ABSTRACT
Tremendous insight into actin-associated proteins has come from careful biochemical and cell biological characterization of their activities and regulation. However, many studies of their cellular behavior have only considered each in isolation. Recent efforts reveal that assembly factors compete for polymerization-competent actin monomers, suggesting that actin is homeostatically regulated. It seems that a major regulatory component is competition between Arp2/3-activating nucleation promoting factors and profilin for actin monomers. The result is differential delivery of actin to different pathways, allowing for simultaneous assembly of competing F-actin structures and collaborative building of higher order cellular structures. Although there are likely to be additional factors that regulate actin homeostasis, especially in a cell type-dependent fashion, we advance the notion that competition between actin assembly factors results in a tunable system that can be adjusted according to extracellular and intracellular cues.
Keywords: actin, Arp2/3 complex, Ena/VASP, formin, motility, profilin
Dynamic actin-based structures play important roles in nearly every aspect of eukaryotic life such as motility, vesicular trafficking and cell division. Though monomeric actin (‘G-actin’) above the critical concentration can spontaneously assemble actin filaments (‘F-actin’) in vitro, it has been appreciated for many years that specific actin assembly factors regulate the generation of F-actin in cells.1 Among these factors is the Arp2/3 complex, which nucleates branched ‘daughter’ filaments from the sides of pre-existing ‘mother’ filaments, giving rise to the branched dendritic networks found at the lamellipodia, phagocytic cup, and endocytic patches.2 Nucleation promoting factors (NPFs) are required for Arp2/3 complex activation and also deliver the initial actin monomers that trigger daughter filament nucleation.1 Although the Arp2/3 complex is the only assembly factor known to generate this type of branched actin, many classes of actin assembly factors generate and/or elongate unbranched actin, including the Ena/VASP,3 formin4 and spire5 protein families. These various nucleation/assembly factors contribute to specific F-actin structures. Fission yeast provides a good example, where the Arp2/3 complex generates endocytic actin patches while formins assemble actin cables and the contractile ring.6 F-actin in mammalian cells is not so starkly segregated, with certain formin7-9 and Ena/VASP10,11 proteins contributing to stress fibers and filopodia while the Arp2/3 complex generates the bulk of the lamellipodial dendritic actin12 (Fig. 1A). In yeast as well as mammals, actin assembly factors are active in the same cytoplasm, and their activities must be precisely balanced to allow normal cellular responses to occur. The biochemical and structural mechanisms of all of these factors, and their contribution to cellular function have been well studied separately, but how they work together in a common cytoplasm is poorly understood.
Figure 1.
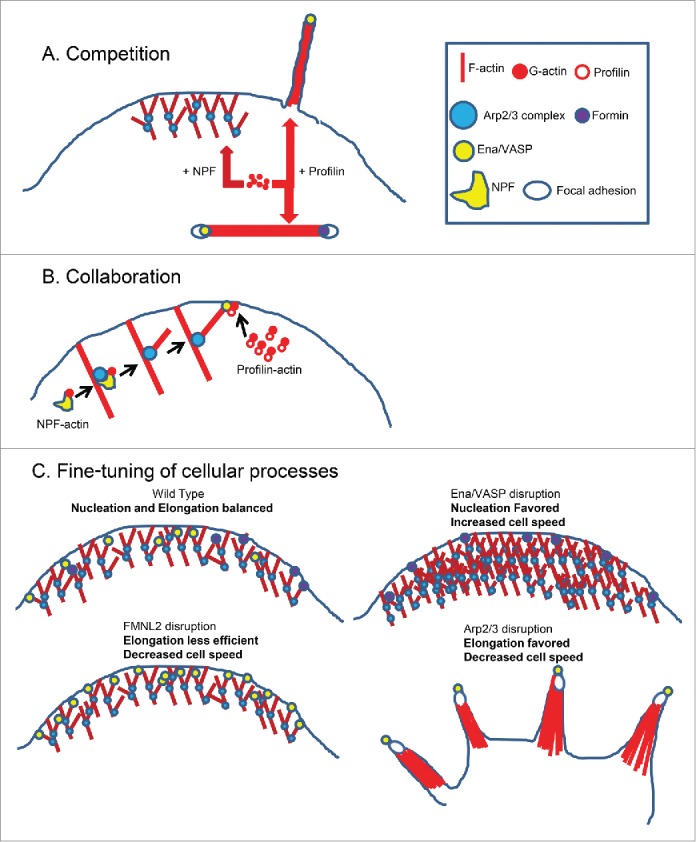
Functional consequences of monomer competition by actin assembly pathways. (A) Monomer competition allows for simultaneous assembly of diverse actin filament populations in the same cytoplasm. NPF-bound actin promotes dendritic actin nucleation and generation of lamellipodia while profilin-actin promotes stress fiber and filopodia elongation. (B) Homeostatic regulation of actin monomers allows distinct assembly pathways to collaboratively build cellular structures. In this example, NPF-actin promotes Arp2/3 complex branch nucleation while Ena/VASP is recruited to the daughter filament barbed end to elongate the growing filament using profilin-actin. (C) Collaboration and competition between actin assembly pathways may fine tune cellular processes by regulating the balance between nucleation of new filaments and elongation