Abstract
The assembly of microtubule-based cellular structures depends on regulated tubulin polymerization and directional transport. Here, we purify and characterize tubulin heterodimers that have human β-tubulin isotype III (TUBB3), and heterodimers with one of two β-tubulin mutations (D417H or R262H). Both point mutations are proximal to the kinesin binding site and have been linked to an ocular motility disorder in humans. Compared to wild-type, microtubules with these mutations have decreased catastrophe frequencies and increased average lifetimes of plus- and minus-end stabilizing caps. Importantly, the D417H mutation does not alter microtubule lattice structure or Mal3 binding to growing filaments. Instead, this mutation reduces the affinity of tubulin for TOG domains and colchicine, suggesting that the distribution of tubulin heterodimer conformations is changed. Together, our findings reveal how residues on the surface of microtubules, distal from the GTP-hydrolysis site and inter-subunit contacts, can alter polymerization dynamics at the plus- and minus-ends of microtubules.
Graphical abstract
Introduction
Heterodimers of α/β-tubulin undergo GTP-dependent polymerization to form microtubules, cytoskeletal filaments essential for diverse cellular processes including neuronal transport, cell migration and cell division (Desai and Mitchison, 1997; Heald and Khodjakov, 2015; Kapitein and Hoogenraad, 2015). The proper organization of microtubules depends on dynamic instability, the stochastic transitions of the microtubule between growth and shrinkage, and on microtubule-associated proteins (MAPs) that can step along these filaments to transport cargoes or regulate filament polymerization dynamics (Heald and Khodjakov, 2015; Nogales and Zhang, 2016). Consistent with these basic functions, mutations in tubulin or MAPs that disrupt MAP-microtubule interactions can lead to defective cytoskeletal architectures and have been linked to disease (Hirokawa et al., 2009; Niwa et al., 2013; Tischfield et al., 2011).
We now have good structural models for tubulin heterodimers, microtubules, and how motor and non-motor MAPs interact with conserved amino acids on the surface of the hollow tube-like filament (Nogales and Zhang, 2016). MAPs such as the end-binding (EB) proteins that regulate microtubule polymerization dynamics bind tubulin subunits at sites proximal to the GTP-binding pocket of tubulin to sense and modulate changes in nucleotide states that directly contribute to polymerization dynamics (Akhmanova and Steinmetz, 2010; Maurer et al., 2012). In contrast, MAPs such as kinesins that move directionally along the microtubule lattice interact with surface-exposed residues that are distal from tubulin’s GTP-binding site and the contact regions between different subunits (Nogales and Zhang, 2016). Based on structural data alone, we would not expect that these surface residues are involved in modulating microtubule polymerization dynamics. However, the effect of mutating residues proximal to the kinesin binding site on parameters of dynamic instability or on tubulin and microtubule conformation is not known.
There are seven α- and eight β-tubulin isotypes in humans (Ludueña and Banerjee, 2008). The distribution of these isotypes varies across different tissues, with β-tubulin isotype III (TUBB3) expression mainly limited to developing and mature neurons (Jiang and Oblinger, 1992). Heterozygous point mutations in TUBB3 have been identified in patients with severe congenital fibrosis of the extraocular muscle type 3 (CFEOM3), an ocular motility disorder (Tischfield et al., 2010). Studies in mice and budding yeast suggest that the phenotypes associated with heterozygous TUBB3 mutations are likely due to reduced binding to kinesins (Niwa et al., 2013; Tischfield et al., 2010). Currently, it is difficult to establish if these mutations directly reduce binding to MAPs. It is also unclear if these mutations in tubulin alter polymerization dynamics. This is in large part due to the challenges in generating human tubulin from recombinant sources.
To examine the contribution of surface residues in TUBB3 to the structural and biochemical properties of microtubules, we devised a strategy to generate recombinant tubulin heterodimers that have human TUBB3. We determined the high-resolution structure, characterized the binding to MAPs and analyzed the polymerization dynamics of wild-type and mutant tubulins. Surprisingly, we find that two disease-related point mutations in TUBB3, D417H and R262H, alter polymerization dynamics at microtubule plus- and minus-ends. Further, we show that mutant tubulin has a lower affinity for both TOG domains and colchicine compared to wild-type tubulin, suggesting that the mutations alter the equilibrium of ‘curved’ and ‘straight’ tubulin heterodimers. Finally, by examining the properties of microtubules assembled from mixtures of wild-type and mutant tubulins, we dissect how the presence of sub-stoichiometric levels of mutant β-tubulin can have distinct effects on MAP binding and dynamic instability.
Results
Purification of recombinant tubulin heterodimers that have wild-type and mutant TUBB3
To generate recombinant human tubulin we first used a recently reported insect cell-based strategy (Minoura et al., 2013). However, in our hands, this immunoprecipitation approach, which generates FLAG-tagged β-tubulin and hexa-histidine-tagged α-tubulin, produced an insufficient yield of pure tubulin. Therefore, to generate recombinant tubulin in sufficient yield and to remove any affinity tags, we significantly redesigned the method, incorporating a cleavable hexa-histidine tag at the C-terminus of β-tubulin, and left α-tubulin untagged. Tagging both α- and β-tubulin substantially reduced the overall protein yield (data not shown). To purify tubulin we designed a three-step purification strategy that employs nickel affinity chromatography, cleavage of the hexa-histidine tag on β-tubulin, and TOG-domain affinity chromatography (Widlund et al., 2012) (Fig. 1A). This protocol typically yielded >95% pure tubulin in amounts sufficient for biochemical and biophysical studies (1.5 mg tubulin per liter of cultured insect cells) (Fig. 1B). We generated recombinant forms of wild-type TUBB3 and two CFEOM3-linked TUBB3 mutants, D417H and R262H (Fig. 1B). Protein immunoblots showed that purified recombinant human tubulin has no detectable hexa-histidine tag (Fig. 1C).
Figure 1. Purification and analyses of recombinant human TUBB3.
A, Schematic detailing the purification of recombinant human tubulin. Typical tubulin purity at key steps is indicated. B, SDS-PAGE analysis of purified tubulin. C, Immunoblot analyses of proteins eluted from nickel affinity and TOG-affinity columns with antibodies against α-tubulin, β-tubulin and C-terminal hexa-histidine tag, as indicated. D, Elution profiles of wild-type TUBB3 (peak volume: 14.4 ml) and bovine tubulin (peak volume: 14.3 ml) from size-exclusion chromatography. E, Elution profiles of TUBB3-D417H (peak volume: 14.3 ml) and TUBB3-R262H (peak volume: 14.3 ml) from size-exclusion chromatography. Bovine tubulin profile is shown as reference. F–H, TIRF-microscopy images of taxol-stabilized wild-type TUBB3 (F), TUBB3-D417H (G), and TUBB3-R262H (H) microtubules. Fluorescently labeled bovine tubulin (~4%) was added to visualize filaments. Scale bars= 2 μm. See also Figure S1.
We next used mass spectrometry to confirm the presence of mutated residues (Fig. S1A–C) and found that purified α/β-tubulin consisted of >99% recombinant TUBB3 and an equimolar mixture of recombinant human (isotype α1B) and endogenous insect α-tubulin (Fig. S1D). As human and insect α-tubulin are ~97% identical by sequence (Fig. S1D), we expect that its presence is not likely to have a substantial impact on our analyses. We believe that our recombinant tubulin purification strategy is particularly useful for directly comparing wild-type and mutant forms of human β-tubulin. Hereafter, to emphasize the specific β-tubulin present in the purified tubulin, we refer to our wild-type tubulin as ‘wild-type TUBB3’ and the mutant forms as ‘TUBB3-D417H’ and ‘TUBB3-R262H’.
