric dTDP‐4‐dehydrorhamnose reductase.
A. Biochemical confirmation that GacA is a functional and metal‐independent dTDP‐4‐dehydrorhamnose reductase. Read‐out of GacA enzyme activity is change in absorbance at 340 nm, indicating oxidation of the GacA cofactor NADPH to NADP +, was observed. GacA is only active in the presence of coupled enzymes SpRmlB and SpRmlC, which provide the substrate for GacA. GacA metal dependence was investigated in presence and absence of 10 mM MgCl2 and 10 mM EDTA.
B. GacA Michaelis‐Menten kinetics were determined for dTDP‐glucose (K m value of 370 μM). The second substrate NADPH was present in excess.
C. Surface plasmon resonance equilibrium fit for the binding of NADPH and NADH to GacA. NADPH and NADH were injected over a concentration range of 4.1 μM to 1,000 μM. Equilibrium affinity fit values are shown in the bottom panel of the figure.
D. Size‐exclusion chromatography (SEC) and SEC‐MALLS of recombinant purified GacA in presence (dashed line) and absence (solid line) of 10 mM MgCl2. All samples reveal the same retention volume, corresponding to a calculated average molecular mass of 27.5 kDa (blue line) demonstrating GacA is a monomer.
Next, we studied Michaelis‐Menten kinetics of GacA. The K m app value for dTDP‐α‐glucose was determined to be 370 μM (Fig. 2B), in agreement to the 110 μM K m value determined for the SeRmlD homologue (Blankenfeldt et al., 2002). Using surface plasmon resonance, we investigated the binding affinity of NADPH and NADH to GacA, which were previously shown to be functional cofactors for RmlD enzymes (Blankenfeldt et al., 2002). Both substrates bind to GacA with binding affinities of K D = 390 ± 2 μM (NADPH) and K D = 110 μM ± 1 μM (NADH) (Fig. 2C).
GacA is active as a monomer
Many SDR family members, including SeRmlD, require dimerization or oligomerization to be functional (Blankenfeldt et al., 2002; Kavanagh et al., 2008). Results from our functional enzymatic assay demonstrated that GacA did not require metal for its activity, suggesting that it might be functional as a monomer. GacA was analyzed by size‐exclusion chromatography (SEC) and SEC‐MALLS and a molecular mass of the elution peak was calculated. The calculated mass of GacA is 27.5 kDa (± 2.4 kDa) (Fig. 2D). This is in good agreement with the theoretical calculated monomeric mass of 32 kDa of the GacA polypeptide sequence and represents the first monomeric RmlD enzyme. We subsequently investigated the effect of Mg2+‐ions on protein size. The purified protein was incubated with and without 10 mM MgCl2 overnight at 4°C and analyzed via SEC in the corresponding buffers (Fig. 2D). Both protein samples show identical elution volumes at 15.8 ml, with an average calculated molecular mass of 27.5 kDa, indicating that no mass change/dimerization occurs in presence of Mg2+ (Fig. 2D).
M. tuberculosis RmlD inhibitors inhibit recombinant GacA
Wang et al. (2011) have identified a series of MtbRmlD inhibitors by virtual screening using the crystal structure of SeRmlD. The sequence identity between MtbRmlD and SeRmlD is 31%, whereas the sequence identity between GacA and MtbRmlD is 36% (Fig. 1). We assessed the inhibitory effect of two of the four previously described inhibitors using enzyme kinetics. Our experiments show that both compounds inhibit the activity of recombinant GacA with an IC50 of approx. 2 μM for compound 3 and approx. 10 μM for compound 2. These data are in good agreement to the MtbRmlD inhibition with corresponding IC50 values of 0.9 μM and 15 μM, respectively (Wang et al., 2011).
GacA crystal structure reveals novel monomeric RmlD form with a conserved catalytic triad
To unravel why GacA functions as a monomer, we employed X‐ray crystallography. Crystals diffracted routinely below 1.2 Å and were subjected to synchrotron data collection. Results were compared with the four GacA (RmlD) homologues that have been deposited in the Protein Data Bank (PDB) (Bernstein et al., 1977) (Supp. Fig. S1): The Gram‐negative SeRmlD (pdb entry 1kbz, 1n2s, 1kc1, 1kc3) (Blankenfeldt et al., 2002), Clostridium acetobutylicum RmlD (CaRmlD; pdb entry 1vl0), the Gram‐positive Bacillus anthracis RmlD (BaRmlD; pdb entry 3sc6) and the archaea Sulfolobus tokodaii RmlD (StRmlD; pdb entry 2ggs).
Consistent with our SEC and activity studies, GacA crystallized as a monomer. The overall structure of the GacA monomer aligns well with all four RmlD structures in the database, with Cα rmsd of 1.4 Å (apo, SeRmlD, 1kbz, Fig. 3A), 1.1 Å (NADPH complex, BaRmlD, 3sc6) and 1.0 Å (NADH complex, CaRmlD, 1vl0) and 1.4 Å (NADPH complex, StRmlD, 2ggs, Supp. Fig. S1). GacA contains the typical α‐helical/β‐sheet arrangement known as Rossmann fold present in SDR domains (Fig. 3A). In agreement with the well‐characterized SeRmlD (Blankenfeldt et al., 2002), the GacA β‐sheet is formed of six β‐strands, in the order 213457 (Fig. 3A). This β‐sheet is flanked by three and four α‐helices on either side, respectively. The previously described prominent kink in α‐helix 4, a characteristic structural feature in SDR enzymes, is also present in GacA and is caused by the conserved Asn78 (Asn81 in SeRmlD) (Figs. 1 and 3B). The second 310‐helix η2 observed in SeRmlD (Blankenfeldt et al., 2002) is missing in GacA due to a shorter loop connecting α‐helix α7 and β‐strand β7 (Figs. 1 and 3A). The α‐helices α1‐5, α7 and α9 form the cofactor binding site, whereas α‐helices α6, α8 and α10 form the substrate‐binding domain (Fig. 3A and B). In comparison with SeRmlD, α8 from GacA lacks two α‐helical turns, due to a deletion of eight amino acids in GacA (Figs. 1 and 3A). Furthermore, we calculated the hydrodynamic radius (Stokes radius) of the soluble GacA and the crystallized monomer. The soluble GacA has a Rh = 26 Å (± 0.3%), in good agreement with the Rh calculated from the obtained crystal structure using HYDROPRO (Rh = 26 Å) (Ortega et al., 2011). No symmetry‐related GacA molecule forms protein–protein interactions via α1 and α10 as observed in SeRmlD (Fig. 3A) (Blankenfeldt et al., 2002). The GacA crystal structure confirms that GacA defines a new class of monomeric RmlD enzymes that do not require metal for enzymatic activity (Fig. 3A).
Figure 3.
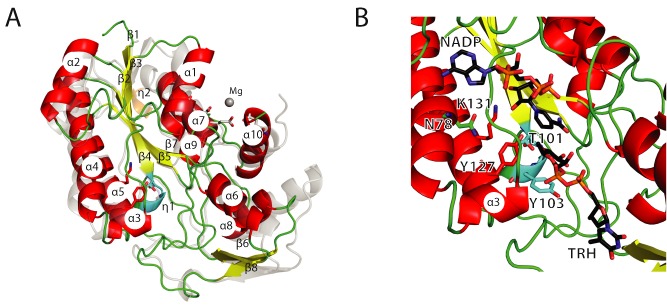
Structural insights into a monomeric Gram‐positive RmlD enzyme.
A. Comparison of the GacA secondary structure elements with SeRmlD [PDB entry 1KBZ (Blankenfeldt et al., 2002)], a dTDP‐4‐dehydrorhamnose reductase. The structures are shown in a cartoon representation. GacA is colored with red helices and yellow strands; SeRmlD is colored in transparent gray. Secondary structure elements are labeled according to Fig. 1. SeRmlD Mg2+‐binding site is shown with the three glutamic acids (gray sticks) co‐ordinating the Mg2+‐ion (gray sphere).
