ABSTRACT
Mitosis entails the bona fide segregation of duplicated chromosomes. This process is accomplished by the attachment of kinetochores on chromosomes to microtubules (MTs) of the mitotic spindle. Once the appropriate attachment is achieved, the spindle assembly checkpoint (SAC) that delays the premature onset of anaphase needs to be silenced for the cell to proceed to anaphase and cytokinesis. Therefore, while it is imperative to preserve the SAC when kinetochores are unattached, it is of paramount importance that SAC components are removed post kinetochore microtubule (kMT) attachment. Precise knowledge of how kMT attachments trigger the removal of SAC components from kinetochores or how the checkpoint proteins feedback in to the attachment machinery remains elusive. This review aims to describe the recent advances that provide an insight into the interplay of molecular events that coordinate and regulate the SAC activity in response to kMT attachment during cell division.
Keywords: checkpoint, kinetochore, KMN, Knl1, mitosis, microtubule, Ndc80, spindle
Abbreviations
- SAC
spindle assembly checkpoint
- kMT
kinetochore microtubule
- MT(s)
microtubule(s)
- Knl1
Kinetochore null 1
- Ndc80
Nuclear division cycle 80
- Mis12
Missegregation 12
- PP
protein phosphatase
- KMN
Knl1-Mis12-Ndc80
- RZZ
Rod1-ZW10-Zwilch
- APC/C
Anaphase promoting complex/cyclosome
- MCC
Mitotic checkpoint complex
- BUB
budding uninhibited by benomyl
- Mps1
Monopolar spindle 1
- CH
Calponin homology
- Mad
Mitotic arrest deficient.
Introduction
Centromeres are discrete compact loci on chromosomes that function as a scaffold for the assembly of mega-molecular protein complexes called kinetochores at the onset of mitosis. The kinetochores comprise of 2 regions; the inner kinetochore that adheres tightly to the centromeric DNA via the highly conserved specialized histone H3 variant (called CENP-A or CenH3) and the outer kinetochore which binds to microtubules (MTs) and facilitates proper chromosome alignment and segregation.1 Certain kinetochore proteins function in the defense against erroneous kinetochore microtubule (kMT) attachments thereby playing a pivotal role in maintaining the fidelity of mitosis.2,3 The outer kinetochore is a dynamic hub comprising of a diverse array of proteins many of which form distinct, evolutionary conserved complexes.1,4,5 The regulatory proteins at kinetochores are involved in circumventing chromosome segregation errors in 2 ways: by facilitating selective stabilization of the bi-oriented kinetochore pairs and by simultaneously destabilizing and eliminating erroneous attachments.6 Besides mediating attachment to the microtubules, kinetochores also orchestrate a highly sophisticated and complex signaling cascade called the spindle assembly checkpoint (SAC), a quintessential fail-safe surveillance mechanism that ensures immaculate chromosomal segregation.6,7
The KMN network - structural and functional components
One of the best studied assembly of kinetochore protein complexes is the KMN network (Knl1, Mis12, and Ndc80), which is vital for the formation of microtubule (MT) binding interface at the mitotic kinetochores.1,3 The components of the KMN network accumulate on kinetochores in prophase and remain stable for the entire duration of mitosis. This is in contrast to the spindle checkpoint components, which are assembled at the kinetochore in high concentrations in the absence of MTs, and their concentration decreases as the number of attached MTs to the kinetochore increases.1,8 The primary activities that are thus attributed to the KMN network are: microtubule attachment and the recruitment of essential SAC components.1,3,5,6 In fact the kinetochore acts as a conduit that can rapidly respond to the changes in microtubule occupancy and effectively switch between a SAC “on-off” state. In this review, we attempt to summarize the recent advances made in understanding the signaling correlates between the kMT attachment status and SAC regulation.