We analyzed the recombinant wild-type and mutant TUBB3 using two approaches. First, size-exclusion chromatography indicated that purified recombinant wild-type and mutant TUBB3 exist as stable dimers in solution with Stokes radii (43 Å) similar to that of bovine tubulin (Fig. 1D and E) purified using standard methods involving polymerization/depolymerization cycles (Al-Bassam et al., 2006; Gell et al., 2011). Second, fluorescence microscopy-based analysis indicated that wild-type TUBB3, TUBB3-D417H and TUBB3-R262H polymerized to form microtubules in the presence of GTP and taxol (Fig. 1F–H).
Disease-related point mutations in TUBB3 reduce binding to motor and non-motor MAPs
To determine effects of the D417H and R262H tubulin mutations on the binding to motor and non-motor MAPs, we first used a TIRF microscopy-based assay (Fig. 2A). We analyzed the association of taxol-stabilized microtubules with GFP-tagged kinesin superfamily proteins that share a motor domain conserved in both structure and sequence (Vale and Fletterick, 1997) (kinesin-1ΔC and kinesin-5, in the presence of 2 mM MgATP), and two non-motor MAPs that have structurally distinct microtubule binding motifs (PRC1-SC and NuMA tail II) (Fig. 2B) (Haren and Merdes, 2002; Subramanian et al., 2010). We found that all of the tested MAPs associate more strongly with wild-type TUBB3 microtubules than with mutant microtubules (Fig. 2C and D, Fig. S2A and B). The GFP fluorescence intensity per micron of microtubule length indicated that under our assay conditions these MAPs have a 5–10 fold reduction in direct association with microtubules polymerized either with TUBB3-D417H or TUBB3-R262H compared to wild-type microtubules (Fig 2E–H).
Figure 2. Point mutations D417H and R262H in TUBB3 reduce the binding of motor and non-motor MAPs to microtubules.
A and B, Schematic for assay (A) and MAP constructs used (B) with microtubule binding regions (black) and GFP (green) highlighted. C and D, Images of microtubule and GFP-tagged MAPs, along with 2-color overlays (microtubule: red; GFP: green), are shown for (C) kinesin-1ΔC-GFP (0.7 nM, 2 mM MgATP) and (D) GFP-PRC1-SC (16 nM). Scale bars = 2 μm E–H, GFP-fluorescence intensity per micron along the microtubules for kinesin-1ΔC (E), kinesin-5 (F), PRC1-SC (G) and NuMA tail II (H) are shown. Data from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (n=62 or greater). I and J, SDS-PAGE analysis of supernatant fractions from microtubule co-sedimentation assays for PRC1-SC (I) kinesin-1ΔC (J). K and L, Band intensities from the gels (I and J) were used to determine fraction protein bound to microtubules and plotted against microtubule concentrations (n=3, mean ± SD). The data were fit to determine the dissociation constants (Kd): PRC1-SC (K), wild-type TUBB3: 160±30 nM; TUBB3-D417H: 370±20 nM; TUBB3-R262H: 300±30 nM, and kinesin-1ΔC (L), wild-type TUBB3: 90±30 nM; TUBB3-D417H: 720±30 nM; TUBB3-R262H: 830±60 nM. See also Figure S2.
We next used a microtubule co-sedimentation assay to measure the apparent dissociation constants (Kd) of wild-type and mutant TUBB3 for PRC1-SC (Fig. 2I) and kinesin-1ΔC (in the presence 2 mM non-hydrolyzable ATP analogue MgAMPPNP) (Fig. 2J). The affinity of wild-type TUBB3 microtubules for PRC1-SC and kinesin-1ΔC is 160±30 and 90±30 nM, respectively. In comparison, microtubules with point mutation D417H in TUBB3 bind PRC1-SC with a 2-fold lower affinity (370±20 nM) and bind kinesin-1ΔC with an 8-fold lower affinity (720±30 nM) (Fig. 2K and L). Furthermore, microtubules polymerized with TUBB3-R262H have a 2-fold lower affinity (300±30 nM) for PRC1-SC and a 9-fold lower affinity (830±60 nM) for kinesin-1ΔC (Fig. 2K and L). Our findings indicate that both D417 and R262 residues contribute to the direct binding of microtubules to motor and non-motor MAPs. These results are largely expected based on available structural models (Gigant et al., 2013) in which both these residues, whose side chains can make electrostatic contacts with each other, are positioned proximal to where these MAPs bind (Fig. S2C).
TUBB3-D417H and TUBB3-R262H microtubules have altered intrinsic polymerization dynamics compared to wild-type microtubules
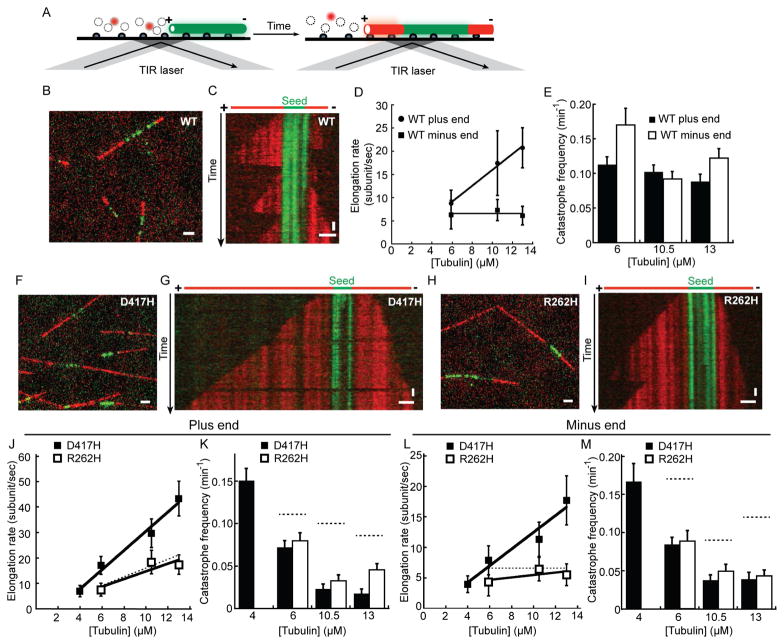
We next employed a TIRF-based single filament assay to analyze polymerization dynamics of wild-type TUBB3 (Fig. 3A). As a template for microtubule formation, we used GMPCPP-stabilized ‘seeds’ generated with wild-type TUBB3 tubulin. In the presence of soluble tubulin and GTP (1 mM), microtubules were observed to grow and shrink at both ends of seeds (Fig. 3B). Kymographs of individual microtubules show a noticeable difference in the length and growth rates of microtubule polymer assembled from one end of the seed compared to the other (Fig. 3C). Hereafter, as per convention, the faster growing end is referred to as the plus-end, and the slower growing end as the minus-end. We next measured key parameters of microtubule dynamic instability, including rate of growth (polymerization rate) and the likelihood of transition from relatively slow growth to rapid shortening (catastrophe frequency) (Desai and Mitchison, 1997) at both plus- and minus-ends.
Figure 3. TUBB3-D417H and TUBB3-R262H have altered intrinsic polymerization dynamics compared to wild-type TUBB3.