of existing filaments. Wild Type cells strike a balance between nucleation and elongation, while being capable of enhancing overall motility upon stimulation. Ena/VASP disruption leads to a denser dendritic actin network, shorter actin branches and enhanced cell motility speed, suggesting that nucleation is favored over branch elongation. Conversely, disruption of the formin FMNL2 makes branch elongation less efficient leading to decreased cell motility speed. Finally, disruption of Arp2/3 complex activity leads to loss of the branched actin network and a shift toward profilin-actin dependence and generation of long, unbranched protrusions containing Ena/VASP and focal adhesion proteins.
The emerging view of the actin cytoskeleton is that it is homeostatically regulated and that all assembly factors in the cell must compete for a finite pool of monomeric actin. Filamentous actin and monomeric actin exist simultaneously in cells, implying an F-actin:G-actin ratio that must be remarkably constant, but could be expected to shift according to extracellular or intracellular cues. As described above, actin is polymerized into actin filaments of varying geometries by assembly factors, crosslinking proteins and monomer-binding proteins (e.g. profilin). All of these processes utilize a common cellular pool of G-actin to make actin filaments. It has long been appreciated that cellular G-actin levels are maintained through an autoregulatory feedback mechanism.13 Furthermore, it has been shown that the G-actin pool can homeostatically regulate cellular signaling based on its size relative to F-actin,14 indicating the importance of a tightly regulated F-actin: G-actin ratio. Insights generated in fission yeast further support the idea that actin assembly is homeostatic. When the Arp2/3 complex is disrupted in this system, endocytic patches are compromised, as expected.15 Surprisingly, actin cable formation is also enhanced, via a compensatory increase in formin activity.15 These data illustrate that actin exists in a finite assembly-competent pool, and that the activities of competing assembly pathways are precisely balanced under normal conditions.
Our studies in mammalian cells reinforced the notion that actin is homeostatically regulated. Genetic ablation of Arp2/3 complex function in fibroblasts, via conditional deletion of the Arpc2 gene (denoted Arpc2−/−), abolished lamellipodia and dramatically changed F-actin organization.16 Arpc2−/− cells protruded via bundled filopodial-like structures that contained core focal adhesion proteins (vinculin), actin assembly factors (Mena, VASP) and bundled actin filaments. Although the organization of actin filaments was dramatically changed, total F-actin levels remained unchanged in Arpc2−/− cells.16 Both formins and Ena/VASP proteins elongate actin filaments more efficiently using profilin-actin,17-20 so we reasoned that disrupting profilin-1 expression via shRNA-mediated knockdown would disrupt both assembly pathways. Profilin-1 depleted Arpc2−/− cells have a significantly reduced spread cell area and a dramatically reduced F-actin level compared to profilin-containing Aprc2−/− cells.16 To our surprise, Arpc2−/− cells treated with the pan-formin inhibitor SMIFH221 maintained overall F-actin levels comparable to both untreated Arpc2−/− and WT cells.16 However, when we disrupted Ena/VASP function via intracellular sequestration (FP4-mito22) in Arpc2−/− cells, there was a striking reduction in both F-actin levels and spread cell area.16 Based on these data, we identified Ena/VASP proteins as the fundamental compensatory mechanism in Arpc2−/− cells, as well as establishing that F-actin in these cells required profilin-bound actin. It is perhaps not so surprising that the cell has a means of compensating for loss of a major cytoskeletal regulator, though we expect to find cell types and/or contexts where compensation does not occur or occurs by different mechanisms.