The Mis12 complex
The 22 nm long, rod shaped Mis12 complex, constituting of 4 subunits, Nnf1, Mis12, Dsn1 and Nsl1 is the fundamental platform for the assembly of outer kinetochore and serves to connect the latter to the inner centromeric DNA.5,6 The C-terminal tail of Nsl1 tethers to the C-termini of both Knl1 and the Spc24/25 subunits of the Ndc80 complex thereby recruiting these 2 components of the KMN network to kinetochores. Although, the Mis12 complex has not been identified to interact with MTs, it has been shown to enhance the MT-binding affinities of the Ndc80 and Knl1 complexes.9 Considering the indispensable role of Mis12 in recruiting other MT-binding proteins to the site of kMT attachment, disruption of its function has been associated with defective chromosomal alignment and segregation.10
The Knl1 complex
The Knl1 complex comprises of 2 subunits: Knl1 and Zwint1. Knl1 is the largest protein of the KMN network that has been linked to 3 crucial mitotic processes: kinetochore assembly, chromosomal segregation and SAC signaling.4-6 The C-terminal globular domain harbors tandem RWD motifs that mediate its interaction with the Mis12 complex, while the coiled coil domain is involved in recruiting Zwint1 (ZW10-interacting protein 1), both of which are essential for proper kinetochore function.5,11 The extreme N-terminus of Knl1 has been shown to possess several other motifs, including RVSF (RVXF) and SILK (S/GILK) that are important in recruiting protein phosphatase 1 (PP1). Aurora B kinase has been implicated in destabilizing the kMT attachment by phosphorylating multiple sites on the KMN network. The recruitment of PP1 phosphatase on the afore-mentioned sites therefore neutralizes the Aurora B kinase activity.12 This region also harbors a stretch of 9 basic amino acid residues that display MT-binding activity.9,13 Importantly, although Knl1 has been shown to directly bind to MTs in vitro, its role in forming force generating end-on kMT attachments has not yet been demonstrated.12
The Ndc80 complex
The Ndc80 complex is the most exhaustively studied component of the KMN network. It is a heterotetramer comprising of Ndc80 or Hec1, Nuf2, Spc24 and Spc25. The Calponin Homology (CH) domain and the charged N-terminal tail of Ndc80 have been attributed to mediate tight interactions of Ndc80 with the negatively charged C-termini of tubulin monomers.3,6 This interaction is debilitated by the multiple Aurora B kinase-mediated phosphorylation on the N-terminal tail of Ndc80.14 Although Knl1 has also been shown to bind to MT in vitro,9,13 the major contributor is Hec1/Ndc80,3,4 since in Caenorhabditis elegans, disruption of Knl1-MT-binding domain did not affect the formation of kMT attachments and chromosome segregation.13
Crosstalk between the KMN network and SAC signaling
Mitotic cells exploit the SAC-signaling pathway to inhibit the progression of chromosomal segregation until each chromosome achieves spindle attachment on either side (bi-orientation).6,7,15 After the last kinetochore is attached to the spindle microtubule, the cells must inactivate the SAC to undergo a transitional switch into anaphase. Therefore, checkpoint silencing requires both MT-attachment dependent cessation of signal generation at kinetochore and also inactivation of the already existing signal in the cytoplasm. Besides providing a multifaceted interface for chromosome-spindle attachment, kinetochores also play a pivotal role in controlling the switch between the SAC sustenance and silencing. The maximal accumulation of SAC proteins during prometaphase at unattached kinetochores and their depletion during metaphase when the kinetochores are optimally attached, point toward the requirement of a sophisticated and dynamic “on-off” signaling switch in response to the kMT attachment status.6,7 However, the downstream mechanism underlying the interplay between attachment of microtubules to kinetochores and checkpoint control remains poorly understood.
The SAC includes several kinase components such as Mps1 (monopolar spindle 1), BUB1 (budding uninhibited by benomyl 1) and Aurora B; non kinase components like Mad1 (Mitotic arrest deficient 1), Mad2, BUB3 and a pseudo-kinase, BUBR1 (BUB1-related 1).6,7,15-17 These SAC proteins are evolutionarily conserved and except for Mps1, all the other components depend on Knl1 for their recruitment to kinetochores.6,12 Mps1, BUB1, Mad1, Mad2, BUBR1 and CDC20 are transiently accumulated in high concentrations at unattached kinetochores and are progressively depleted from the site upon microtubule attachment.6,7 Mps1-mediated phosphorylation has been shown to essentially recruit all the other SAC components to unattached kinetochores.18-20 Mad2, BUBR1 and BUB3 generate an inhibitory mitotic checkpoint complex (MCC) with CDC20, which is an essential cofactor for APC/C (Anaphase Promoting Complex/Cyclosome), the key E3 ubiquitin ligase that promotes anaphase entry. The incorporation of CDC20 into MCC delays anaphase and prevents precocious mitotic exit.21
The KMN network serves as the major scaffold for kinetochore assembly and microtubule binding. The components of the KMN network have also now been recognized as a signaling framework for the SAC and studies have conclusively shown that the disruption of this network abolishes recruitment of SAC proteins to kinetochores.5,6,17 SAC proteins were initially thought to reside in the peripheral corona region of the kinetochore which is distally placed to the MT attachment sites on the outer plate. However, biochemical evidence and nanometer-scale mapping studies have shown that these SAC proteins in fact are in close proximity to the core kinetochore MT attachment sites constituted by the KMN network during metaphase with active kinetochore SAC signaling.22 An emerging paradigm is that the kinetochore proteins that possess MT binding activity also play a role in regulating SAC. Consistent with this hypothesis, the earliest event in the SAC activation is Mps1-mediated recognition and phosphorylation of threonine residue within conserved MELT (M[ED][ILVM][ST]) repeats on Knl1.6,23-25 Intriguingly, Plk1 is shown to be an auxiliary kinase that can phosphorylate Knl1 MELT repeats in cooperation with Mps1 to maintain SAC in mammalian cells.26 Concurrently, an independent study demonstrates that Plk1 phosphorylates MELT motifs in C. elegans that naturally lacks a recognizable gene for Mps1. This implies that functions commonly attributed to Mps1 in vertebrates are effectively substituted and executed by Plk1 in this nematode.27 The Mps1-phosphorylated Knl1 (MELTP) has been shown to generate a docking site for BUB3. Consequently, the BUB1 kinase piggybacks on BUB3, in turn recruiting BUBR1, as demonstrated by studies in both yeast and human systems.6,11,15,16 These molecular events are illustrated in Fig. 1. Recently, Zhang et al. reported that BUB1 via its R1LM domain can recruit BUBR1 to kinetochores through a direct interaction independent of BUB328 and this was substantiated by another study from the Musacchio lab.29 Although, BUB3 has been identified as the primary protein that interacts with the MELTP, studies by Primorac et al. have demonstrated cooperative effect of BUB1 on BUB3-MELTP complex formation.30 In turn, BUB1 is involved in recruiting downstream SAC proteins like Mad1, Mad2, BUBR1 and Cdc20 to kinetochores,6,7,31 underscoring the crucial role of Knl1 as a primary launching pad for SAC activation (Fig. 1).