B. Active site view with cartoon and stick representation of GacA in complex with superimposed ligands NADPH and dTDP‐L‐rhamnose (TRH) from the ternary SeRmlD complex [PDB entry 1KC3 (Blankenfeldt et al., 2002)]. The conserved catalytic residues T101, Y127 and K131 (GacA) and N78 are shown as sticks, color‐coded according to Fig. 1.
We superimposed the ternary SeRmlD complex (Blankenfeldt et al., 2002) onto the GacA crystal structure to investigate active site sequence conservation (Figs. 1 and 3B). The GacA crystal structure is in the ‘open’ conformation, allowing binding of the cofactor NADPH and the acceptor substrate dTDP‐4‐dehydrorhamnose. All active site residues are conserved with the SeRmlD (Fig. 1, green triangles). The catalytic triad identified for SeRmlD (Blankenfeldt et al., 2002), consisting of T104, Y128 and K132, occupies identical conformations in GacA (T101, Y127 and K131, Figs. 1 and 3B), suggesting that GacA is a functional monomeric RmlD homologue using a conserved catalytic mechanism.
RmlD enzymes from Gram‐positive bacteria are monomers due to lack of a conserved RmlD dimerization motif
To investigate the molecular basis for the novel monomeric RmlD form, we focused on the amino acids at the SeRmlD dimerization interface. From the published SeRmlD structure (Blankenfeldt et al., 2002), there appear to be two critical parameters for SeRmlD dimerization: (i) the three glutamate residues E15, E190 and E292 at the SeRmlD dimerization interface that help co‐ordinate the Mg2+‐ion (Figs. 1 and 4A) and (ii) a shortened α1‐helix caused by a proline residue (P22, SeRmlD) that allows binding of the second SeRmlD monomer via tight protein–protein interactions (Fig. 4A). To experimentally confirm the contribution of these two parameters to SeRmlD dimerization, we cloned, expressed and purified the wild‐type RmlD enzyme from S. enterica and designed a triple‐mutant (3M), in which the putative key residues for SeRmlD dimerization (PVG, E, E) are replaced with the corresponding residues in the monomeric GacA (ERGV, R, Q). SEC analysis revealed that the SeRmlD triple mutant is a monomeric protein, identical to GacA (Supp. Fig. S2A), whereas wild‐type SeRmlD runs at a shorter retention time, in agreement to its higher dimeric mass (Blankenfeldt et al., 2002). Furthermore, EDTA treatment of wild‐type SeRmlD does not affect the retention time (Supp. Fig. S2A), suggesting that the removal of the Mg2+‐ion does not disrupt dimerization. The peak fractions were analyzed using fingerprint mass‐spectrometry and contained the correct protein.
Figure 4.
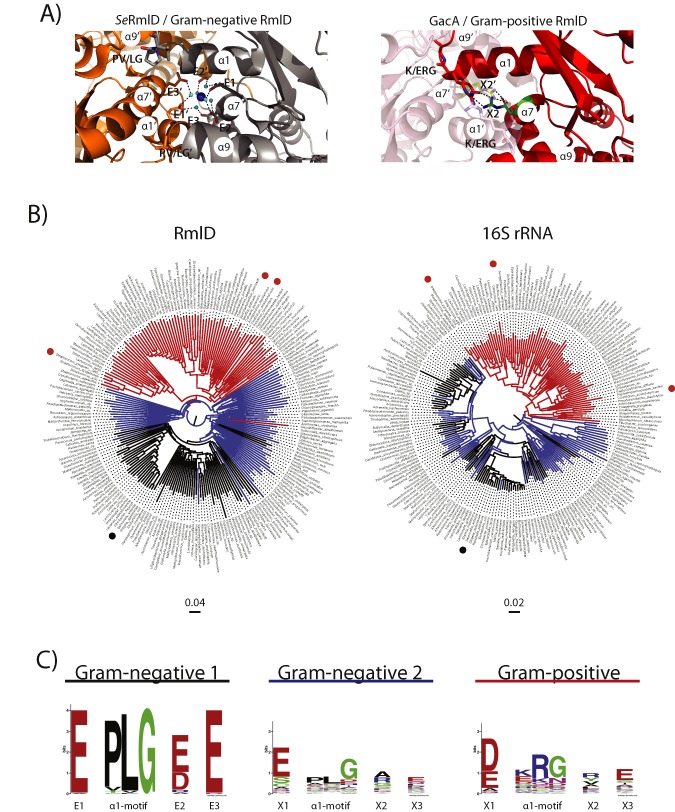
GacA represents a new class of monomeric RmlD enzymes.
A. Structural insight in dimerization interface of RmlD reveals steric hindrances that would prevent homo‐dimerization in Gram‐positive RmlD enzymes. Left: View of the SeRmlD homo‐dimer Mg2+‐binding site, representing Gram‐negative RmlD enzymes (Blankenfeldt et al., 2002). The E1‐E/D2‐E3 motif co‐ordinates the Mg2+‐ion. The α1‐helix contains the PLG motif, which allows dimerization, as the P introduces a turn into the α1‐helix. Right: Hypothetical model of a GacA homo‐dimer based on the SeRmlD homo‐dimer (red and transparent cartoon representation). The K/ERG motif prevents dimerization, as it extends the α1‐helix by half a turn, which would clash with the α7′‐helix of the second molecule. The X2 side‐chain (R189) points into the Mg2+‐binding pocket and prevents dimerization (salmon and red arginine side‐chains). Large, basic or hydrophobic residues, mainly R or Y, replace the E2 motif in Gram‐positive RmlD homologues. A salt bridge between GacA E21 from the ERG motif and X2 (R189) stabilizes the tertiary structure of the α1‐ and α7‐helices, which is located at the corresponding position of the Gram‐negative Mg2+‐binding site (green and yellow side‐chains).
B. Neighbor‐Joining trees of 213 RmlD orthologous sequences (left) and their corresponding 16S rRNA sequences (right). Gram‐positive bacteria are colored in red, Gram‐negative that contain the conserved dimerization sequence logo E1‐PLG‐E/D2‐E3 are colored in black (‘Gram‐negative 1’) and Gram‐negative bacteria that lack one or both of the dimerization criteria are colored in blue (‘Gram‐negative 2’). S treptococcus pyogenes (GAS), C lostridium acetolyticum and B acillus anthracis are marked with red dots, S almonella enterica with a black dot.
C. Left: Sequence logo of dimerization interface for Gram‐negative RmlD enzymes. 135 (putative) orthologous RmlD sequences from Gram‐negative bacteria (Supp. Table S1) were analyzed for their E1, E2, E3 and α1‐helix motifs, which is critical for Mg2+‐binding and dimerization as described by Blankenfeldt et al. (Blankenfeldt et al., 2002). Seventy RmlD homologues contain the ‘Gram‐negative 1’ motif E‐PLG‐E/D‐E (Supp. Table S1). Sixty‐five RmlD homologues are lacking these conserved residues (‘Gram‐negative 2’) and are therefore expected to function as monomers (Supp. Table S2). Right: Seventy‐eight RmlD sequences from (putative) Gram‐positive bacteria (Supp. Table S3) were analyzed for the same motifs. The Gram‐positive RmlD homologues lack a distinctive motif and therefore lack the ability to co‐ordinate a divalent metal ion. The ‘Gram‐negative 1’ α1‐helix PLG‐motif is replaced in Gram‐positive RmlD sequences by a K/ERG motif.
GacA lacks both of the confirmed critical dimerization parameters as it does not contain all conserved negatively charged E residues and has an extended α1‐helix due to the ERG motif that occupies the putative Mg2+‐binding pocket (Fig. 4A). Interestingly, the GacA structure reveals that a salt bridge is introduced (E21 and R189), stabilizing the tertiary structure of the α1‐ and α7‐helices, which could contribute to the fact that GacA functions as a monomer (Fig. 4A). We have modeled an artificial GacA dimer (Fig. 4A), which further reveals structural features that support why GacA functions as a monomer. The large, basic R22 at the end of the α1‐helix points with its side‐chain into the modeled second monomer. Furthermore, the extended α1‐helix would clash with the α7‐helix of the second GacA molecule (Fig. 4A). These features are incompatible with homo‐dimerization, but most likely stabilize the monomeric enzyme.