Upon examining our data further, a second idea emerged based on our observation that profilin-1 depletion in otherwise normal WT cells elicited changes consistent with enhanced Arp2/3 complex function. These included a larger lamellipodia, higher levels of Arp2/3 complex at the cell edge, and a wider band of both actin and Arp2/3 complex at the lamellipodia.16 Important observations made via in vitro TIRF assays,23 biochemical studies24,25 and microinjection26 experiments through the years indicated that the Arp2/3 complex nucleates daughter filaments less efficiently in the presence of profilin-actin. With these studies in mind, we wondered whether increasing profilin levels would introduce changes consistent with Arp2/3 complex inhibition. Our cells were not amenable to stable profilin overexpression so instead we acutely increased profilin-1 concentration via microinjection. Exogenous profilin disrupted lamellipodia and decreased Arp2/3 complex localization to the cell periphery long after the initial injection.16 We also observed that the F-actin generated in response to profilin microinjection tended to be in the form of stress fibers rather than protrusive structures.16 Together these data imply a redistribution of monomeric actin from the lamellipodia to internal stress fibers. Interestingly, Arpc2−/− cells were largely resistant to profilin microinjection, as expected, since it is already strongly reliant upon profilin-actin to generate F-actin.16
In a companion paper, Suarez et al. found that antagonism between profilin and the Arp2/3 complex regulated actin monomer usage and, therefore, the balance between formin- and Arp2/3 complex-generated actin structures. They showed that fission yeast overexpressing profilin could not form Arp2/3 complex-dependent endocytic patches but still competently generated formin-dependent actin structures.27 Conversely, they showed that increasing actin (and thereby decreasing the relative amount of profilin-actin) enhanced patch formation and blocked formin-generated structures.27 These experiments, together with our studies in fibroblasts, supported the overall insight that profilin and the Arp2/3 complex are mutually antagonistic.
With these results in hand, the authors of this study used elegant TIRF microscopy and biochemical assays to gain greater mechanistic insight into this relationship. They confirmed that profilin suppresses Arp2/3 complex branch nucleation and that this behavior was dependent only upon profilin's ability to bind free monomeric actin.27 However, purified formin could only robustly generate F-actin in the presence of the Arp2/3 complex if profilin was present.27 Furthermore, both profilin's poly-proline binding and actin-binding activities were required for formin activity in these assays.27 Finally, and importantly, the authors also demonstrated that nucleation promoting factors (NPFs) and profilin compete with one another for free actin.27 It is noteworthy that data generated from in vitro TIRF microscopy, fission yeast, and mammalian cell culture have all converged to argue for an antagonistic relationship between profilin and the Arp2/3 complex.
Our work and that of Suarez et al. indicate that the competition between profilin and NPFs for actin monomers is an ancient and fundamental mechanism that solves the problem of dividing the pool of actin monomers among distinct F-actin assembly pathways (Fig. 1A). However, yeast lack Ena/VASP homologues and respond to loss of Arp2/3 complex function by inducing formin-based actin assembly,15 while F-actin in our spread Arpc2−/− cells is largely indifferent to the formin inhibitor SMIFH2.16 These findings argue that each model system, and potentially each cell type, possesses a unique strategy for distributing actin monomers. Though there are generalizable lessons to be learned about the interplay among actin regulatory proteins, the field will need to bear in mind that the insights generated from different cells and contexts may yield slightly different results.
So what advantage does monomer competition confer to eukaryotic cells? The most obvious interpretation of these findings suggests that different pools of polymerization competent monomers can be added differentially to distinct F-actin populations (Fig. 1A). Keeping these pathways segregated based on the type of actin required for optimal activity (e.g., NPF- or profilin-bound) confers specificity to each process as well as reserving a subpopulation of the total actin monomer pool for assembly by a given pathway. The cellular outcome of this is that distinct pools of F-actin can be built simultaneously by competing actin assembly pathways in the same cytoplasm.