Figure 1.
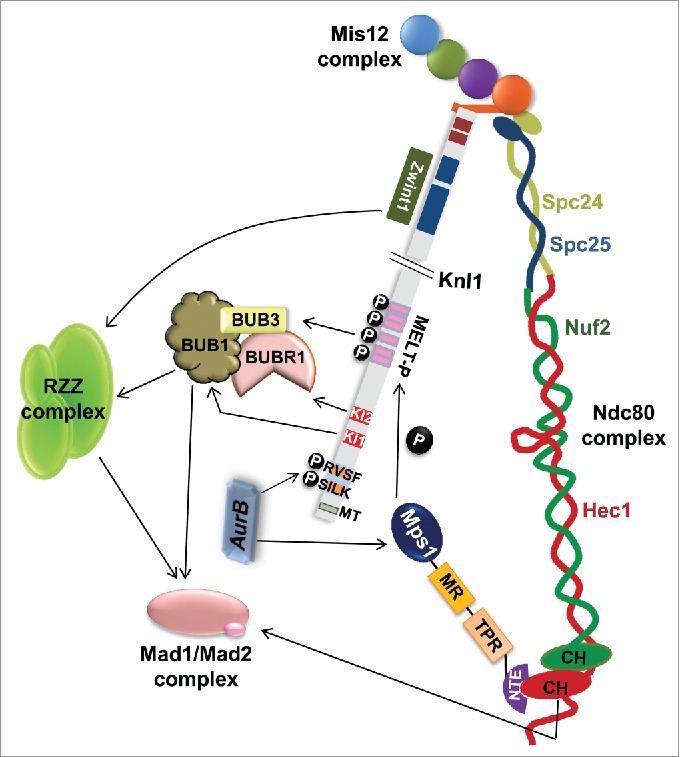
Crosstalk between the components of the KMN network and recruitment of the spindle assembly checkpoint (SAC) proteins. The key motifs involved in human Knl1 (hKnl1) function are indicated in the schematic of the protein. Briefly, the amino acid residues corresponding to each illustrated motif are: MT binding region (1-68), SILK (25-28), RVSF (58-61), KI1 (176-187), KI2 (212-223), MELT repeats (151, 762-1154), coiled-coil Zwint1 binding region (1904-2316), RWD repeats/Mis12 binding region (2106-2316). During prometaphase, when kinetochores are unattached to the microtubules, the SAC is “on.” Aurora B Kinase phosphorylates SILK and RVSF motifs at the N-terminus of Knl1 and this phosphorylation abrogates PP1 binding to Knl1. Mps1, the principal SAC kinase contains an N-terminal extension (NTE), a tetratricopeptide region (TPR) domain, a conserved middle region (MR) and a C-terminal kinase domain. Both NTE (depicted) and MR (not depicted) regions of Mps1 bind to the Ndc80 complex independently. The Aurora B kinase activity also promotes the recruitment of Mps1. Mps1 docked on Ndc80 via its N-terminal extended region (NTE), phosphorylates MELT repeats on Knl1 converting Knl1 into a dockyard for SAC components. The phosphorylated MELT-P recruits BUB3 that in turn recruits BUB1 and BUBR1 complex. Robustness in recruiting these SAC components around MELT repeats is further achieved by a direct interaction between KI1 and KI2 motifs of Knl1 with BUB1 and BUBR1, respectively. BUB1-BUB3 recruited on to the MELT repeats seems to be instrumental in recruiting the RZZ complex thereby driving the recruitment of Mad1/Mad2 to unattached kinetochores. Ndc80 has also been shown to positively influence the localization of Mad1/Mad2 to the unattached kinetochores in prometaphase.