A, Schematic for assay used to analyze polymerization dynamics of single microtubules. Tubulin (red) incorporates at plus- and minus-ends of GMPCPP-stabilized microtubule ‘seeds’ (green). B and C, TIRF-microscopy image (B) and kymograph (C) of dynamic microtubules growing from GMPCPP seeds in the presence of 10.5 μM wild-type TUBB3. D, Elongation rates of wild-type TUBB3 at plus- (●) and minus-ends (■) across three different tubulin concentrations. E, Catastrophe frequency of wild-type TUBB3 at microtubule plus- and minus-ends. Data acquired from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (n=57 or greater). F–I, TIRF-microscopy images (F and H) and kymographs (G and I) of dynamic microtubules growing from GMPCPP seeds in the presence of TUBB3-D417H (F and G) or TUBB3-R262H (H and I) (10.5 μM). J–M, Analyses of dynamic instability parameters at plus- (J and K) and minus-ends (L and M) of TUBB3-D417H (■) and TUBB3-R262H (□) microtubules: elongation rate (J and L) and catastrophe frequency (K and M) are shown. For comparison, the dashed-lines indicate the elongation rates and the catastrophe frequency of wild-type TUBB3. Data from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (n=41 or greater). Elongation rates were fitted with the linear equation as a function of the free tubulin concentration. Assuming a Poisson distribution, the standard deviations of catastrophe frequency were calculated as (observed catastrophe frequency)/(number of events counted)0.5. Scale bars: horizontal= 2 μm, vertical= 60 sec. See also Table S1.
The polymerization rate from plus-ends increased with tubulin concentration and could be fit to a line (Oosawa, 1970), whose slope and intercept suggested the apparent association (k+) and dissociation (k-) rate constants of tubulin subunits (k+=1.7±0.15 μM−1s−1 and k-=1.1±1.5 s−1) in the 1-D model (Fig. 3D). By contrast, the minus-end polymerization rate of wild-type TUBB3 was relatively constant across the range of tubulin concentrations tested (Fig. 3D). The catastrophe frequency was 0.1±0.01 min−1 for plus-ends and 0.09±0.01 min−1 for minus-ends at a tubulin concentration (10.5 μM) close to physiologic levels (Fig. 3E). While variation in catastrophe frequency was observed at different tubulin concentrations, the scatter in these data did not allow for the establishment of a strong correlation between catastrophe frequency and concentration. Together, these data demonstrate wild-type TUBB3’s dynamic instability and yield the key parameters, which thus far have not been available for any purified human tubulin isotype.
We next examined the dynamics of mutant TUBB3 microtubules. At equivalent time intervals, TUBB3-D417H and TUBB3-R262H assembled longer microtubule extensions from GMPCPP-stabilized seeds compared to wild-type TUBB3 across different tubulin concentrations (Fig. 3F–I). We first focused on plus-end dynamic instability parameters. For TUBB3-D417H, polymerization rates at plus-ends of microtubules were ~1.7-fold faster than those for wild-type TUBB3 (10.5 μM), with a 2-fold higher k+ and a 7-fold higher k- (Fig. 3J). Remarkably, these microtubules were substantially more stable, undergoing catastrophe ~4-fold less frequently than did microtubules assembled from wild-type TUBB3 (10.5 μM, Fig. 3K). In the case of TUBB3-R262H, the polymerization rates at plus-ends were similar to those measured for wild-type TUBB3 (Fig. 3J). The catastrophe frequency at microtubule plus-ends was 3-fold lower than wild-type TUBB3 (10.5 μM, Fig. 3K). Based on the model developed by Leibler and colleagues (Verde et al., 1992) and these measurements, we estimate that TUBB3-D417H microtubules would be, on average, ~7 times longer than wild-type, and TUBB3-R262H microtubules would be ~3 times longer than wild-type (detailed calculation is provided in Supplemental information).
D417H and R262H mutations in TUBB3 not only affect microtubule polymerization dynamics at the plus-ends but also alter filament dynamics at the minus-ends. The growth rates of TUBB3-D417H microtubule minus-ends were substantially faster than those we measured for wild-type TUBB3 and varied with tubulin concentration (k+=1.4±0.23 μM−1s−1 and k-=1.2±2.1 s−1, Fig. 3L). Furthermore, catastrophe frequency was ~2-fold lower for TUBB3-D417H compared to wild-type tubulin (10.5 μM, Fig. 3M). In the case of TUBB3-R262H microtubule minus-ends, the catastrophe frequency was 2-fold lower compared to wild-type TUBB3 (10.5 μM, Fig. 3M,). However, substantial differences in minus-end polymerization rates were not observed between wild-type TUBB3 and TUBB3-R262H (Fig. 3L). Table S1 summarizes these data. Together, our findings reveal that mutations near the kinesin-binding site can directly alter microtubule dynamics at both filament ends.
Analyses of wild-type and TUBB3-D417H tubulin conformations in the microtubule lattice
In order to investigate the effects of disease-related mutations on microtubule structure, we characterized our recombinant wild-type and TUBB3-D417H tubulin using cryo-EM reconstructions. Using protocols previously described (Alushin et al., 2014), we obtained a reconstruction to 3.8 Å resolution of 13-protofilament wild-type TUBB3 microtubules in the GDP-state bound to kinesin-1 motor domains (the kinesin density helps distinguish between α- and β-tubulin during image alignment) (Fig. 4A). Comparison with structures at similar resolution of porcine brain tubulin (Zhang et al., 2015) did not reveal significant differences.
Figure 4. Analysis of wild-type and TUBB3-D417H tubulin conformations in polymeric and soluble forms.
A, Densities for a wild-type TUBB3 tubulin heterodimer and bound kinesin motor domain, segmented from the cryo-EM reconstructions of kinesin-decorated microtubules. B, Absorbance at 350 nm, or turbidity, for wild-type TUBB3 (blue) and TUBB3-D417H (red) polymerization. Data from six independent experiments were analyzed to determine averages and SD (gray area). C, Densities for a TUBB3-D417H tubulin heterodimer and bound kinesin motor domain, segmented from the cryo-EM reconstructions of kinesin-decorated microtubules. A and C are shown at equivalent density thresholds. D, Cα trace of wild-type TUBB3 (grey) and TUBB3-D417H (blue) atomic models. Mutated D417 is shown in red. The RMSD between Cα positions of wild-type and mutant tubulin heterodimer is indicated. E, Schematic for assay used to analyze Mal3-GFP (green) binding growing microtubule ends (red). F and G, Overlaid TIRF-microscopy images of Mal3-GFP (green) and dynamic X-rhodamine labeled wild-type TUBB3 (H) and TUBB3-D417H (I) microtubules (red) are shown. Scale bars= 5 μm. H, Tip-aligned and averaged Mal3-GFP intensity profile at the growing plus-ends of wild-type TUBB3 (black dots) and TUBB3-D417H (red dots) microtubules when tubulin concentrations are the same. I, Distribution of growth speeds for Mal3-GFP tagged growing microtubule plus-ends (wild-type TUBB3, n=21; TUBB3-D417H, n=25). J, Tip-aligned and averaged Mal3-GFP intensity profile at growing microtubule plus-ends (wild-type TUBB3, n=34, black dots; TUBB3-D417H, n=29, red dots). Tubulin concentrations were adjusted to achieve similar filament growth speeds. K and L, Averaged Mal3-GFP intensity profiles shown in (J) were further analyzed by fitting to a two-step model with binding kinetics as previously described (Maurer et al., 2014). (K) Mal3-GFP binding densities from deconvolved model fitting. (L) Individual maturation rates from convolved model fitting. M, Fraction of unbound tubulin in the GST-TOG1/2 pull-down assay (n=3, mean ± SEM) were used to determine the dissociation constants (Kd) (wild-type TUBB3 (blue, Kd=0.4±0.1μM) and TUBB3-D417H (blue, Kd=1.2±0.3 μM). N, SDS-PAGE analysis of the supernatant (s) and pellet (p) fractions from the microtubule sedimentation assays of wild-type TUBB3 and TUBB3-D417H in the presence of colchicine. O, Band intensities from the gels (N) were used to determine fraction of soluble tubulin (n=3, mean ± SD). See also Figure S3.