The second potential consequence of monomer competition is that it allows branched and unbranched actin assembly pathways to operate collaboratively in the same cellular compartment (Fig. 1B). It is tempting to speculate that competition between profilin and NPFs for actin monomers may tune the ratio of branch elongation to branch nucleation. The high local concentration of profilin feeding into these elongating branches may effectively block further NPF activity after the initial nucleation event, thereby spatially constraining the actively elongating actin filaments from daughter filament nucleation. The lamellipodial actin network is mainly composed of branched actin polymerized by the Arp2/3 complex.12 However, both Ena/VASP28,29 and the formin FMNL230 have also been shown to localize to the lamellipodial edge. FMNL2 has been observed via in vitro TIRFM to elongate Arp2/3 complex-generated daughter filaments in a profilin-dependent fashion.30 Ena/VASP proteins antagonize capping protein, a relationship that affects both the length and density of actin filaments in the dendritic network.28 Recent work investigating the regulation of actin dynamics in dendritic spines revealed that the Arp2/3 complex and VASP/FMNL2 are spatially segregated,31 supporting a model wherein the former nucleates new actin filaments and the latter factors elongate the resulting daughter filaments. Presumably both activities are required to form dendritic spines of the correct size and shape.
Finally, monomer competition may allow for the fine-tuning of cellular processes (Fig. 1C). Lamellipodial protrusion rates can vary dramatically between cell types, or within the same cell when faced with different contextual cues. For example, keratocytes protrude much more rapidly (2.4 microns/min)32 compared to fibroblasts (ranging from 0.08 to 0.89 microns/min),32,33 despite the structure and function of the lamellipodia being nearly identical across much of the metazoan lineage. Disruption of Ena/VASP activity yields a denser dendritic network with shorter filaments, and cell speed is enhanced.28 Conversely, disruption of FMNL2 negatively impacts lamellipodial protrusion and cell speed.30 As branch nucleation requires NPF-actin and filament elongation is more efficiently accomplished by profilin-actin, subtle alterations in the balance of NPF-bound actin to profilin-bound actin may fine-tune protrusion rates by adjusting the balance of filament nucleation and elongation at the leading edge. The observations yielded by Suarez et al.27 and Rotty et al.16 suggest that this balance is tunable, and that the set point may be different in distinct cell types based on the operant factors in a particular cell. Since we know that a balance is struck between profilin- and NPF-dependent actin assembly pathways in these systems, a fascinating future direction will be defining the mechanical, cytoskeletal and signaling pathways that collaborate to tune the balance between them.
Our data agrees well with the emerging view of actin as a homeostatic system. However, our work has focused exclusively upon actin assembly, and we recognize that other factors will also affect actin homeostasis. When discussing dynamic actin structures, it is important to realize that assembly and disassembly are fundamentally linked. F-actin disassembly and/or severing via ADF/cofilin yields polymerization competent monomers12,34 as well as yielding free polymerization-competent barbed ends.35,36 The role of the cofilin phosphatase Slingshot37,38 and the coronin39 family of proteins may be major factors in sensing F-actin and in regulating the size of the G-actin pool. Furthermore, the activities of cofilin and the Arp2/3 complex are synergistic34,40 and both are required for proper lamellipodia function,41-43 so it seems that polymerization-competent actin generated by disassembly near membranes may be primed for the nucleation of new daughter filaments. In fact, this ‘recycling’ has recently been directly observed.44 In addition to disassembly, F-actin bundling (via fascin, filamin, α-actinin, etc.), myosin contractility and actin monomer sequestration (via thymosin-β4) can all be expected to affect G-actin levels and F-actin production. Indeed, thymosin- β4-bound actin has been shown to contribute differentially to the lamellipodial actin network through formins, revealing an additional axis of competition for G-actin.44 Furthermore, certain isoforms of the F-actin binding protein tropomyosin increase F-actin levels in adipose cells, indicating that the F-actin to G-actin ratio can potentially be modulated by F-actin binding proteins as well as actin assembly factors.45 However, this effect may depend upon the ability of different tropomyosin isoforms to bind preferentially to F-actin generated by distinct formin isoforms.46 This combined activity seems to impart specific characteristics to the resulting tropomyosin-bound actin filament, implying that not all actin filaments are functionally equivalent (for more extensive review, see Gunning et al.47). Each of these actin-binding proteins is itself regulated by upstream signaling pathways, as well as spatial cues that recruit them to particular locations within the cell. Many of these factors will vary (in expression as well as activity) based on the type of cell being assayed. Understanding the actin cytoskeleton at a systematic level represents both a challenge and opportunity for the field moving forward. There can be little doubt that a global understanding of actin regulation will yield significant cell biological insights, as well as pointing us toward the interactions, crosstalk and feedback mechanisms that might be leveraged for the treatment of pathophysiological conditions.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
FUNDING
This work was supported by a NIH grant to JEB (GM111557) and support from HHMI to JEB. JDR was supported by a NIH NRSA fellowship (GM109664).