Knl1 carries up to 6-7 MELT motifs in yeast and up to 19 in humans though not all of them are found to be active.11,16,23 Loss of activity in the repeats is attributed to either degeneration or vast variation from the consensus in the 3 most important motifs, MELT, TΩ (Ω = aromatic amino acid) and SHT (located C-terminal of MELT). Recent studies by Vleugel et al. has provided remarkable insight into the complexity in recognition of these MELT motifs and have also shown that Mps1-mediated phosphorylation of SHT motif encompassed within the MELT repeat augments SAC proficiency.23 Nonetheless, it is noteworthy that SHT phosphorylation by Mps1 requires prior phosphorylation on MELT motifs.23 The redundancy and necessity of multiple motifs i.e how many MELT repeats or whether all the MELT repeats are involved in recruiting BUB1 and BUB3 still remains enigmatic. It has been speculated that since binding affinity of BUB1:BUB3 to an individual MELTP is very high, each MELT repeat can potentially recruit a BUB1:BUB3 complex and therefore multiple BUB1:BUB3 complexes can concurrently bind to multiple MELTP repeats at a given time.23,24,30 Variations in the level of phosphorylation of different active repeats is likely to function as a governing factor or may provide a subtle layer of regulation in controlling the strength of SAC response but this hypothesis needs to be substantiated experimentally. In addition, the MELT repeats function cooperatively with the 2 N-terminally located, 10-12 amino acids long, and closely related yet distinct KI (Lys-Ile) motifs, KI1 (KIDTTSFLANLK) and KI2 (KIDFNDFIKRLK) to directly recruit BUB1 and BUBR1, respectively, by binding to their tetratricopeptide repeat (TPR) domains to ensure robust SAC activity (Fig. 1).11,16
The role of Knl1 in recruiting other essential SAC components like Mad1 and Mad2 to the kinetochores, most likely through indirect pathways, has also been recently unraveled.12 There has been no evidence demonstrating direct binding of Mad1 to Knl1. In fact, Mad1 requires both BUB1 and the RZZ (Rod-ZW10-Zwilch) checkpoint complex but no evidence of direct interaction between Mad1/2 and RZZ exists.32,33 In addition, the Knl1 binding protein, Zwint1, upon phosphorylation by Aurora B Kinase has been shown to promote recruitment of the RZZ complex along with the dynein motor to facilitate SAC signaling and kinetochore motility (Fig. 1).33-36 However, Zwint1 independent recruitment of the RZZ complex has been reported in multiple studies.22,28 Earlier, recruitment of the RZZ complex through a direct interaction between Zwint1 and ZW10 of the RZZ complex was proposed to be the mechanism. But, normal kinetochore localization of ZW10 mutants defective in Zwint1 binding supported the existence of an alternate mechanism for RZZ recruitment.35 The surrogate mechanism is accomplished by a central region in BUB1 that could efficiently recruit the RZZ complex to kinetochores (Fig. 1).28 But direct interaction between Bub1 and RZZ has not been reported so far. Because, the RZZ complex has been shown to contribute to the proper localization of Mad1/Mad2 complex to unattached kinetochores during prometaphase, the findings of Zhang et al. revealing BUB1 and not Zwint1 dependent RZZ recruitment could explain BUB1's contribution in Mad1/Mad2 localization.28 In fact, the deletion of RZZ recruiting region in BUB1 (437-521 amino acids) prevented both BUB1-dependent RZZ and Mad1/2 localization to kinetochores.28 The central region in BUB1 that is identified to be important for recruiting RZZ corresponds to the same region in yeast BUB1 that can directly bind to Mad1/2 and localize it to kinetochores as demonstrated by the Biggins laboratory.37 Further, Mps1 has also been shown to phosphorylate BUB1 to promote Mad1 recruitment to kinetochores in budding yeast.37,38 Previously, a direct interaction between BUB1 and Mad1 in both budding yeast and nematodes was also shown.37,39 Therefore, Zhang et al. conjectures that the yeast BUB1-Mad1/2 interaction evolved to BUB1-RZZ-Mad1/2 in higher and complex eukaryotes.28 Though this study implies that Mad1 is absolutely dependent on BUB1 for recruitment to kinetochores, recent study from the Kops group demonstrates that in human cells while BUB1 is needed for timely Mad1 recruitment, it is not absolutely required for Mad1 kinetochore localization.40 Therefore, it is evident that the mechanism of Mad1 kinetochore binding in human cells is at least in part different from that in budding yeast and nematodes, where BUB1 is the primary kinetochore receptor for Mad1.