We next examined the structure of TUBB3-D417H microtubules. We found that TUBB3-D417H (20 μM), in the presence of 5% (w/v) glycerol and GTP (1 mM), failed to generate microtubules detectable by cryo-EM analysis. To examine this further we compared wild-type and mutant tubulin polymerization using a turbidity-based assay (Gaskin et al., 1974). In the presence of GTP (1 mM) and the highest concentrations of recombinant tubulins (26 μM) that could be reliably achieved, we observed a time-dependent increase in turbidity, indicating polymerization (Fig. 4B). We observed a shorter time-lag in the increase in turbidity for mutant compared to wild-type tubulin, suggesting that TUBB3-D417H assembles filaments faster than wild-type tubulin. If nucleation is more efficient for TUBB3-D417H than wild-type, at similar tubulin concentrations TUBB3-D417H may assemble filaments too short to be readily detected in cryo-EM preparations. We therefore used wild-type GMPCPP-stabilized ‘seeds’ to generate TUBB3-D417H microtubules for cryo-EM studies and obtained a structure to 3.7 Å resolution (Fig. 4C).
The kinesin-1 density in the reconstruction of mutant microtubules is much weaker than for wild-type, consistent with the mutant tubulin’s reduced affinity for kinesin (Fig. 2L and 4C). However, we were able to detect sufficient bound kinesin to allow accurate alignment of α- and β-tubulin for TUBB3-D417H microtubules. At this resolution we can observe extra density for the histidine 417 side chain at the surface of mutant microtubules with respect to the wild-type aspartate 417 (Fig. S3A). The Cα-atoms root-mean-square deviation (RMSD) between wild-type and TUBB3-D417H microtubule structures is small (~0.36 Å), which indicates that point mutation D417H in β-tubulin did not induce substantial structural changes in microtubule lattice parameters or in the overall tubulin subunit structure within the polymer (Fig. 4D). As the effects on tubulin polymerization of the R262H mutation are less significant than what we measured for the D417H mutation we predict that this mutation would also not alter the structure of tubulin in the microtubule lattice.
Analyses of the binding of EBs to the growing wild-type and TUBB3-D417H microtubule ends
To determine if the D417H mutation impacts the end regions of growing filaments, we employed an established TIRF-based assay and subpixel-precision image analysis on averaged fluorescence intensity profiles to examine the interaction of GFP-tagged end-binding proteins (EBs) with microtubules (Akhmanova and Steinmetz, 2010; Maurer et al., 2014). Mal3, a fission yeast EB1 homolog, autonomously interacts with an extended region at growing microtubule ends and accelerates the consecutive conformational transitions that occur as the polymer end matures (Maurer et al., 2014). Dynamic wild-type and TUBB3-D417H microtubules were observed to grow off the ends of GMPCPP-stabilized wild-type ‘seeds’ in the presence of GFP-tagged Mal3 (Fig. 4E). Automated tracking of individual microtubule ends showed a noticeable difference in the length and growth rates of microtubule polymer assembled from one end of the seeds compared to the other. We observed Mal3-GFP decoration at the growing plus- and minus-ends of wild-type and TUBB3-D417H microtubules (Fig. 4F and G). The comet-like Mal3-GFP distribution was slightly longer for TUBB3-D417H than for wild-type microtubules (Fig. 4H), which can be due to the ~20% faster elongation rate of mutant microtubules compared to wild-type at equal tubulin concentrations under these assay conditions (Fig. 4I).
We adjusted the concentration of mutant tubulin so that the microtubule growth rates would be within a similar range. Under these conditions, the Mal3-GFP fluorescence intensity profile was indistinguishable between wild-type and mutant microtubules (Fig. 4J). The deconvolved peak intensity indicates that Mal3-GFP binds at the same density to the tips of wild-type and mutant microtubules, suggesting the binding affinity remains unchanged (Fig. 4K). This finding agrees with the fact that D417 is far from the site of microtubule interaction described for Mal3 and EB3 in cryo-EM studies (Maurer et al., 2012; Zhang et al., 2015). Furthermore, using convolved model fitting to the averaged fluorescence intensity profile, based on methods described previously (Maurer et al., 2014), we showed that mutation D417H does not affect the maturation rates of growing filament ends (Fig. 4L). These data indicate that the D417H tubulin mutation suppresses the binding of kinesins and non-motor microtubule crosslinkers to filaments but not the binding of EB proteins to growing microtubule ends.
Analyses of soluble wild-type and TUBB3-D417H tubulin conformations
To characterize the distribution of tubulin heterodimer conformations, which can be ‘curved’ or ‘straight’ in solution (Brouhard and Rice, 2014), we examined binding to TOG domains and colchicine, both of which preferentially bind to ‘curved’ αβ-tubulin heterodimers (Ayaz et al., 2014; Ayaz et al., 2012; Ravelli et al., 2004). The D417 residue in β-tubulin is distal from the tubulin-TOG binding site and the colchicine-binding site and is unlikely to directly disrupt these interactions (Fig. S3B–D).
We first examined the binding of wild-type TUBB3 and TUBB3-D417H to GST-tagged TOG domains. Pull-down assays revealed that the binding affinity of wild-type TUBB3 (Kd=0.4±0.1 μM) for TOG1/2 domain is comparable to what has been measured between yeast tubulin and TOG domains (Ayaz et al., 2014; Ayaz et al., 2012). Importantly, this affinity is ~3-fold tighter than for TUBB3-D417H (Kd=1.2±0.3 μM) (p< 0.05) (Fig. 4M). Second, we analyzed colchicine binding by measuring dose-dependent inhibition of microtubule polymerization. We find that the polymerization of TUBB3-D417H is more resistant to colchicine than wild-type TUBB3. In particular, the amount of tubulin that did not polymerize (i.e. fraction of soluble tubulin) is ~2-fold higher for wild-type TUBB3 than for TUBB3-D417H at each of the colchicine concentrations examined (7–35 μM, Fig. 4N and O). These data suggest that colchicine has a higher affinity for wild-type than for mutant tubulin. As an additional test we used allocolchicine, a colchicine analogue whose fluorescence increases when it binds tubulin heterodimers (Ravelli et al., 2004; Rice et al., 2008). We find that the polymerization of TUBB3-D417H is more resistant to allocolchicine than wild-type TUBB3 (Fig. S3E). While we estimate a ~2-fold lower affinity for TUBB3-D417H using fluorescence-based analyses, we cannot achieve high statistical significance in this experiment (P value: <0.1, Fig. S3F). This is most likely due to a low signal to noise ratio, as the fluorescence increase of allocolchicine upon binding TUBB3 is ~3-fold lower than it is upon binding bovine tubulin (Fig. S3G). Together, these data indicate that the D417H mutation in TUBB3 alters the distribution of tubulin heterodimer conformations, reducing the effective concentration of ‘curved’ heterodimer in solution.