We analyzed whether the lack of structural dimerization features is unique to GacA or are present in RmlD enzymes from other species. Using two independent psi‐BLAST runs, one starting with SeRmlD as a representative of dimeric RmlD enzymes and the other starting with GacA as a representative of monomeric RmlD enzymes, we identified 213 bacterial (putative) RmlD homologues, including 78 from Gram‐positive and 135 from Gram‐negative species (Suppl. Tables S1–S3). All full‐length RmlD protein sequences as well as the corresponding 16S rRNA DNA sequences of the same 213 species were aligned and used to build bootstrapped Neighbor‐Joining trees (Fig. 4B). Interestingly, RmlD sequences from all Gram‐positive bacteria, except Desulfitibacter alkalitolerans, clustered separately from the RmlD sequences of Gram‐negative bacteria. We aimed to determine whether the observed phylogenetic split was associated with sequence differences at the dimerization interface. Therefore, we aligned the 135 Gram‐negative and 78 Gram‐positive RmlD sequences corresponding to the critical negatively charged amino acids and the PLG sequence identified at the SeRmlD dimerization interface (Supp. Tables S1–S3). Interestingly, the RmlD sequences from Gram‐negative species could be divided in two groups. The group named ‘Gram‐negative 1’ contained the fully conserved dimerization site represented as an E1‐PLG‐E/D2‐E3 motif (Fig. 4C ‘Gram‐negative 1’) and similar to the previously described metal‐dependent dimeric SeRmlD (Supp. Table S1). This strongly suggests that RmlD proteins from this subset of Gram‐negative species form dimers. The remaining ∼ 45% of Gram‐negative RmlD enzymes lacked one or both of the critical dimerization parameters (‘Gram‐negative 2’; Fig. 4C, Supp. Table S2) and likely function as monomers similar to GacA. Superimposing the presence/absence of the dimerization motif on the 16S rRNA tree suggests that Gram‐negative bacteria have gained or lost the RmlD dimerization motif multiple times during evolution (Fig. 4B). Strikingly, RmlD sequences from all Gram‐positive species also lacked a conserved ‘Gram‐negative 1’ dimerization motif (Fig. 4C ‘Gram‐negative 1’ and ‘Gram‐positive’). For example, most Gram‐positive RmlD enzymes replaced the negatively charged glutamate at E2 with a large positively charged arginine or aromatic tyrosine substitute (X2 in Fig. 4C) and the position at E3 is also substituted in Gram‐positive bacteria compared with ‘Group 1’ Gram‐negative species (X3 in Fig. 4C). In addition, RmlD in Gram‐positive bacteria have an extended α1‐helix by half a turn due to lack of the PLG motif. This is illustrated in the artificial GacA dimer that we modeled based on the SeRmlD homo‐dimer (Fig. 4A) as well as in the deposited RmlD crystal structures from the Gram‐positive species C. acetobutylicum CaRmlD (1VL0.pdb, Suppl. Fig. S1) and B. anthracis BaRmlD (3SC6.pdb, Suppl. Fig. S1). These enzymes have not been biochemically characterized; however, both proteins did not crystallize as dimers. Furthermore, these two enzymes are similar to the GacA structure and primary sequence (Fig. 1) containing large residues at the end of the α1‐helix and missing the three conserved negatively charged E1‐E/D2‐E3 residues to accommodate a metal ion, hindering the SeRmlD typical dimerization (Supp. Fig. S1).
Overall, we confirmed that RmlD dimerization requires specific structural features that are absent in GacA. Additionally, comprehensive sequence analysis suggests that GacA is representative of a new class of monomeric RmlD enzymes that is present in all Gram‐positive species and a subset of Gram‐negative species.
GacA is an essential gene for GAS during growth in rich medium
To confirm the function of GacA in GAS, we attempted to generate mutants in GAS by plasmid insertion. However, we were unable to obtain mutants on multiple attempts, suggesting that gacA is essential for GAS (van Sorge et al., 2014). Additional proof for potential essentiality of gacA was investigated as part of a larger screen for essential genes in GAS using the mariner transposon Krmit (Le Breton et al., 2015). Saturated mutant libraries were produced and analyzed by Tn‐seq to identify the insertion sites within each mutant pool that survive growth in THY rich medium at 37°C. Chromosomal position and abundance of Tn‐seq reads were mapped to the gacA genome sequence and a Bayesian statistical analysis was performed to identify regions with limited Krmit insertions compared with surrounding sequences indicative of gene essentiality (Tables 1 and 2). For known essential genes dnaG and rpoD < 100 insertions per kB are observed, whereas 10‐ to 50‐fold more insertions are observed for non‐essential control genes M5005_Spy_0601 and emm1 (Fig. 5A–F, Tables 1 and 2). Insertions for gacA demonstrate that gacA is indeed essential in the GAS strains 5448 (M1T1) and NZ131 (M49) when growing in rich media. These data are in agreement with a previous study conducted on Mycobacterium smegmatis (Ma et al., 2002), where rmlD was shown to be essential for mycobacterial growth. To validate gacA essentiality in an independent manner, we employed a previously published conditionally lethal approach that takes advantage of a theophylline‐sensitive synthetic riboswitch functional in GAS (Le Breton et al., 2015). In the presence of theophylline, which results in expression of gacA, the GAS gacAi strain was normally viable, whereas lack of theophylline significantly compromised growth of the bacteria (Fig. 5B). Visual inspection of the inducible gacA mutant bacteria (without theophylline) using scanning electron microscopy (SEM) indeed shows aberrant cell morphology, which defects in cell separation resulting in long chains and aberrant septum placement resulting in irregularly shaped cocci (Fig. 5C). These data underpin the critical role of rhamnose production in GAS physiology.
Table 1.
Bayesian analysis of Tn‐seq data from GAS 5448 and NZ131
| Genea | GAS 5448 | GAS NZ131 | ||||
|---|---|---|---|---|---|---|
| Locusb | Zbarc | Scored | Locuse | Zbarc | Scored | |
| Spy0601 | 0 | NE | Spy0610 | 0 | NE | |
| gacA | Spy0602 | 1 | E | Spy0611 | 1 | E |
| dnaG | Spy0599 | 1 | E | Spy0608 | 1 | E |
| rpoD | Spy0600 | 1 | E | Spy0609 | 1 | E |
| emm | Spy1719 | 0 | NE | Spy1671 | 0.57 | NE |
When available, gene name is provided.
Spy numbers from the MGAS5005 genome.
Essentiality values as determined by the Bayesian analysis. Genes with a Z value over 0.992 is classified as essential, genes with Z value under 0.03 are non‐essential.
Essentiality score. E, essential; NE, non‐essential.
Spy numbers for the M49 NZ131 GAS genome sequence.
Table 2.
Insertion analysis of Tn‐seq data from GAS 5448
| Locus | Gene | Total number of insertions per gene | Gene size (kb) | Total insertions per gene per kb |
|---|---|---|---|---|
| Spy0599 | dnaG | 86 | 1.812 | 47.46 |
| Spy0600 | rpoD | 86 | 1.107 | 77.69 |
| Spy0601 | 409 | 0.336 | 1217.26 | |
| Spy0602 | gacA | 85 | 0.852 | 99.77 |
| Spy1719 | emm1 | 7877 | 1.452 | 5424.93 |
| Average | 5297 | 0.851 | 6224.44 |
Figure 5.