Besides RZZ, the Ndc80 complex has also been shown to contribute to the proper localization of Mad1/Mad2 complex to kinetochores (Fig. 1).41,42 Fairly recently in nematodes, the Rod subunit of the RZZ complex was shown to interact directly with the microtubule binding N-terminus of Ndc80 but this association seems to be dispensable for RZZ and Mad1/Mad2 recruitment in human cells.43 Interestingly, it has been reported that Mps1 is recruited to kinetochores in an Aurora dependent manner, where it binds to the Calponin Homology (CH) domain of Ndc80 to initiate SAC signaling.31 It is important to reiterate here that these CH domains are the regions on Ndc80 that are involved in binding to microtubules.3,5,6 Assuming the close proximity of Knl1 and Ndc80 at kinetochores, the docking of Mps1 on Ndc80 is likely to facilitate its interaction with the MELT repeats on Knl1 N-terminus. Indeed, 2 recent reports further establish the direct relationship between attachment and SAC by precisely demonstrating a direct binding of Ndc80 to Mps1 via 2 independent interactions, which were inhibited in the presence of microtubules (Fig. 1).44 However, this inhibition was not absolutely competitive in nature because the Mps1 interface on Hec1 is in close proximity but not identical to the microtubule interface.45 These studies thus point toward a delicate regulation by Aurora B, which on one hand phosphorylates the Ndc80 basic tail and weakens the MT binding while on the other hand, augments Mps1 binding to Nuf2 that in turn promotes Knl1 phosphorylation and BUB1 recruitment.44,45 In accordance with these observations, a recent study demonstrates that microtubule attachment factually induces physical separation between Mps1 kinase (docked on CH domain of Ndc80) and phosphodomain of Spc105 (Knl1 ortholog in yeast) to abrogate MELT phosphorylation and SAC silencing.46
Integration of the KMN network with SAC silencing
Although SAC signaling is essential to provide sufficient time for all the chromosomes to attain bi-orientation, it is equally important that the SAC signaling cascade is promptly extinguished upon kMT attachment.6,15 kMT attachment triggers removal of enriched Mad1, Mad2, BUB1, BUBR1, BUB3 and Mps1 from kinetochores, resulting in SAC silencing. This is supported by the observation that constitutive targeting of Mad1 to kinetochores preserves SAC activity even after chromosome bi-orientation.47 Because phosphoregulation is the central mechanism that sustains SAC activity, it is reasonable to argue that SAC inactivation would depend on recruitment of a phosphatase. This was indeed supported by the observation that deletion of the R1LM domain of BUB1 that obliterated BUB1-dependent BUBR1 kinetochore localization did not prevent SAC activity but instead strengthened it.28 This led to the identification of a role for B56-PP2A phosphatase in SAC silencing.6,12 The B56 regulatory subunit of PP2A binds to a short 17 amino acid conserved region within BUBR1 referred to as the kinetochore attachment regulatory domain (KARD) that is phosphorylated by Plk1 and Cdk1.11,17 It remained counterintuitive how SAC signal maintains its proficiency when it also simultaneously recruits its antagonizing phosphatase (PP2A) until a 2-step SAC silencing mechanism was unveiled. These studies propose that the PP2A does not inhibit the SAC directly but counteracts the Aurora B activity to promote recruitment of PP1, another phosphatase that binds to Knl1 via the conserved SILK/RVSF motifs.48 PP1 can only associate with Knl1 once the Aurora B-phosphorylated PP1-binding sites SSILK and RVSF on the N-terminus of Knl1 are dephosphorylated. Once PP1 is docked on unphosphorylated RVSF/SSILK motifs, it dephosphorylates Knl1.49 Thus, PP2A bound to BUBR1 dephosphorylates Aurora B sites on Knl1 thereby allowing PP1 binding to Knl1 to mediate dephosphorylation of MELT motifs and transition from SAC “+” to SAC “-” state.48 Recent study by Nijenhuis et al. further suggests that the kinetochore localized PP1 promotes removal of kinetochore-PP2A-B56 by dephosphorylating MELT and KARD motifs.48 Similarly, Mps1-mediated phosphorylation of MELT repeats activates SAC and at the same time sets the way for SAC silencing by recruiting PP2A-B56 via BUBR1. PP2A therefore plays a key role in regulating 2 major events: controlling the formation of stable kMT attachments by opposing Aurora B activity and also initiating SAC silencing by opposing Mps1 activity.48,50 Hence spatiotemporal juxtaposition should exist between the 2 phosphatases (PP1 and PP2A) and 2 kinases (Aurora B and Mps1) to elicit a robust SAC signal. The PP2A-B56 promotes PP1 recruitment to kinetochores (by inhibiting Aurora B activity) that subsequently quenches the SAC by delocalizing PP2A from kinetochores. Contrary to these findings, Espert et al. demonstrates the exclusive and key role of PP2A-B56 in opposing both Aurora B and Mps1 activity in mammals.49 Here, Knl1 is shown to be the direct substrate for PP2A-B5649 and not PP1 as observed in yeast.51
Interestingly, the phosphorylation of BUBR1 by Plk1 and Cdk1 exclusively happens at unattached tensionless kinetochores so that Aurora B activity can be neutralized but how these kinases monitor and relay the signal in response to the kMT attachment status is still unknown. Also, the kinetochore pool of Plk1 kinase and the activity of Aurora B kinase are down-regulated in response to kMT attachment. In summary, Knl1 is involved in localizing proteins that augment (BUB1) and neutralize (PP1 via RVSF/SILK motifs and PP2A via BUBR1) the SAC activity (Figs. 1 and 2). While the Knl1-PP1 interaction is disrupted by Aurora B-mediated phosphorylation of the SILK/RVSF motifs,52 the Knl1-BUB1 interaction is promoted by Mps1-dependent phosphorylation of the MELT residues.6,11,12,15
Figure 2.