TUBB3-D417H and TUBB3-R262H mutations increase the stability of ‘end-caps’ at both ends of microtubules
Microtubule dynamic instability at plus-ends has been proposed to depend on a GTP ‘cap’ that stabilizes the ends of growing filaments (Bowne-Anderson et al., 2013; Carlier et al., 1984). In current models, the loss of GTP-bound tubulin subunits at filament ends exposes GDP-tubulin subunits in the microtubule lattice and promotes catastrophe (Bowne-Anderson et al., 2013). Direct measurements of the size of the ‘end-stabilizing cap’ cannot be easily made and indirect analyses are necessary. In particular, the stability of the ‘end-stabilizing’ cap can be estimated by measuring the time lag before polymerizing microtubules undergo catastrophe when the tubulin concentration is rapidly reduced (Walker et al., 1991). We therefore adapted this assay to determine whether TUBB3-D417H and TUBB3-R262H impact ‘end-stabilizing cap’ stability. We rapidly perfused the experimental chamber with tubulin-free buffer to decrease tubulin concentration by 85% in 2–3 seconds and used time-lapse TIRF microscopy to monitor microtubule length (Fig. 5A, Fig. S4A). Kymographs of individual microtubules revealed a time lag before catastrophe upon dilution (Fig. 5B–G). The observed time lag in our assays is within the range previously reported for tubulin purified from porcine brain (Walker et al., 1991). For microtubules generated with 13 μM wild-type tubulin we observed a 9.9±4.2 sec delay before catastrophe at the plus-ends (Fig. 5H). Our data suggest an ‘end-stabilizing cap’ also exists at growing minus-ends, as an average delay of 9.1±5.7 sec is observed before catastrophe (Fig. 5I). Both D417H and R262H mutations in tubulin increase the time lag before catastrophe at both plus- and minus-ends of filaments (TUBB3-D417H: plus-end, 16.3±9.8 sec; minus-end, 13.7±11 sec and TUBB3-R262H: plus-end, 14.3±6 sec; minus-end, 12.5±7.2 sec) by ~1.5-fold compared to wild-type TUBB3 (Fig. 5H and I). In contrast, the observed depolymerization rates, which correspond to the dissociation rate constant of GDP-tubulin, are the same for wild-type TUBB3 and the mutant construct (Fig. S4B and C, Table S1). These findings suggest that the TUBB3-D417H and TUBB3-R262H mutations alter dynamic instability by increasing the stability of ‘caps’ at both ends of microtubules.
Figure 5. TUBB3-D417H and TUBB3-R262H are more resistant to dilution-induced catastrophe than wild-type TUBB3 at both ends of microtubules.
A, Schematic detailing the assay used to analyze the dilution-induced catastrophe of single microtubules. B–G, Time-lapse sequences acquired during microtubule depolymerization (B–D) and corresponding kymographs (E–G) of wild-type TUBB3 (B and E), TUBB3-D417H (C and F) and TUBB3-R262H (D and G). H and I, Time lag before the initiation of microtubule catastrophe at plus- (H) and minus-ends (I). Data from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (Wild-type TUBB3: n=80; TUBB3-D417H n=65; TUBB3-R262H n=69). Scale bars: horizontal= 2 μm, vertical= 5 sec. See also Figure S4 and Table S1.
Analyses of mixtures of wild-type and mutant tubulins
To test whether these TUBB3 mutations have dose-dependent effects on microtubule function, we mixed wild-type and mutant tubulins and examined both binding to MAPs and assembly dynamics. First, we compared MAP binding between ‘mixed’ mutant and wild-type microtubules. At different ratios of TUBB3-D417H or TUBB3-R262H and wild-type tubulin, microtubules readily polymerized in the presence of taxol (Fig. 6A and B). We added GFP-tagged kinesin-1ΔC to these microtubules and measured the fluorescence intensity per unit filament length. The average GFP fluorescence intensity on ‘mixed’ microtubules was lower than that observed for microtubules assembled from wild-type TUBB3, and decreased further with increasing amounts of TUBB3-D417H in the polymer (Fig. 6C). We also examined the binding of GFP-tagged PRC1-SC to these microtubules and observed fluorescence intensities that were lower than what we observed for wild-type tubulin filaments and higher than what we observed for TUBB3-D417H filaments (Fig. 6D). A similar trend was observed for TUBB3-R262H and wild-type mixed microtubules (Fig. 6C and D). Next, we measured dynamic instability parameters in single filament assays. We observed dynamic microtubule extensions at both plus- and minus-ends of GMPCPP-seeds (Fig. 6E and F). The catastrophe frequency at filament plus-ends was ~0.11 min−1 at equal ratios of TUBB3-D417H and wild-type TUBB3, and ~0.09 min−1 at equal ratios of TUBB3-R262H and wild-type microtubules (10.5 μM total tubulin concentration). These values are similar to those measured for microtubules assembled with wild-type TUBB3 (~0.10 min−1, 10.5 μM total tubulin) (Fig. 6G). The catastrophe frequency at the minus-ends of mixed microtubules was also similar to wild-type alone (Fig. 6H). Together, our data suggest that the presence of wild-type tubulin can suppress the altered catastrophe frequencies but only partially recover the reduction in MAPs binding due to these point mutations in TUBB3.
Figure 6. Analyses of MAPs binding and polymerization dynamics of microtubules polymerized from mixtures of wild-type and mutant tubulin.
A and B, Images of microtubules polymerized from equal ratios of TUBB3-D417H or TUBB3-R262H and wild-type TUBB3, associated GFP-tagged MAPs, along with 2-color overlays (microtubule: red; GFP: green), are shown for (A) kinesin-1ΔC (0.7 nM, 2 mM MgATP) and (B) PRC1-SC (16 nM). C and D, GFP-fluorescence intensity of kinesin-1ΔC (C) and PRC1-SC (D) per micron of taxol-stabilized microtubules polymerized from tubulin with different ratios of wild-type and mutant tubulin. Data from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (kinesin-1ΔC: n=64 or greater; PRC1-SC: n=66 or greater). E and F, Kymographs of dynamic microtubule extensions from GMPCPP seeds at equal ratios of TUBB3-D417H and wild-type TUBB3 (E) and at equal ratios of TUBB3-R262H and wild-type TUBB3 (F). G and H, Analyses of catastrophe frequency at the growing plus- (G) and minus-ends (H) of microtubules. Wild-type and mutant tubulin were mixed in equal ratios while the total tubulin concentration was kept at 10.5 μM. For comparison dashed-lines indicate catastrophe frequency values for microtubules assembled with wild-type (red), TUBB3-D417H (blue) or TUBB3-R262H (orange) tubulin (see Fig 3). Data from three independent experiments were pooled for each condition and analyzed to determine averages and SD (error bars) (n=43 or greater). Assuming a Poisson distribution, the standard deviations of catastrophe frequency were calculated as (observed catastrophe frequency)/(number of events counted)0.5. Scale bars: horizontal= 2 μm, vertical= 60 sec.
Discussion
Our studies indicate that mutations proximal to the TUBB3 kinesin binding site alter polymerization dynamics. In particular, these mutants suppress catastrophe frequency by magnitudes similar to what is achieved by regulatory proteins or microtubule stabilizing drugs (Mohan et al., 2013; Wieczorek et al., 2015). Structural and mutagenesis studies have indicated that conformational changes in tubulin’s intermediate domain are coupled to GTP hydrolysis upon microtubule polymerization (Geyer et al., 2015; Nogales et al., 1998; Ravelli et al., 2004). In contrast, the effects of mutating residues located in tubulin’s kinesin binding site on microtubule dynamics have not been explored. Our studies reveal a largely unexpected mechanism by which residues located on the surface of the microtubule lattice, distal from the nucleotide-binding site and from longitudinal and lateral contacts between subunits can impact polymerization dynamics.