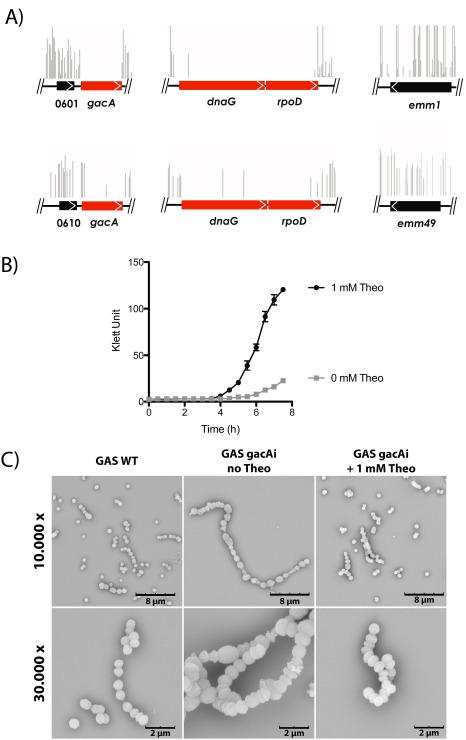
GacA is essential for GAS growth in vitro.
A. Essentiality of gacA in GAS 5448 and NZ131 as determined by Tn‐seq. Complex Krmit transposon libraries were generated in the GAS strains 5448 and NZ131 and analyzed during growth in rich medium (THY) at 37°C. Tn‐seq analyses mapped the location and relative abundance of K rmit insertions (vertical lines) to the relevant GAS genome sequences. Using a Bayesian statistical analysis, gac A was determined to be essential (red arrows) based on limited transposon insertions (vertical lines) similar to the known essential genes dna G and rpo D. Insertions for non‐essential genes Spy_0601/0610 and emm from each strain are shown for comparison.
B. Conditional interference with GacA expression results in severe growth attenuation of GAS. Growth parameters as followed by Klett measurements of the GAS 5448 gacAi mutant in THY rich medium in the presence (1 mM, black circles, GacA expression) or absence (gray squares, no GacA expression) of theophylline.
C. Representative SEM images of GAS wild‐type strain and the generated riboswitch strain gacAi in absence and presence of 1 mM of theophylline (Theo).
GAS GacA can functionally replace S. mutans RmlD
To confirm the role of GacA in dTDP‐L‐rhamnose production in live bacteria, we made use of heterologous expression. It was previously shown that classical targeted disruption of rmlD in S. mutans (SmRmlD) is feasible (Tsukioka et al., 1997; Nakano and Ooshima, 2009). Rhamnose is incorporated in the serotyping cell wall‐anchored carbohydrate composed of rhamnose decorated with a glucose side‐chain (Nakano and Ooshima, 2009). Consequently, disruption of rmlD results in complete loss of rhamnose and glucose from the cell wall but also significantly attenuates growth (Tsukioka et al., 1997; Nakano and Ooshima, 2009). SmRmlD and GacA are 82% identical (234 out of 284 residues), suggesting that they catalyze the same enzymatic reaction. We constructed a S. mutans rmlD mutant strain (SMUΔrmlD) by replacing rmlD in frame with an erythromycin resistance cassette. Similar to disruption of gacA in GAS, loss of RmlD significantly affected cell morphology (Fig. 6A), bacterial growth (Fig. 6B) and resulted in complete loss of rhamnose (Fig. 6C) as previously published (Tsukioka et al., 1997). We next complemented SMUΔrmlD with GAS gacA on a complementation plasmid (SMUΔrmlD + pGacA). Expression of GacA restored S. mutans growth (Fig. 6B), rhamnose production (Fig. 6C) and almost completely restored cell appearance (Fig. 6A). These results demonstrate that gacA can functionally replace rmlD in S. mutans, supporting the role of GacA as a dTDP‐4‐dehydrorhamnose reductase.
Figure 6.
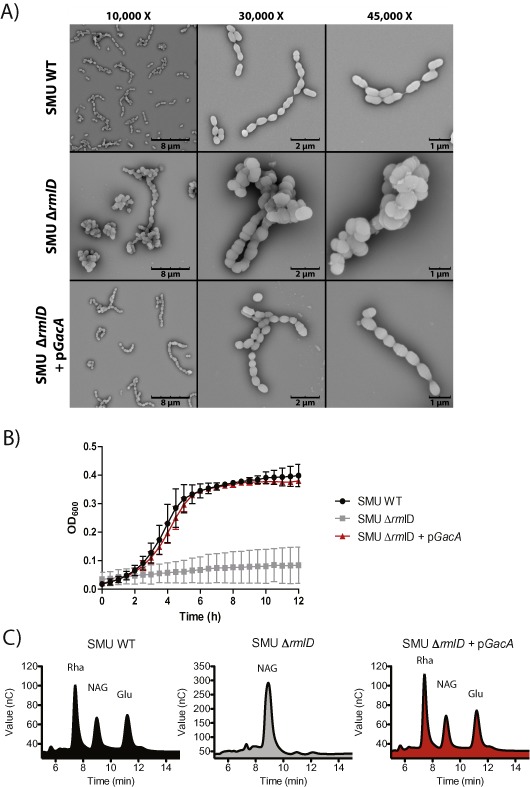
GacA functionally replaces S. mutans RmlD in vivo.
A. Representative SEM images of S. mutans Xc (SMU) wild‐type (WT), SMUΔrmlD and SMUΔrmlD + pGacA. Cells of the wild‐type strain appear as short chains with division in a single plane, whereas the Δrml D mutant forms clumps with long chains of swollen cocci and aberrant multi‐directional cell division. This aberrant morphology is almost completely restored by introduction of GAS gac A on an expression plasmid.
B. Representative growth curves (mean ± SD, n = 3) of SMU WT (black circles), SMU ΔrmlD (grey squares) and SMU ΔrmlD + pGacA (red triangles) complemented strains cultured at 37°C without 5% CO2 for 12 h. (C) Analysis of monosaccharide composition of SMU WT (black), SMU ΔrmlD (grey) and SMU ΔrmlD + pGacA (red) by chromatography. Rha, rhamnose; NAG, GlcNAc; Glu, glucose.
Conclusions
We have demonstrated through genetics, biochemistry, functional and structural analysis that GacA is the dTDP‐4‐dehydrorhamnose reductase RmlD homologue in GAS and is critical for normal growth. GacA represents the first structurally and biochemically characterized Gram‐positive RmlD homologue. In contrast to the published RmlD structure from Gram‐negative S. enterica (Blankenfeldt et al., 2002), GacA is representative of a new class of RmlD enzymes that are functional as a monomer. Bioinformatics analysis of 213 RmlD sequences complemented with experimental evidence from wild‐type and mutated SeRmlD, we have identified two sequence features that characterize the (putative) metal‐dependent dimeric RmlDs that are not conserved in monomeric RmlDs. Interestingly, dimeric RmlD enzymes cluster within Gram‐negative species, whereas monomeric RmlD enzymes are predominantly present in Gram‐positive bacteria. SeRmlD was described as a metal‐dependent enzyme as addition of EDTA reduced activity by about 70% (Graninger et al., 1999; Blankenfeldt et al., 2002). However, it remained unknown whether absence of Mg2+ results in loss of dimerization. It was speculated that the presence of Mg2+ stabilizes cofactor binding (Blankenfeldt et al., 2002). Our data show that the EDTA treated SeRmlD wild‐type enzyme remains dimeric (Supp. Fig. S2A), suggesting that the removal of Mg2+‐ion disturbs the proper tertiary structure of each monomer and therefore inactivates the enzyme.
From targeted mutagenesis attempts and Tn‐Seq studies, we conclude that gacA is essential for growth of GAS in rich medium, in agreement to a previous study that suggested that gacA might be essential (van Sorge et al., 2014). We validated that gacA is essential by a riboswitch‐inducible expression system resulting in attenuated growth and severe cellular abnormalities in the absence of gacA. Additional experiments in S. mutans confirm the critical role of GacA in rhamnose biosynthesis as gacA could functionally replace rmlD in S. mutans resulting in restored rhamnose in cell wall, growth and morphology.