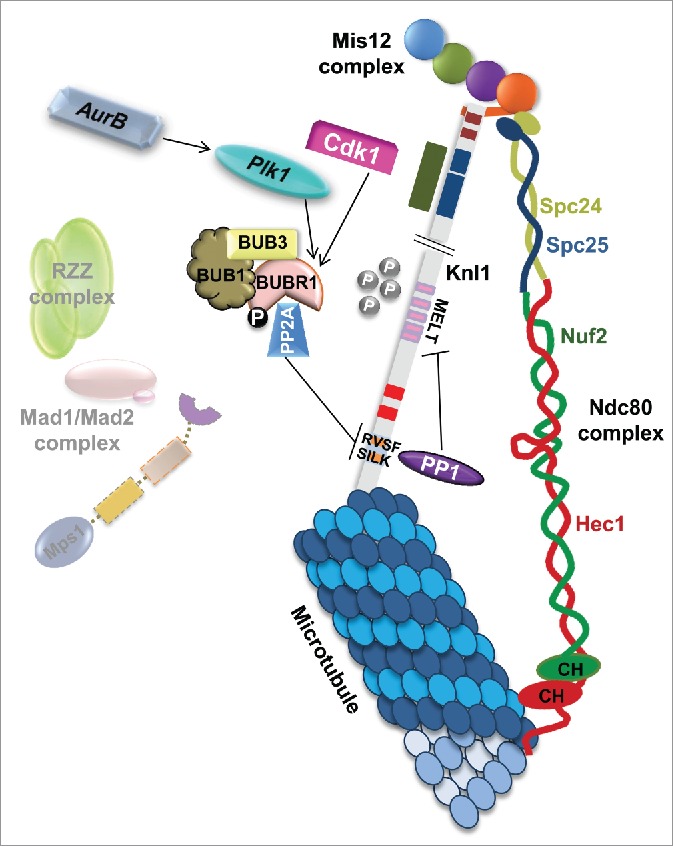
Spindle assembly checkpoint (SAC) inactivation in response to kinetochore microtubule attachment. In the presence of kinetochore microtubules attached end-on (to both Knl1 and Ndc80), Aurora B activates Plk1, which triggers PP2A binding to BUBR1 on phosphorylated KARD motif and causes the dephosphorylation of the RVSF/SILK motifs, thereby alleviating the inhibition of PP1 binding on to the Knl1. The binding of PP1 dephosphorylates the MELT repeats and the SAC components are released from the KMN network, extinguishing the SAC to an “off” state. Cdk1 also phosphorylates BUBR1 and augments PP2A binding to BUBR1. Further, in the presence of microtubules, Mps1 can no longer bind to Ndc80 and the phosphoswitch-mediated signaling subsides. For details on the amino acid map of all the Knl1 motifs, please refer to Fig. 1 legend.
Dynein motor-mediated removal of checkpoint proteins has been identified as the major SAC inactivation pathway as kinetochore-localized dynein/dynactin is instrumental in silencing the checkpoint by transporting the checkpoint proteins from MT-attached kinetochores to the spindle poles. 6,7 The RZZ complex is bound to the dynein motor through an adaptor protein called Spindly.53 Spindly is recruited to kinetochores by the RZZ complex and there is evidence for a weak interaction between them in both C. elegans and human cells.54 Although Dynein and Spindly are responsible for stripping the SAC components leading to SAC inactivation, the precise mechanism is unclear. The fact that Dynein is involved during the earlier steps where it is implicated in lateral capture of microtubules and chromosome alignment and is required later for stripping the checkpoint proteins, when appropriate end-on attachments are achieved,55 indicate the existence of a spatial and temporal control of dynein function. Even in the absence of Spindly, the silencing of checkpoint following attachment indicates the occurrence of a dynein-independent mechanism of silencing.54 The kinetochore dynein/dynactin motor is absent in fungi characterized by closed mitosis and it is completely lost in higher plants. Therefore in cases where Spindly is either deleted or the kinetochore dynein/dynactin complex is absent naturally, the KMN network which is the key platform providing core microtubule binding is speculated to play an important role in checkpoint silencing.