In current models, ‘curved’ tubulin heterodimers in solution undergo structural transitions during polymerization to adopt a ‘straight’ form within the microtubule lattice. These transitions within newly incorporated tubulin subunits at growing filament ends generate and subsequently destroy Mal3 binding sites (Maurer et al., 2014). Within this framework, we can dissect how TUBB3 mutations impact polymerization dynamics. Our structural studies indicate that the mutations are not likely to cause substantial changes within the microtubule lattice. In addition, the Mal3-GFP binding profiles at growing ends of mutant microtubules are unchanged relative to wild-type when polymerization rates are matched. Therefore, neither the formation nor the loss of Mal3 binding site are likely impacted by these mutations. We find that the D417H mutation in TUBB3 likely reduces the fraction of ‘curved’ versus ‘straight’ tubulin heterodimer relative to wild-type tubulin. As polymerization requires conversion of tubulin from the ‘curved’ to the ‘straight’ conformation at the growing microtubule ends, the higher levels of ‘straight’ heterodimers of mutant tubulin could increase the filament growth rates. Further, the propensity to adopt the ‘straight’ conformation may also account for the observed increase in the lifetime of the end-stabilizing cap and decrease in catastrophe frequencies.
The minimum requirement for preventing the rapid disassembly of a microtubule has been proposed to be a single layer of GTP-bound tubulin subunits at the filament end (Bowne-Anderson et al., 2013; Caplow and Shanks, 1996; Drechsel and Kirschner, 1994). In this model, each of the ~13 protofilaments must be ‘capped’ and the loss of GTP-tubulin at a single protofilament can destabilize the ‘cap’ (Caplow and Shanks, 1996; Drechsel and Kirschner, 1994). Accordingly, we hypothesize that mutant tubulins must form a complete ‘ring’ at the end of the polymer to suppress microtubule catastrophe frequency. When mutant and wild-type tubulin copolymerize, the presence of wild-type tubulin at the ends of a subset of the ~13 protofilaments can lower the stability of the ‘cap’ and thereby increase the catastrophe frequency. Our findings support this hypothesis as we find that adding equal amounts of wild-type tubulin increases the catastrophe frequencies of D417H and R262H microtubules to that measured for wild-type microtubules alone.
In contrast to the all-or-nothing effect on polymerization dynamics, the impact on MAP binding when mixing wild-type tubulin with mutant forms is dose-dependent, as MAP affinities decrease with increasing amounts of mutant tubulins in the polymer. The concentration of tubulin in cells is estimated to be ~10 μM, of which up to ~80% can be in the polymerized form (Olmsted, 1981; Ostlund et al., 1979, 1980). Based on simple binding principles, the fraction of microtubule-bound MAPs that have sub-micromolar binding constants (e.g. kinesin) to wild-type, would be reduced by only a very small amount (~5%) for mutant microtubules. A more substantial difference in the fraction bound to wild-type versus mutant microtubules would be observed for MAPs with weak microtubule-binding affinities (details on these calculations are provided in Experimental Procedures). Therefore, we posit that these mutations in tubulin would cause the associated phenotypes in only those cells where the functions of weakly binding MAPs are critical. Additional cell biological studies are needed to establish if this hypothesis can help explain disease phenotypes (Niwa et al., 2013; Tischfield et al., 2010).
Our findings reveal that disease-related tubulin mutations alter polymerization dynamics at not only the plus-ends but also the minus-ends of microtubules. The regulation of microtubule plus-ends by MAPs, such as the EB (end-binding) proteins, has been extensively studied (Galjart, 2010; Howard and Hyman, 2007). In contrast, the regulation and function of microtubule minus-end polymerization in cells are poorly understood. It has been generally accepted that γ-tubulin caps and stabilizes microtubule minus-ends in cells (Kollman et al., 2011). However, it is now becoming clear that MAPs, such as katanin and spastin, sever existing filaments to remodel microtubule networks (Roll-Mecak and McNally, 2010). The fate of these new minus-ends is unclear and we do not know if and when they are capped and stabilized. One clue comes from recent studies of calmodulin-regulated spectrin-associated proteins (CAMSAPs), MAPs that selectively bind microtubule minus-ends, showing that newly generated minus-ends in human cells are not immediately capped but can grow (Jiang et al., 2014). Further, it has been shown that minus-end binding proteins have important roles in the development and maintenance of axons and dendrites in neurons (Yau et al., 2014). Together, these data suggest that regulation of microtubule minus-end polymerization plays a key role in regulating microtubule organization. It is likely that access to recombinant human tubulin and different mutant forms should help dissect how these microtubule minus-end binding proteins interact with the ‘end-stabilizing cap’ to control microtubule minus-end polymerization in different cellular contexts.
Experimental Procedures
Purification of recombinant human tubulin
The cDNA encoding Homo sapiens α-tubulin 1B (NP_006073.2) and β-tubulin 3 (NP_006077.2) were cloned into pFastBac Dual vector (Life Technologies). For affinity purification, a sequence encoding a Tobacco Etch Virus (TEV) protease site and hexahistidine-tag was fused to the 3′ end of the β-tubulin TUBB3 cDNA sequence. We used the Bac-to-Bac system (Life Technologies) to generate recombinant baculovirus. HiveFive cells (Life Technologies), grown to 3.0–3.5 × 106 cells/ml in Sf-900 II SFM (Life Technologies 10902–096) supplemented with 1X antibiotic-antimyocotic (Life Technologies 15240–062), were infected with P3 viral stocks. Cells were cultured in suspension at 27 °C and harvested at 60 hours after infection. The following steps were done on ice or at 4 °C. We lysed cells in an equal volume of lysis buffer (50 mM HEPES, 20 mM imidazole, 100 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 0.1 mM GTP, 3 U/ml benzonase, 1X protease inhibitor Roche Complete EDTA-free, pH 7.2) by dounce homogenizer (20 strokes) and centrifuged the homogenate at 55,000 rpm in a Ti70 rotor (Beckman Coulter) for 1 hr. The supernatant was then filtered through a 0.22 μm Millex-GP PES membrane (Millipore SLGP033RS) and loaded at 0.8 ml/min onto a 1 ml HisTrap HP column (GE life science 17–5247–01) pre-equilibrated with lysis buffer. The column was washed with 25 ml lysis buffer and then eluted with nickel elution buffer (1X BRB80 (80 mM PIPES, 1mM MgCl2, 1mM EGTA), 500 mM imidazole, 0.1 mM GTP, 1 mM DTT, pH 6.8). The fractions containing proteins were pooled, diluted 6-fold with TOG-column buffer (1X BRB80, 1 mM DTT, 0.2 mM GTP, pH 6.8), mixed with 4 mg TEV protease and incubated for 1 hr on ice. The TEV-digested protein solution was loaded at 0.8 ml/min onto a tandem chromatography consisting of a 1 ml HiTrap SP Sepharose FF column (GE life science 17–5054–01) and a 1 ml TOG-affinity column (Widlund et al., 2012) and washed with 10 ml TOG-column buffer. The 1 ml HiTrap SP Sepharose FF column was removed and the 1 ml TOG-affinity column was washed with 25 ml of wash buffer 1 (1X BRB80 1 mM DTT, 0.1 mM GTP, 10 mM MgCl2, 5 mM ATP, pH 6.8), 25 ml of wash buffer 2 (1X BRB80, 1 mM DTT, 0.1 mM GTP, 0.1 % Tween-20, 10% glycerol, pH 6.8) and 10 ml of TOG-column buffer. The tubulin was eluted with TOG-elution buffer (1X BRB80, 500 mM (NH4)2SO4, 1 mM DTT, 0.2 mM GTP, pH 6.8). The eluate containing tubulin was pooled, quickly exchanged into storage buffer (1X BRB80, 5% glycerol, 1 mM DTT, 0.2 mM GTP, pH 6.8), and concentrated to at least 1.5 mg/ml with an Amicon Ultra 50K MWCO centrifugal filter (Millipore UFC901024). The purified tubulin was snap frozen in liquid nitrogen and stored at −80 °C.