References
- 1.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 2010; 11:237-51; PMID:20237478; http://dx.doi.org/ 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 2013; 14:7-12; PMID:23212475; http://dx.doi.org/ 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- 3.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol 2003; 19:541-64; PMID:14570581; http://dx.doi.org/ 10.1146/annurev.cellbio.19.050103.103356 [DOI] [PubMed] [Google Scholar]
- 4.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci 2013; 126:1-7; PMID:23516326; http://dx.doi.org/ 10.1242/jcs.107250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renault L, Bugyi B, Carlier M-F. Spire and Cordon-bleu: multifunctional regulators of actin dynamics. Trends Cell Biol 2008; 18:494-504; PMID:18774717; http://dx.doi.org/ 10.1016/j.tcb.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 6.Kovar DR, Sirotkin V, Lord M. Three's company: The fission yeast actin cytoskeleton. Trends Cell Biol 2011; 21:177-87; PMID:21145239; http://dx.doi.org/ 10.1016/j.tcb.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol 2005; 7:619-25; PMID:15908944; http://dx.doi.org/ 10.1038/ncb1266 [DOI] [PubMed] [Google Scholar]
- 8.Skau CT, Plotnikov SV, Doyle AD, Waterman CM. Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly. Proc Natl Acad Sci U S A 2015; 112:E2447-56; PMID:25918420; http://dx.doi.org/ 10.1073/pnas.1505035112 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol 2006; 173:383-94; PMID:16651381; http://dx.doi.org/ 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a Relative of VASP and Drosophila Enabled, Is Implicated in the Control of Microfilament Dynamics. Cell 1996; 87:227-39; PMID:8861907; http://dx.doi.org/ 10.1016/S0092-8674(00)81341-0 [DOI] [PubMed] [Google Scholar]
- 11.Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena Is Required for Neurulation and Commissure Formation. Neuron 1999; 22:313-25; PMID:10069337; http://dx.doi.org/ 10.1016/S0896-6273(00)81092-2 [DOI] [PubMed] [Google Scholar]
- 12.Svitkina TM. Arp2/3 Complex and Actin Depolymerizing Factor/Cofilin in Dendritic Organization and Treadmilling of Actin Filament Array in Lamellipodia. J Cell Biol 1999; 145:1009-26; PMID:10352018; http://dx.doi.org/ 10.1083/jcb.145.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leavitt J, Ng SY, Aebi U, Varma M, Latter G, Burbeck S, Kedes L, Gunning P. Expression of transfected mutant beta-actin genes: alterations of cell morphology and evidence for autoregulation in actin pools. Mol Cell Biol 1987; 7:2457-66; PMID:3614198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell 2002; 13:4167-78; PMID:12475943; http://dx.doi.org/ 10.1091/mbc.02-05-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol 2014; 24:579-85; PMID:24560576; http://dx.doi.org/ 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell 2015; 32:54-67; PMID:25543281; http://dx.doi.org/ 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol 2002; 4:626-31; PMID:12134165 [DOI] [PubMed] [Google Scholar]
- 18.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol 2002; 4:260-9; PMID:11875440; http://dx.doi.org/ 10.1038/ncb718 [DOI] [PubMed] [Google Scholar]
- 19.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 2006; 124:423-35; PMID:16439214; http://dx.doi.org/ 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- 20.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 2010; 191:571-84; PMID:21041447; http://dx.doi.org/ 10.1083/jcb.201003014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol 2009; 16:1158-68; PMID:19942139; http://dx.doi.org/ 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB. Negative Regulation of Fibroblast Motility by Ena/VASP Proteins. Cell 2000; 101:717-28; PMID:10892743; http://dx.doi.org/ 10.1016/S0092-8674(00)80884-3 [DOI] [PubMed] [Google Scholar]
- 23.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 2000; 404:1007-11; PMID:10801131; http://dx.doi.org/ 10.1038/35010008 [DOI] [PubMed] [Google Scholar]
- 24.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A 1999; 96:3739-44; PMID:10097107; http://dx.doi.org/ 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol 2003; 13:1000-8; PMID:12814545; http://dx.doi.org/ 10.1016/S0960-9822(03)00383-X [DOI] [PubMed] [Google Scholar]