Concluding remarks and perspectives
Dysfunctional SAC and consequential premature APC/C activation leads to aneuploidy, a hallmark of many tumors.56 Our understanding of the integration and coordination between kMT attachment and SAC signaling is still very primitive, and definitely warrants extensive deliberation. Although the exact nature of SAC signaling is poorly understood, several studies have postulated that the signal is emanated at the centromere/kinetochore where tension is generated upon amphitelic attachment. How such a mechanical sensory signal is transduced into a biochemical signaling cascade remains enigmatic; however, it seems to be mediated by phosphorylation. Delineating the fine-tuned balance, flux and coordination of kinase and phosphatase activities at the kinetochore upon MT binding would likely be able to provide answers to this intriguing phenomenon. Until recently, the only study that directly correlates MT binding and SAC activity was in C. elegans by Espeut et al., wherein the worms harboring a Knl1 mutant defective in MT binding, exhibited delay in anaphase onset.13 However, the fate of checkpoint proteins at the kinetochore was not evaluated and the phenomenon may not be completely reflective of the scenario in vertebrates. Two contemporary studies demonstrating kinetochore recruitment of Mps1, a master SAC initiator, via Ndc80 in the absence of microtubules further contribute radically to this field. Nonetheless, there exist important yet unanswered questions that remain tantalizing areas for further research in attachment responsive SAC activity. Some of these questions include: (a) what is the precise molecular link between the Ndc80 complex, the key kMT attachment pivot and SAC activity? It seems that although Ndc80 is the primary contributor in binding to MTs, the major framework for recruiting checkpoint proteins, kinases and phosphatases is provided by Knl1 (except for Mps1); (b) what is the direct or indirect role of the KMN network in SAC silencing in the absence of dynein/dynactin motor machinery?; and (c) does the “Constitutive centromere-associated network” (CCAN)57,58 have any role in distinguishing attached vs unattached kinetochores and regulating the SAC? This possibility stems from the observation that cells devoid of CENP-I lost Mad1 from the kinetochores with immature attachments.59 The CCAN therefore has been alluded in lengthening the half-life of Mad1 at kinetochores and also in inhibiting the dynein-mediated stripping on laterally attached MTs by an unknown mechanism.59. The next few years of research will impart various degrees of nuances and answers to these and many other intriguing yet unresolved questions and promise to be an exciting time for mitosis investigators.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Acknowledgments
Authors would like to thank Sana Afreen, Suchithra Sheshadrinathan, and the other members of the Varma lab for their valuable input and critical reading of the manuscript.
FUNDING
The funding from the National Cancer Institute (R00CA178188) to D.V. is also acknowledged.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008; 9:33-46; PMID:18097444; http://dx.doi.org/ 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 2.Bakhoum SF, Compton DA. Kinetochores and disease: keeping microtubule dynamics in check! Curr Opin Cell Biol 2012; 24:64-70; PMID:22196931; http://dx.doi.org/ 10.1016/j.ceb.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLuca JG, Musacchio A. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol 2012; 24:48-56; PMID:22154944; http://dx.doi.org/ 10.1016/j.ceb.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J 2009; 28:2511-31; PMID:19629042; http://dx.doi.org/ 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varma D, Salmon ED. The KMN protein network–chief conductors of the kinetochore orchestra. J Cell Sci 2012; 125:5927-36; PMID:23418356; http://dx.doi.org/ 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 2013; 14:25-37; PMID:23258294; http://dx.doi.org/ 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012; 22:R966-80; PMID:23174302; http://dx.doi.org/ 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 8.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 2001; 12:1995-2009; PMID:11451998; http://dx.doi.org/ 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006; 127:983-97; PMID:17129783; http://dx.doi.org/ 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 10.Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 2006; 173:9-17; PMID:16585270; http://dx.doi.org/ 10.1083/jcb.200509158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghongane P, Kapanidou M, Asghar A, Elowe S, Bolanos-Garcia VM. The dynamic protein Knl1 - a kinetochore rendezvous. J Cell Sci 2014; 127:3415-23; PMID:25052095; http://dx.doi.org/ 10.1242/jcs.149922 [DOI] [PubMed] [Google Scholar]
- 12.Caldas GV, DeLuca JG. KNL1: bringing order to the kinetochore. Chromosoma 2014; 123:169-81; PMID:24310619; http://dx.doi.org/ 10.1007/s00412-013-0446-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol 2012; 196:469-82; PMID:22331849; http://dx.doi.org/ 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006; 127:969-82; PMID:17129782; http://dx.doi.org/ 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- 15.Stukenberg PT, Burke DJ. Connecting the microtubule attachment status of each kinetochore to cell cycle arrest through the spindle assembly checkpoint. Chromosoma 2015; PMID:25917595 [DOI] [PubMed] [Google Scholar]