Cryo-EM microscopy
We prepared cryo-EM grids for the analysis of wild-type TUBB3 and TUBB3-D417H microtubules as described previously (Zhang et al., 2015). Details of the experimental procedures are presented in the Supplemental Experimental Procedures.
Microtubule co-sedimentation assay
Taxol stabilized microtubules were polymerized from purified recombinant tubulin. Purified kinesin-1ΔC was spun in TLA120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 4 °C. Pre-clarified PRC1-SC (300 nM) kinesin-1ΔC (200 nM) was incubated with increasing concentrations of microtubules (40 μl reaction volume) for 30 mins at room temperature in assay buffer (1XBRB80, 1mM MgCl2, 5% sucrose, 1 mM TCEP, 0.25 mg/ml BSA and 10 μM Taxol. 2 mM AMPPNP was supplemented in the assays containing kinesin-1ΔC) and then subjected to sedimentation in TLA 120.1 rotor at 90000 rpm for 10 mins at 25 °C (Beckman Coulter). The amount of protein in pellet and supernatant was analyzed by SDS-PAGE and the coomassie-stained bands were quantified (LI-COR Odyssey) to calculate the depletion of PRC1-SC and kinesin-1ΔC from the supernatant, which gives the fraction protein bound. The fraction protein bound (y) from the mean of three independent experiments was plotted against microtubule concentration (x). We used Kaleidagraph to fit the hyperbolic titration curves with the following equation to determine the dissociation constants (Kd).
Microtubule turbidity assay
Solutions of 26 μM tubulin in assay buffer (1X BRB80, 5% (v/v) glycerol, 1 mM GTP and 1 mM TCEP) were prepared and pre-clarified by high-speed centrifugation in TLA120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 4 °C. Pre-clarified tubulin (30 μl reaction volume) was transferred to a warm (37 °C) 348-well plate and each reaction was covered by 15 μl of fluorescence-free immersion oil (Cargille Laboratories Inc. 16212) to prevent evaporation. The absorbance at 350 nm was measured every 15 sec for 3.5 hr at 37 °C.
The binding of allocolchicine to tubulin
Allocolchicine was synthesized by following a published method (Fernholz, 1950). Purified tubulin was pre-clarified by high-speed centrifugation in TLA120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 4 °C. Pre-clarified tubulin (3 μM) was mixed with increasing concentration of allocolchicine (0, 1.3, 2.6, 3.9, 5.2, 7.8, 15.6, 31.2 or 62.4 μM) in assay buffer (1X BRB80, 5% (v/v) glycerol, 1 mM GTP and 1 mM TCEP) to a final 35 μl reaction volume and incubated at room temperature for 2 hr. For each sample, the fluorescence intensities at 400 nm were collected using excitation at 310 nm. The measured fluorescence intensities at 400 nm were plotted against allocolchicine concentration. To determine the affinity of tubulin for allocolchicine, the unnormalized equilibrium binding curves from three independent experiments were fit with the following equation using Kaleidagraph:
where Kd represents the dissociation constant for allocolchicine binding, Fmax represents the fluorescence intensity at plateau, and Fbackground represents the background fluorescence intensity.
Glutathione S-transferase (GST) pull-down assay
Purified tubulin and GST-TOG1/2 was pre-clarified by high-speed centrifugation in TLA120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 4 °C. Solutions of pre-clarified tubulin (0.3 μM) containing 0–5 μM GST-TOG1/2 were prepared in assay buffer (1X BRB80, 5 % (v/v) glycerol, 1 mM GTP and 1 mM TCEP). The reactions (40 μl reaction volume) were incubated at 4 °C for 15 mins. The reactions were then mixed with buffer-equilibrated glutathione magnetic beads (Pierce 88821) at 4 °C for another 1 hr. After sepa rating the supernatant from the magnetic beads using a magnet, 20 μl of the supernatant was mixed with 5 μl of 5X SDS-Sample buffer. The magnetic beads were rinsed twice with 100 μl assay buffer and mixed with 50 μl of 1X SDS-sample buffer. The supernatant and the bead-bound protein fractions were analyzed by SDS-PAGE and band intensities were quantified using ImageJ. To calculate the dissociation constant (Kd), data from three independent experiments were fit to the following equation using Kaleidagraph:
Microtubule sedimentation assay in the presence of colchicine
Purified tubulin was pre-clarified by centrifugation in TLA120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 4 °C. Colchici ne (Sigma C9754) was dissolved in DMSO (>99.7%, Sigma D2650) to prepare 100 mM stock solutions. Solutions (40 μl reaction volume) of pre-clarified tubulin (10 μM) containing 0–35 μM colchicine or 3 % DMSO were prepared in assay buffer (1X BRB80, 33.33% (v/v) glycerol, 1 mM GTP and 1 mM TCEP). The reactions were incubated at room temperature for 30 min, followed by another 30 min incubation at 37 °C, and then subjected to high-speed centrifugation in TLA 120.1 rotor (Beckman Coulter) at 90000 rpm for 10 mins at 30°C. The supe rnatant was carefully aspirated and saved for SDS-PAGE analysis. The pellet was gently rinsed twice with 40 μl warm wash buffer (1X BRB80, 60% (v/v) glycerol and 1 mM TCEP), and then suspended in 1X sample buffer for SDS-PAGE analysis.
Microtubule sedimentation assay in the presence of allocolchicine
The experiments were performed at 13 μM tubulin and 60 μM allocolchicine with the same procedures as described in the assay with colchicine.
Estimating the fraction of microtubule-bound MAPs in vivo
In cells, the concentration of tubulin is ~10 μM and up to 80% can be in the polymerized form. To estimate the fraction of microtubule-bound MAPs in vivo, we used the following simple binding principle that describe the dissociation constant (Kd):
When the number of binding sites on microtubules is much larger than the [MAPs], we make the simplifying assumption that [MT]0-[MAPs_MT]=[MT]0. The equation is then simplified to:
When [MT]0 is 10 μM and the Kd of MAPs for wild-type microtubule is 0.1 μM and for mutant microtubule is 0.5 μM, the fraction of microtubule-bound MAPs on wild-type and mutant filaments is 0.99 and 0.95, respectively.
Supplementary Material
Highlight.