In this study, we have identified GacA as an attractive drug target for the development of novel antimicrobial compounds against GAS. More importantly, these inhibitors could serve as lead compounds to inhibit L‐rhamnose biosynthesis in other bacteria. The dTDP‐L‐rhamnose biosynthesis is an interesting target for the development of new drugs since (i) the pathway affects either the viability or virulence of many bacteria, including Mycobacterium spp. (Ma et al., 2002), Pseudomonas spp. (Engels et al., 1985) and E. faecalis (Teng et al., 2009) and (ii) the pathway does not exist in humans, reducing the risk of side‐effects by off‐target effects. Known MtbRmlD inhibitors inhibit recombinant GAS GacA in the mid‐ to low micromolar range in kinetic assays. However, we were unable to demonstrate a reliable effect of these compounds on GAS growth as the compounds are highly water insoluble. This implies that these compounds are not suitable as a starting point for structure‐based drug design on the basis of their chemical properties. The genetic, biochemical and structural data presented here on GAS GacA forms the framework for future screenings to identify novel inhibitors that target GAC biosynthesis in GAS and dTDP‐L‐rhamnose biosynthesis through related RmlD enzymes in other human pathogens.
Experimental procedures
GacA cloning, expression and purification
Full‐length gacA (accession number AAZ51220.1; M5005_Spy_0602) was PCR amplified from GAS genomic DNA (M1T1 5448) and cloned by restriction‐free cloning into a modified pGEX vector, with an N‐terminal hexa‐histidine‐GST‐tag followed by a PreScission Protease cleavage site (His6GST‐GacA). His6GST‐GacA was transformed in E. coli BL21(DE3) cells and recombinant protein expression was induced with 0.5 mM isopropyl‐1‐thio‐β‐D‐galactopyranoside for 18 h. Cells were harvested by centrifugation and resuspended in buffer A (50 mM Tris‐base, pH 8.0, 250 mM NaCl), supplemented with 10% glycerol, 0.5 complete Protease Inhibitor Cocktail Tablets (Roche) and 2 mM Tris (2‐Carboxyethyl)‐phosphine hydrochloride (TCEP). All purification steps were carried out at 4°C. Cells were disrupted, and the supernatant from two subsequent centrifugation runs at 15 000× g for 20 min and 100 000× g for 1.5 h was adjusted to 10 mM imidazole. The sample was passed over a 5 ml His‐trap column HP charged with Co2+, washed with 10 column volumes (CV) of 25 mM imidazole in buffer A, and the protein was eluted with a gradient over 10 CV of 500 mM imidazole in buffer A. The eluted protein was concentrated using a 50 k Da MW cutoff concentrator and passed over a desalting column, equilibrated in buffer A. The His6GST‐tag was cleaved using PreScission Protease overnight, passed over a 5 ml His‐trap column equilibrated in buffer A supplemented with 20 mM imidazole, the flow through was collected and concentrated using a 10 k Da MW cutoff concentrator and injected into a Superdex 75 26/60 column equilibrated in TBS‐buffer, supplemented with 0.2 mM TCEP. The fractions containing GacA were collected and concentrated to 25 mg ml−1. The purified protein was confirmed by tryptic fingerprint mass spec (University of St. Andrews).
Enzymatic activity of GacA
To analyze GacA enzyme kinetics, we cloned and expressed GAS homologues of rmlB (SpRmlB; accession number AAZ51354.1; M5005_Spy_0736) and rmlC (SpRmlC; accession number AAZ51353.1; M5005_Spy_0735) using a modified pET vector, harboring an N‐terminal octa‐histidine tag. The RmlB and RmlC‐fusion proteins were expressed and purified as described for GacA. The assay was performed following the protocol from Sivendran et al. (2010) with the following changes: the assay buffer system contained 25 mM Tris‐base (pH 7.5), 150 mM NaCl, 0.1 mM NADPH and 2 pM GacA. SpRmlB and SpRmlC were added to the assay in 25‐fold molar excess relative to GacA. The assay was started with the addition of 400 μM dTDP‐D‐glucose. Michaelis‐Menten kinetics for GacA for dTDP‐glucose was calculated using a concentration range from 0.025 to 1 mM of dTDP‐glucose. The oxidation of NADPH to NADP+ was measured by the change in intrinsic absorbance at 340 nm using a SpectraMax M2 plate reader. Background absorbance was subtracted and data interpreted using the Michaelis‐Menten model in GraphPad Prism (GraphPad Software, Inc., 7825 Fay Avenue, Suite 230, La Jolla, CA 92037 USA). RmlD inhibitors 2 and 3 (Sigma) described for M. tuberculosis (Wang et al., 2011) were dissolved to 10 mM stock concentration in 100% DMSO, diluted in enzyme assay buffer and added at the same time‐point as the GacA enzyme. All reactions contained a final concentration of 2% DMSO.
SeRmlD cloning, expression and purification
Full‐length rmlD (GI 16420628) was PCR amplified from S. enterica genomic DNA (LT2) using the procedure described for gacA. Point mutations were inserted using standard mutagenesis procedures. Wild type and mutant enzymes were expressed and purified as described for GacA. EDTA treatment was performed by incubation of SeRmlD (0.1 mg ml−1) with 10 mM EDTA for 30 min at room temperature, followed by a concentration‐step and size‐exclusion chromatography in buffer A containing 10 mM EDTA.
Surface plasmon resonance (SPR) experiments
Recombinant, purified GacA was chemically biotinylated and captured on a streptavidin surface of a Biacore T200 instrument (GE‐Healthcare) at densities of 3 k–4 k RU. To stabilize captured protein over time, all experiments were run at 10°C. Ligands were injected over captured protein at flow rate 30 μl min−1 in running buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.05%Tween, 3% DMSO), with each compound injected in duplicates in concentration series 4–1000 μM. Association was measured for 60 s and dissociation for 120 s. All data were double referenced for blank injections of buffer and biotin‐blocked Streptavidin surface. Scrubber 2 (BioLogic Software) was used to process and analyze the data.
Crystallization and structure determination
Vapor diffusion sitting‐drop crystallization was carried out at 20°C. Crystals appeared after mixing equal amounts of protein and crystallization buffer containing 12.5% w/v PEG 1000, 12.5% w/v PEG 3350, 12.5% v/v MPD, 0.02 M of each carboxylic acid (0.2 M sodium formate, 0.2 M ammonium acetate, 0.2 M trisodium citrate, 0.2 M sodium potassium L‐tartrate, 0.2 M sodium oxamate) and 0.1 M MES/imidazole pH 6.5 (Gorrec, 2009) after 1 day. Crystals were flash frozen into liquid nitrogen prior to data collection. Data were collected at beamline ID23‐1 at ERSF and processed with iMOSFLM (Battye et al., 2011). Data collection statistics are summarized in Table 3. On the basis of the high‐resolution dataset, the structure was solved ab initio using the program ‘Acorn’ in the CCP4 suite (McCoy et al., 2007). The initial model was used for autobuilding in Phenix (Adams et al., 2010). With the exception of the first and last residue, 288 residues out of 290 residues were built, and the structural model was refined in iterative cycles using Coot (LMB Cambridge, UK) (Emsley et al., 2010) and Refmac (Murshudov et al., 1997) to the statistics shown in Table 3. The final model was refined to 1.1 Å resolution with an Rfactor of 12.9% and Rfree of 15.5%. The Ramachandran plot revealed that 98% of all residues are in favored regions, with no outliers as calculated by MolProbity (Lovell et al., 2003). The co‐ordinates and structure factors have been deposited with the RCSB Protein Data Bank with PDB ID code 4WPG.
Table 3.
Crystallographic data collection and refinement statistics
| Spacegroup | P1 |
|---|---|
| Unit cell dimension (Å) | 36.48 45.98 48.87 |
| Resolution range (last shell) (Å) | 43.8–1.1 (1.139–1.1) |
| Unit cell angles (°) | 66.3 81.21 95.27 |
| Unique reflections | 108172 (9823) |
| Completeness (%) | 94.0 (85.5) |
| I/σI | 22.5 (5.7) |
| Wilson B‐factor | 12.20 |
| Rfactor | 0.129 (0.175) |
| Rfree | 0.155 (0.205) |
| Number of atoms | 5062 |
| Macromolecules | 2314 |
| Ligands | 21 |
| Water | 468 |
| Protein residues | 287 |
| Rms bond length (Å) | 0.023 |
| Rms bond angles (°) | 2.08 |
| Ramachandran favored (%) | 98 |
| Ramachandran outliers (%) | 0 |
| Average B‐factor | 18.10 |
| Macromolecules | 16.10 |
| Solvent | 27.90 |
I/σI, intensity divided by standard deviation of intensity, averaged over all measurement.