- 26.Cao L, Babcock GC, Rubeinstein PA, Wang Y. Effects of profilin and profilinactin on antin structure and function in living cells. J Cell Biol 1992; 117:1023-9; PMID:1577865; http://dx.doi.org/ 10.1083/jcb.117.5.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell 2015; 32:43-53; PMID:25543282; http://dx.doi.org/ 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al.. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 2002; 109:509-21; PMID:12086607; http://dx.doi.org/ 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- 29.Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell Biol 2003; 13:386-92; PMID:12837609; http://dx.doi.org/ 10.1016/S0962-8924(03)00130-2 [DOI] [PubMed] [Google Scholar]
- 30.Block J, Breitsprecher D, Kühn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al.. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol 2012; 22:1005-12; PMID:22608513; http://dx.doi.org/ 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, et al.. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J 2014; 33:2745-64; PMID:25293574; http://dx.doi.org/ 10.15252/embj.201488837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theriot JA, Mitchison TJ. Comparison of actin and cell surface dynamics in motile fibroblasts. J Cell Biol 1992; 119:367-77; PMID:1400580; http://dx.doi.org/ 10.1083/jcb.119.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cramer LP. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr Biol 1999; 9:1095-105; PMID:10531004; http://dx.doi.org/ 10.1016/S0960-9822(99)80478-3 [DOI] [PubMed] [Google Scholar]
- 34.Ichetovkin I, Grant W, Condeelis J. Cofilin Produces Newly Polymerized Actin Filaments that Are Preferred for Dendritic Nucleation by the Arp2/3 Complex. Curr Biol 2002; 12:79-84; PMID:11790308; http://dx.doi.org/ 10.1016/S0960-9822(01)00629-7 [DOI] [PubMed] [Google Scholar]
- 35.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol 2007; 365:1350-8; PMID:17134718; http://dx.doi.org/ 10.1016/j.jmb.2006.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrianantoandro E, Pollard TD. Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin. Mol Cell 2006; 24:13-23; PMID:17018289; http://dx.doi.org/ 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 37.Howell M, Brickner H, Delorme-Walker VD, Choi J, Saffin J-M, Miller D, Panopoulos A, DerMardirossian C, Fotedar A, Margolis RL, et al.. WISp39 binds phosphorylated Coronin 1B to regulate Arp2/3 localization and Cofilin-dependent motility. J Cell Biol 2015; 208:961-74; PMID:25800056; http://dx.doi.org/ 10.1083/jcb.201410095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of Actin Reorganization by Slingshot, a Family of Phosphatases that Dephosphorylate ADF/Cofilin. Cell 2002; 108:233-46; PMID:11832213; http://dx.doi.org/ 10.1016/S0092-8674(01)00638-9 [DOI] [PubMed] [Google Scholar]
- 39.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B Coordinates Arp2/3 Complex and Cofilin Activities at the Leading Edge. Cell 2007; 128:915-29; PMID:17350576; http://dx.doi.org/ 10.1016/j.cell.2007.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci 2004; 117:3499-510; PMID:15252126; http://dx.doi.org/ 10.1242/jcs.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machesky LM, Insall RH. Scar1 and the related Wiskott–Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol 1998; 8:1347-56; PMID:9889097; http://dx.doi.org/ 10.1016/S0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- 42.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 2012; 148:973-87; PMID:22385962; http://dx.doi.org/ 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol 2007; 177:465-76; PMID:17470633; http://dx.doi.org/ 10.1083/jcb.200610005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitriol EA, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ. Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. Cell Rep 2015; 11:433-45; PMID:25865895; http://dx.doi.org/ 10.1016/j.celrep.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kee AJ, Yang L, Lucas CA, Greenberg MJ, Martel N, Leong GM, Hughes WE, Cooney GJ, James DE, Ostap EM, et al.. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic 2015; 16:691-711; PMID:25783006; http://dx.doi.org/ 10.1111/tra.12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson M, East DA, Mulvihill DP. Formins determine the functional properties of actin filaments in yeast. Curr Biol 2014; 24:1525-30; PMID:24954052; http://dx.doi.org/ 10.1016/j.cub.2014.05.034 [DOI] [PubMed] [Google Scholar]
- 47.Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci 2015; 128:2965-74; PMID:26240174; http://dx.doi.org/ 10.1242/jcs.172502 [DOI] [PubMed] [Google Scholar]