TUBB3 point mutations near the kinesin binding site change polymerization dynamics
Mutations alter dynamic instability parameters and end-stabilizing cap lifetimes
TUBB3 point mutation alters the distribution of tubulin conformations in solution
3.8 Å cryo-EM structures of microtubules with wild-type and mutant human TUBB3
Acknowledgments
This research was supported by the NIH (GM65933, PI: TMK). S.C.T. acknowledges support from the Leukemia & Lymphoma Society (CDP-5307-14). R.E.K. was supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship (DRG-2118-12) and by a Charles H. Revson Foundation Senior Fellowship in Biomedical Science. S.F. was supported by a NIH NRSA fellowship (F32-GM099380). E.N. is a Howard Hughes Medical Institute Investigator. C.D., N.I.C. and T.S. acknowledge support by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust. C.D. and T.S. also acknowledge funding from FP7 ERC grant 323042. We also thank Brain Chait (Rockefeller University) for access to mass spectrometry instruments and Luke Rice (UT Southwestern) for helpful discussions.
Footnotes
Author contributions
S.C.T., M.C.P. and T.M.K. designed experiments and prepared the manuscript. S.C.T., R.E.K. and M.C.P. generated and characterized recombinant tubulins. S.C.T. and M.C.P. examined polymerization dynamics and binding to MAPs other than TOG domain and Mal3. R.E.K did the mass spectrometry analyses. S.F. assisted with the quantification of dynamic instability parameters. S.C.T. did experiments involving colchicine and TOG domains. S.C.H. and E.N. carried out the cryo-EM studies. C.D. and N.I.C. and T.S. did the experiments involving Mal3.
Accession numbers
Cryo-EM maps and atomic models for the wild-type TUBB3 microtubules (EMDB: 8094; PDB: 5IJ0) and the TUBB3-D417H mutant microtubules (EMDB:8095; PDB:5IJ9) have been deposited.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Steinmetz MO. Microtubule +TIPs at a glance. Journal of cell science. 2010;123:3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. The Journal of cell biology. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Munyoki S, Geyer EA, Piedra FA, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, Rice LM. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife. 2014;3:e03069. doi: 10.7554/eLife.03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne-Anderson H, Zanic M, Kauer M, Howard J. Microtubule dynamic instability: a new model with coupled GTP hydrolysis and multistep catastrophe. Bioessays. 2013;35:452–461. doi: 10.1002/bies.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM. The contribution of alphabeta-tubulin curvature to microtubule dynamics. The Journal of cell biology. 2014;207:323–334. doi: 10.1083/jcb.201407095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplow M, Shanks J. Evidence that a single monolayer tubulin-GTP cap is both necessary and sufficient to stabilize microtubules. Molecular biology of the cell. 1996;7:663–675. doi: 10.1091/mbc.7.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Hill TL, Chen Y. Interference of GTP hydrolysis in the mechanism of microtubule assembly: an experimental study. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:771–775. doi: 10.1073/pnas.81.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annual review of cell and developmental biology. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Kirschner MW. The minimum GTP cap required to stabilize microtubules. Current biology : CB. 1994;4:1053–1061. doi: 10.1016/s0960-9822(00)00243-8. [DOI] [PubMed] [Google Scholar]
- Fernholz H. *Uber Die Umlagerung Des Colchicins Mit - Natriumalkoholat Und Die Struktur Des Ringes-C. Liebigs Ann Chem. 1950;568:63–72. [Google Scholar]
- Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Current biology : CB. 2010;20:R528–537. doi: 10.1016/j.cub.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Gaskin F, Cantor CR, Shelanski ML. Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J Mol Biol. 1974;89:737–755. doi: 10.1016/0022-2836(74)90048-5. [DOI] [PubMed] [Google Scholar]
- Gell C, Friel CT, Borgonovo B, Drechsel DN, Hyman AA, Howard J. Purification of tubulin from porcine brain. Methods in molecular biology. 2011;777:15–28. doi: 10.1007/978-1-61779-252-6_2. [DOI] [PubMed] [Google Scholar]
- Geyer EA, Burns A, Lalonde BA, Ye X, Piedra FA, Huffaker TC, Rice LM. A mutation uncouples the tubulin conformational and GTPase cycles, revealing allosteric control of microtubule dynamics. Elife. 2015;4 doi: 10.7554/eLife.10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Pluckthun A, Wang C, Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nature structural & molecular biology. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Haren L, Merdes A. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. Journal of cell science. 2002;115:1815–1824. doi: 10.1242/jcs.115.9.1815. [DOI] [PubMed] [Google Scholar]
- Heald R, Khodjakov A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. The Journal of cell biology. 2015;211:1103–1111. doi: 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nature reviews Molecular cell biology. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Current opinion in cell biology. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AF, Heck AJ, Hoogenraad CC, et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Jiang YQ, Oblinger MM. Differential regulation of beta III and other tubulin genes during peripheral and central neuron development. Journal of cell science. 1992;103(Pt 3):643–651. doi: 10.1242/jcs.103.3.643. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Hoogenraad CC. Building the Neuronal Microtubule Cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nature reviews Molecular cell biology. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludueña RF, Banerjee A. The Isotypes of Tubulin. In: Fojo T, editor. The Role of Microtubules in Cell Biology, Neurobiology, and Oncology. Humana Press; 2008. pp. 123–175. [Google Scholar]
- Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Current biology : CB. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S, Muto E. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 2013;587:3450–3455. doi: 10.1016/j.febslet.2013.08.032. [DOI] [PubMed] [Google Scholar]
- Mohan R, Katrukha EA, Doodhi H, Smal I, Meijering E, Kapitein LC, Steinmetz MO, Akhmanova A. End-binding proteins sensitize microtubules to the action of microtubule-targeting agents. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8900–8905. doi: 10.1073/pnas.1300395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Takahashi H, Hirokawa N. beta-Tubulin mutations that cause severe neuropathies disrupt axonal transport. The EMBO journal. 2013;32:1352–1364. doi: 10.1038/emboj.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nogales E, Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr Opin Struct Biol. 2016;37:90–96. doi: 10.1016/j.sbi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB. Tubulin pools in differentiating neuroblastoma cells. The Journal of cell biology. 1981;89:418–423. doi: 10.1083/jcb.89.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa F. Size distribution of protein polymers. J Theor Biol. 1970;27:69–86. doi: 10.1016/0022-5193(70)90129-3. [DOI] [PubMed] [Google Scholar]
- Ostlund RE, Jr, Leung JT, Hajek SV. Biochemical determination of tubulin-microtubule equilibrium in cultured cells. Analytical biochemistry. 1979;96:155–164. doi: 10.1016/0003-2697(79)90568-2. [DOI] [PubMed] [Google Scholar]
- Ostlund RE, Jr, Leung JT, Hajek SV. Regulation of microtubule assembly in cultured fibroblasts. The Journal of cell biology. 1980;85:386–391. doi: 10.1083/jcb.85.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Current opinion in cell biology. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Cederquist GY, Gupta ML, Jr, Engle EC. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr Opin Genet Dev. 2011;21:286–294. doi: 10.1016/j.gde.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annual review of cell and developmental biology. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. The Journal of cell biology. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RA, Pryer NK, Salmon ED. Dilution of individual microtubules observed in real time in vitro: evidence that cap size is small and independent of elongation rate. The Journal of cell biology. 1991;114:73–81. doi: 10.1083/jcb.114.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, Drechsel DN. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Molecular biology of the cell. 2012;23:4393–4401. doi: 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nature cell biology. 2015;17:907–916. doi: 10.1038/ncb3188. [DOI] [PubMed] [Google Scholar]
- Yau KW, van Beuningen SF, Cunha-Ferreira I, Cloin BM, van Battum EY, Will L, Schatzle P, Tas RP, van Krugten J, Katrukha EA, et al. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 2014;82:1058–1073. doi: 10.1016/j.neuron.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.