Rms is root‐mean‐square deviation from ideal value.
Neighbor‐Joining tree construction
To construct midpoint‐rooted Neighbor‐Joining trees of RmlD sequences, we used two‐independent psi‐BLAST runs using SeRmlD and GacA as the query sequences. We obtained 135 Gram‐negative RmlD homologues of SeRmlD using an E‐value cutoff of > 7e−58, and we obtained 78 Gram‐positive homologues of GAS GacA using a cutoff of > 1e−48. All hypothetical sequences and all RmlD sequences for which no corresponding 16S rRNA sequences were available were rejected. We used only one representative for each bacterium, avoiding the use of multiple strain variants. Multiple sequence alignments of RmlD protein sequences and 16S rRNA sequences were constructed using ClustalW2 (McWilliam et al., 2013). Neighbor‐Joining trees were built from the RmlD and 16S rRNA alignments using ClustalX (2000 bootstraps were run) (Larkin et al., 2007). Neighbor‐Joining trees were displayed using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). All bacteria analyzed were assigned to the corresponding monomer/dimerization motif groups and were marked with related colors.
Tn‐seq to identify genes essential for GAS growth in THY
The pKRMIT plasmid contains a mariner mini‐transposon named Krmit (Kanamycin‐resistant element for massive identification of transposants) modified for Tn‐seq (Le Breton et al., 2015) and was used to perform saturating transposition for random mutagenesis in GAS 5448 and NZ131 as previously described for the mariner transposon Oskar (Le Breton and McIver, 2013; Le Breton et al., 2013). Tn‐seq (van Opijnen and Camilli, 2010) was performed as described recently (Le Breton et al., 2015) with the following primers oKrmit‐Tnseq2 (5′‐CAAGCAGAAGACGGCATACGAAGCGCCTACG‐AGGAATTTGTATCG‐3′) and oAdapterPCR (5′‐ACACTCTTTCCCTACACGACGCTCTT‐CCGATCT‐3′), resulting in the production of 176 bp Krmit insertion tags. Quality and yield of the resulting tags was assessed using a NanoDrop spectrophotometer (Thermo Scientific) and an Agilent Bioanalyzer. Krmit insertion tags were analyzed by Illumina sequencing (50 nt single end reads) on a HiSeq 1500 platform in the Institute for Bioscience and Biotechnology Research (IBBR) Sequencing Facility located at the University of Maryland, College Park. The quality of read datasets (Sanger FastQ format) was determined using FastQC (Ramirez‐Gonzalez et al., 2013); data were filtered and trimmed using Biopieces (biopieces.org) to select for reads containing the Tn‐seq barcodes and Krmit ITR ends. Reads were then de‐multiplexed and count tables generated using SamTools (Li et al., 2009) and HTseq (Chandramohan et al., 2013). Reads were mapped to the GAS 5448 or NZ131 genome using Bowtie (Langmead et al., 2009) and data relevant to the gacA‐L locus visualized using the Integrative Genomics Viewer (IGV) browser (broadinstitute.org/igv/home). Gene essentiality was determined using a Bayesian statistical model based on the Metropolis‐Hastings algorithm using the Python script (saclab.tamu.edu/essentiality) developed by DeJesus et al. (2013).
Construction of a conditionally lethal gacA mutant
A GAS 5448 merodiploid mutant, gacAi, was constructed using the pSinS/pHplK system for gacA gene expression to be under the control of a theophylline‐dependent riboswitch (Topp et al., 2010) as recently described (Le Breton et al., 2015). Succinctly, a ca. 600‐nu fragment of the 5′‐end of the gacA gene was amplified using primers oGacA‐1 (5′‐CCCTGCTAAGGAGGTAACAACAAGATGATTTTAATTACAGGAAGCAATGG‐3′) and oGacA‐2 (5′‐cccGGATCCGTCAAATAACACATGAATTCTGC‐3′) and subsequently fused by SOE‐PCR to the Psag promoter along with the synthetic riboswitch E as previously described (Le Breton et al., 2015). The resulting PCR product was then cloned into the BamHI site of the pSinS plasmid producing pGacAi and mutation carried out as described (Le Breton et al., 2015), creating the GAS 5448 gacAi mutant; and the junction between the Psag promoter, the synthetic riboswitch E and the gacA gene in the GAS 5448 gacAi mutant was verified by DNA sequencing (data not shown).
Bacterial strains and growth conditions
Streptococcus mutans Xc is a serotype C wild‐type strain (Koga et al., 1989) and was kindly provided by Dr. Y Yamashita (Kyushu University, Japan). S. mutans was routinely cultured in Todd‐Hewitt Broth (THB; Oxoid) or on THB agar at 37°C with 5% CO2. GAS strain 5448 is a representative of the serotype M1T1 clone (Kansal et al., 2000) and GAS NZ131 (Simon and Ferretti, 1991) is an invasive strain of the M49 serotype. GAS was grown in THB (Becton Dickinson) supplemented with 1% yeast extract (THY) or on THY agar at 37°C. When required, growth medium was supplemented with 10 μg ml−1 erythromycin (Erm) or 3 μg ml−1 chloramphenicol (Cm) for S. mutans or with 300 μg ml−1 kanamycin (Km) or 100 μg ml−1 spectinomycin (Spec) for GAS. For cloning purposes, E. coli strain MC1061 was grown in Luria–Bertani (LB; Oxoid) or on LB agar with ampicillin (100 μg ml−1, Amp), Erm (500 μg ml−1) or Cm 10 μg ml−1.
Genetic manipulation of S. mutans
To confirm the function of GacA in the production of dTDP‐L‐rhamnose in bacteria, we heterologously expressed GAS gacA (M5005_Spy_0602; accession number AAZ51220.1) in a S. mutans rmlD deletion mutant (SMUΔrmlD), which is unable to produce dTDP‐L‐rhamnose (Tsukioka et al., 1997). For complementation, full‐length gacA was amplified from the GAS 5448 chromosome using primers XbaI_gacAF 5′‐GCTCTAGAATGATTTTAATTACAGGAAGCAATGGTC‐3′ and BamHI_gacAR 5′‐CGCGGATCCTACTTACTTTTTCAGTCCTTGTTGGT‐3′ and cloned into expression vector pDC123 using XbaI and BamHI restriction sites, yielding pGacA. pGacA was transformed into S. mutans wild‐type and selected for Cm resistance. The presence of pGacA was confirmed by PCR analysis. Subsequently, rmlD was knocked out by precise in frame allelic replacement of rmlD with an Erm resistance gene in S. mutans + pGacA. Briefly, 700 bp immediately upstream of rmlD was amplified with the primers rmlDupF, 5′‐CGCAGCAAGCAGTTACGTGATTTTGTTGAAG‐3′, and rmlDupR+erm 5′‐GTTTTGAGAATATTTTATATTTTTGTTCATTATTTTTTCTCCTTTAAAAAGCTTTATTACTATTACC‐3′, and 674 bp immediately downstream of rmlD was amplified with the primers rmlDdownF+erm 5′‐AGTTATCTATTATTTAACGGGAGGAAATAATATTTTAGCAAA‐GAAGGACAGGTTTAAACC‐3′, and rmlDdownR, 5′‐CTGAAGGTGATAA‐ATCCGTGCCATA‐3′. The rmlDupR+erm and rmlDdownF+erm primers were constructed with 30 bp 5′ extensions (underlined) corresponding to the 5′ and 3′ ends of the erm gene respectively. The upstream and downstream PCR fragments were combined with the 738 bp amplicon of the erm gene [amplified off the pDCerm plasmid (Jeng et al., 2003)] as templates in a second round of PCR using primers rmlDupF and rmlDdownR. The resultant PCR amplicon, containing an in frame substitution of rmlD with erm, was transformed into S. mutans + pGacA and selected for Erm and Cm resistance as previously described (Perry et al., 1983), to yield S. mutans ΔrmlD + pGacA (SMUΔrmlD + pGacA). PCR analysis was used to confirm the deletion of rmlD.
Scanning Electron Microscopy (SEM)
Overnight cultures of S. mutans strains were diluted and grown to mid‐log phase (S. mutans wild‐type and S. mutans ΔrmlD + pGacA OD600 of 0.3, S. mutans ΔrmlD OD600 of 0.15). GAS WT strains were grown in THY medium overnight. GAS 5448 gacAi were selected from a plate containing Spec and 2 mM theophylline and grown overnight cultures in the presence of different concentrations of theophylline (0, 0.5, 1, 1.5 and 2 mM) in THY. All cultures reached an OD600 of 0.5/0.6 except for strains without theophylline, which reached OD600 of 0.35. Cultures were diluted in THY with or without theophylline to an OD600 of 0.06 and grown to midlog phase. Samples were washed, fixed and dehydrated as described previously (Garufi et al., 2012), mounted onto 12.5 mm specimen stubs (Agar scientific, Stansted, Essex, UK) and coated with gold to 1 nm using a Quorum Q150R S sputter coater at 20 mA. Visual examination was performed with a Phenom PRO desktop SEM (Phenom‐World BV). The SEM was operated with an acceleration voltage of 10 kV.
S. mutans growth curves
Growth curves of S. mutans Xc wild‐type, Δ rmlD and Δ rmlD + pGacA were obtained after dilution of an overnight culture to OD600 to 0.025 in THB. Optical density was recorded every 30 min over 12 h at 37°C without 5% CO2 in a 100 Honeycombe plate using a Bioscreen C MBR machine (Growth Curves AB Ltd, Oy, Finnland).
Carbohydrate analysis of S. mutans strains
For the isolation of cell wall carbohydrates in S. mutans, 2 l bacterial cultures were centrifuged (8000× g, 30 min, 4°C) and washed with ice‐cold water. Five grams of wet bacterial cells was resuspended in 0.1 M citrate buffer (pH 4.6, 4°C) and disrupted with a bead‐beater (Biospec). The bacterial lysate was centrifuged (1000× g, 5 min, 4°C), the white suspension was isolated and centrifuged (35 000× g, 30 min, 4°C) to collect the cell walls. The white pellet was washed with citrate buffer, resuspended in 0.1 M sodium acetate (pH 4.6) containing 4% sodium dodecyl sulfate (SDS) and boiled for 1 h at 100°C with agitation. The suspension was centrifuged (35 000× g, 30 min, 21°C) and washed with SuperQ to remove SDS. The pellet was resuspended in 10 ml MilliQ and treated with RNase (10 μg ml−1) and DNase (10 μg ml−1, 37°C, 2 h) and afterward with pronase E (100 μg ml−1, 37°C for 24 h). Cell wall carbohydrates were extensively washed with SuperQ (35 000× g for 30 min), treated with trypsin (100 μg ml−1, 37°C, 2 h), washed with SuperQ and lyophilized before the sample was subjected to hydrolysis with TFA according to published procedures (Fan et al., 1994). Carbohydrate analysis was performed on a Dionex ICS‐3000 Ion Chromatography System (Dionex / Thermo Scientific, 1228 Titan Way, P.O. Box 3603, Sunnyvale, CA, 94088‐3603, United States) using a CarboPac PA20 (Dionex / Thermo Scientific, 1228 Titan Way, P.O. Box 3603, Sunnyvale, CA, 94088‐3603, United States) 3 × 150 mm column, equipped with a ICS‐3000 Electrochemical Detector (Dionex / Thermo Scientific, 1228 Titan Way, P.O. Box 3603, Sunnyvale, CA, 94088‐3603, United States). The ‘Carbohydrates (Standard Quad)’ waveform was used for detection. Eluent was run at 92% H2O and 8% 0.2M NaOH(aq) for 25 min and increased to 100% 0.2 M NaOH over a 2 min period and held at this concentration for 10 min. The concentration was then returned to 92% H2O and 8% 0.2 M NaOH(aq) over a 2 min period and held at this concentration for 11 min before a new sample was injected. The flow rate was 0.5 ml min−1.
Supporting information
Supporting information
Acknowledgements
This work was supported by a Sir Henry Wellcome Postdoctoral Fellowship (092193) to HCD, a Wellcome Trust Senior Fellowship (WT087590MA) and MRC Programme Grant (M004139) to DvA, an NWO‐VIDI grant (91713303) to NvS and a NIH/NIAID Grant (AI047928) to KSM.
The authors thank Drs. Ashton T. Belew and Najib El‐Sayed for support in bioinformatics design and implementation for the Tn‐seq analysis. The authors acknowledge the ESRF ID23‐1 for synchrotron beam time and Dr. Stephen Carr for assistance with SEC‐MALLS. The authors acknowledge Dr. Mark de Been for assistance to compose the Neighbor‐Joining trees and Miss Tonia Aristotelous for assistance in performing the SPR experiments. The authors thank Emrul Islam for his assistance in generating the gacAi plasmids and strains and Dr. Konstantinos Beis and Prof. So Iwata for suggestions and support.
PDB accession code: Co‐ordinates and structure factors have been deposited with the Protein Data Bank (PDB entry 4WPG).
Conflict of Interest: The authors declare to have no conflict of interest.
References
- Adams, P.D. , Afonine, P. , Bunkóczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye, T.G. , Kontogiannis, L. , Johnson, O. , Powell, H.R. , and Leslie, A.G. (2011) iMOSFLM: a new graphical interface for diffraction‐image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis, K. , Allard, S.T. , Hegeman, A.D. , Murshudov, G. , Philp, D. , and Naismith, J.H. (2003) The structure of NADH in the enzyme dTDP‐d‐glucose dehydratase (RmlB). J Am Chem Soc 125: 11872–11878. [DOI] [PubMed] [Google Scholar]
- Bernstein, F.C. , Koetzle, T.F. , Williams, G.J. , Meyer, E.F., Jr , Brice, M.D. , Rodgers, J.R. , et al (1977) The Protein Data Bank: a computer‐based archival file for macromolecular structures. J Mol Biol 112: 535–542. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt, W. , Asuncion, M. , Lam, J.S. , and Naismith, J.H. (2000) The structural basis of the catalytic mechanism and regulation of glucose‐1‐phosphate thymidylyltransferase (RmlA). The EMBO Journal 19: 6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt, W., I. , Kerr, D. , Giraud, M.F. , McMiken, H.J. , Leonard, G. , Whitfield, C. , et al (2002) Variation on a theme of SDR. dTDP‐6‐deoxy‐L‐ lyxo‐4‐hexulose reductase (RmlD) shows a new Mg2+‐dependent dimerization mode. Structure 10: 773–786. [DOI] [PubMed] [Google Scholar]
- Caliot, E. , Dramsi, S. , Chapot‐Chartier, M.P. , Courtin, P. , Kulakauskas, S. , Pechoux, C. , et al (2012) Role of the group B antigen of Streptococcus agalactiae: a peptidoglycan‐anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis, J.R. , Steer, A.C. , Mulholland, E.K. , and Weber, M. (2005) The global burden of group A streptococcal diseases. Lancet Infect Dis 5: 685–694. [DOI] [PubMed] [Google Scholar]
- Chandramohan, R. , Wu, P.Y. , Phan, J.H. , and Wang, M.D. (2013) Benchmarking RNA‐Seq quantification tools. Conf Proc IEEE Eng Med Biol Soc 2013: 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan, J.E. , Kindt, T.J. , and Krause, R.M. (1978) Structure of the streptococcal groups A, A‐variant and C carbohydrates. Immunochem 15: 755–760. [DOI] [PubMed] [Google Scholar]
- DeJesus, M.A. , Zhang, Y.J. , Sassetti, C.M. , Rubin, E.J. , Sacchettini, J.C. , and Ioerger, T.R. (2013) Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics 29: 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C. , Beis, K. , Giraud, M.F. , and Blankenfeldt, W. , Allard, S. , Major, L.L. , Kerr, I.D. , Whitfield, C. , and Naismith, J.H. (2003) A structural perspective on the enzymes that convert dTDP‐d‐glucose into dTDP‐l‐rhamnose. Biochem Soc Trans 31: 532–536. [DOI] [PubMed] [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W.G. , and Cowtan, K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. , Endert, J. , Kamps, M.A. , and van Boven, C.P. (1985) Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa . Infect Immun 49: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J.Q. , Namiki, Y. , Matsuoka, K. , and Lee, Y.C. (1994) Comparison of acid hydrolytic conditions for Asn‐linked oligosaccharides. Anal Biochem 219: 375–378. [DOI] [PubMed] [Google Scholar]
- Garufi, G. , Hendrickx, A.P. , Beeri, K. , Kern, J.W. , Sharma, A. , Richter, S.G. , et al (2012) Synthesis of lipoteichoic acids in Bacillus anthracis . J Bacteriol 194: 4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud, M.F. , and Naismith, J.H. (2000) The rhamnose pathway. Curr Opin Struct Biol 10: 687–696. [DOI] [PubMed] [Google Scholar]
- Gorrec, F. (2009) The MORPHEUS protein crystallization screen. J Appl Crystallogr 42: 1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graninger, M. , Nidetzky, B. , Heinrichs, D.E. , Whitfield, C. , and Messner, P. (1999) Characterization of dTDP‐4‐dehydrorhamnose 3,5‐epimerase and dTDP‐4‐dehydrorhamnose reductase, required for dTDP‐L‐rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J Biol Chem 274: 25069–25077. [DOI] [PubMed] [Google Scholar]
- Jeng, A. , Sakota, V. , Li, Z. , Datta, V. , Beall, B. , and Nizet, V. (2003) Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin‐binding protein, SfbX. J Bacteriol 185: 1208–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansal, R.G. , McGeer, A. , Low, D.E. , Norrby‐Teglund, A. , and Kotb, M. (2000) Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun 68: 6362–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh, K.L. , Jornvall, H. , Persson, B. , and Oppermann, U. (2008) Medium‐ and short‐chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65: 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, T. , Asakawa, H. , Okahashi, N. , and Takahashi, I. (1989) Effect of subculturing on expression of a cell‐surface protein antigen by Streptococcus mutans . J Gen Microbiol 135: 3199–3207. [DOI] [PubMed] [Google Scholar]
- Kornfeld, S. , and Glaser, L. (1961) The enzymatic synthesis of thymidine‐linked sugars. J Biol Chem 236: 1791–1794. [PubMed] [Google Scholar]
- Lancefield, R.C. (1933) A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med 57: 571–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. , and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Le Breton, Y. , and McIver, K.S. (2013) Genetic manipulation of Streptococcus pyogenes (the Group A Streptococcus, GAS). Curr Protoc Microbiol 30: Unit 9D 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton, Y. , Mistry, P. , Valdes, K.M. , Quigley, J. , Kumar, N. , Tettelin, H. , and McIver, K.S. (2013) Genome‐wide identification of genes required for fitness of group A Streptococcus in human blood. Infect Immun 81: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton, Y. , Belew, A.T. , Valdes, K.M. , Islam, E. , Curry, P. , Tettelin, H. , et al (2015) Essential genes in the core genome of the human pathogen Streptococcus pyogenes . Scientific Reports 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell, S.C. , Davis, I. , Arendall, W.B., 3rd , de Bakker, P.I. , Word, J.M. , Prisant, M.G. , et al (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 15: 437–450. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Pan, F. , and McNeil, M. (2002) Formation of dTDP‐rhamnose is essential for growth of mycobacteria. J Bacteriol 184: 3392–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, M. (1952) The lysis of group A hemolytic streptococci by extracellular enzymes of Streptomyces albus. II. Nature of the cellular substrate attacked by the lytic enzymes. J Exp Med 96: 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J. , Grosse‐Kunstleve, R.W. , Adams, P.D. , Winn, M.D. , Storoni, L.C. , and Read, R.J. (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson, D.F. , Manning, P.A. , and Morona, R. (1994) Characterization of the dTDP‐rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri . Mol Microbiol 11: 281–292. [DOI] [PubMed] [Google Scholar]
- McWilliam, H. , Li, W. , Uludag, M. , Squizzato, S. , Park, Y.M. , Buso, N. , et al (2013) Analysis tool web services from the EMBL‐EBI. Nucleic Acids Res 41: W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona, J.K. , Morona, R. , and Paton, J.C. (1997) Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol 23: 751–763. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N. , Vagin, A. , and Dodson, E.J. (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Nakano, K. , and Ooshima, T. (2009) Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol 4: 891–902. [DOI] [PubMed] [Google Scholar]
- van Opijnen, T. , and Camilli, A. (2010) Genome‐wide fitness and genetic interactions determined by Tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol 1: Unit1E 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, A. , Amoros, D. , and Garcia de la Torre, J. (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic‐ and residue‐level models. Biophys J 101: 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazur, J.H. , and Shuey, E.W. (1961) The enzymatic synthesis of thymidine diphosphate glucose and its conversion to thymidine diphosphate rhamnose. J Biol Chem 236: 1780–1785. [PubMed] [Google Scholar]
- Perry, D. , Wondrack, L.M. , and Kuramitsu, H.K. (1983) Genetic transformation of putative cariogenic properties in Streptococcus mutans . Infect Immun 41: 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez‐Gonzalez, R.H. , Leggett, R. , Waite, D. , Thanki, A. , Drou, N. , Caccamo, M. , and Davey, R. (2013) StatsDB: platform‐agnostic storage and understanding of next generation sequencing run metrics. F1000Res 2: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, D. , and Ferretti, J.J. (1991) Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett 66: 219–224. [DOI] [PubMed] [Google Scholar]
- Sivendran, S. , Jones, V. , Sun, D. , Wang, Y. , Grzegorzewicz, A.E. , Scherman, M.S. , et al (2010) Identification of triazinoindol‐benzimidazolones as nanomolar inhibitors of the Mycobacterium tuberculosis enzyme TDP‐6‐deoxy‐d‐xylo‐4‐hexopyranosid‐4‐ulose 3,5‐epimerase (RmlC). Bioorg Med Chem 18: 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge, N.M. , Cole, J.N. , Kuipers, K. , Henningham, A. , Aziz, R.K. , Kasirer‐Friede, A. , et al (2014) The classical lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15: 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe, I.C. , Black, G.W. , and Harrington, D.J. (2008) Bioinformatic insights into the biosynthesis of the Group B carbohydrate in Streptococcus agalactiae . Microbiol 154: 1354–1363. [DOI] [PubMed] [Google Scholar]
- Teng, C.H. , Cai, M. , Shin, S. , Xie, Y. , Kim, K.J. , Khan, N.A. , et al (2005) Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect Immun 73: 2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, F. , Singh, K.V. , Bourgogne, A. , Zeng, J. , and Murray, B.E. (2009) Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis . Infect Immun 77: 3759–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp, S. , Reynoso, C.M. , Seeliger, J.C. , Goldlust, I.S. , Desai, S.K. , Murat, D. , et al (2010) Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl Environ Microbiol 76: 7881–7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukioka, Y. , Yamashita, Y. , Nakano, Y. , Oho, T. , and Koga, T. (1997) Identification of a fourth gene involved in dTDP‐rhamnose synthesis in Streptococcus mutans . J Bacteriol 179: 4411–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Hess, T.N. , Jones, V. , Zhou, J.Z. , McNeil, M.R. , and McCammon, J.A. (2011) Novel inhibitors of Mycobacterium tuberculosis dTDP‐6‐deoxy‐L‐lyxo‐4‐hexulose reductase (RmlD) identified by virtual screening. Bioorg Med Chem Lett 21: 7064–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier, C. , and Peschel, A. (2008) Teichoic acids and related cell‐wall glycopolymers in Gram‐positive physiology and host interactions. Nat Rev Microbiol 6: 276–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