- 16.Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol 2015; 2:e970484; PMID:NOT_FOUND; http://dx.doi.org/ 10.4161/23723548.2014.970484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma 2013; 122:135-58; PMID:23512483; http://dx.doi.org/ 10.1007/s00412-013-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol 2010; 190:89-100; PMID:20624902; http://dx.doi.org/ 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol 2010; 190:25-34; PMID:20624899; http://dx.doi.org/ 10.1083/jcb.201002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol 2010; 190:73-87; PMID:20624901; http://dx.doi.org/ 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci 2013; 38:302-11; PMID:23598156; http://dx.doi.org/ 10.1016/j.tibs.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 22.Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, Lawrimore J, Desai A, Salmon ED. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol 2013; 202:735-46; PMID:23979716; http://dx.doi.org/ 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vleugel M, Tromer E, Omerzu M, Groenewold V, Nijenhuis W, Snel B, Kops GJ. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol 2013; 203:943-55; PMID:24344183; http://dx.doi.org/ 10.1083/jcb.201307016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Lischetti T, Nilsson J. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J Cell Sci 2014; 127:871-84; PMID:24363448; http://dx.doi.org/ 10.1242/jcs.139725 [DOI] [PubMed] [Google Scholar]
- 25.Caldas GV, DeLuca KF, DeLuca JG. KNL1 facilitates phosphorylation of outer kinetochore proteins by promoting Aurora B kinase activity. J Cell Biol 2013; 203:957-69; PMID:24344188; http://dx.doi.org/ 10.1083/jcb.201306054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Schubert C, Cubizolles F, Bracher JM, Sliedrecht T, Kops GJ, Nigg EA. Plk1 and Mps1 Cooperatively Regulate the Spindle Assembly Checkpoint in Human Cells. Cell Rep 2015; 12:66-78; PMID:26119734; http://dx.doi.org/ 10.1016/j.celrep.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 27.Espeut J, Lara-Gonzalez P, Sassine M, Shiau AK, Desai A, Abrieu A. Natural Loss of Mps1 Kinase in Nematodes Uncovers a Role for Polo-like Kinase 1 in Spindle Checkpoint Initiation. Cell Rep 2015; 12:58-65; PMID:26119738; http://dx.doi.org/ 10.1016/j.celrep.2015.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Lischetti T, Hayward DG, Nilsson J. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat Commun 2015; 6:7162; PMID:26031201; http://dx.doi.org/ 10.1038/ncomms8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overlack K, Primorac I, Vleugel M, Krenn V, Maffini S, Hoffmann I, Kops GJ, Musacchio A. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife 2015; 4:e05269; PMID:25611342; http://dx.doi.org/ 10.7554/eLife.05269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife 2013; 2:e01030; PMID:24066227; http://dx.doi.org/ 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol 2015; 25:21-8; PMID:25220181; http://dx.doi.org/ 10.1016/j.tcb.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 32.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 2005; 15:856-61; PMID:15886105; http://dx.doi.org/ 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- 33.Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR, 3rd, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 2005; 169:49-60; PMID:15824131; http://dx.doi.org/ 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr DA, Saffery R, Li Z, Simpson AE, Choo KH, Yen TJ, Goldberg ML. HZwint-1, a novel human kinetochore component that interacts with HZW10. J Cell Sci 2000; 113 (Pt 11):1939-50; PMID:10806105 [DOI] [PubMed] [Google Scholar]
- 35.Famulski JK, Vos L, Sun X, Chan G. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J Cell Biol 2008; 180:507-20; PMID:18268100; http://dx.doi.org/ 10.1083/jcb.200708021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Hu X, Ding X, Dou Z, Yang Z, Shaw AW, Teng M, Cleveland DW, Goldberg ML, Niu L, et al.. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J Biol Chem 2004; 279:54590-8; PMID:15485811; http://dx.doi.org/ 10.1074/jbc.M407588200 [DOI] [PubMed] [Google Scholar]
- 37.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Gen dev 2014; 28:140-52; PMID:24402315; http://dx.doi.org/ 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 2014; 204:647-57; PMID:24567362; http://dx.doi.org/ 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol 2000; 10:675-8; PMID:10837255; http://dx.doi.org/ 10.1016/S0960-9822(00)00515-7 [DOI] [PubMed] [Google Scholar]
- 40.Vleugel M, Hoek T, Tromer E, Sliedrecht T, Groenewold V, Omerzu M, Kops GJ. Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J Cell Sci 2015; PMID:26148513 [DOI] [PubMed] [Google Scholar]
- 41.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 2002; 297:2267-70; PMID:12351790; http://dx.doi.org/ 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- 42.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol 2008; 18:1778-84; PMID:19026543; http://dx.doi.org/ 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 2013; 342:1239-42; PMID:24231804; http://dx.doi.org/ 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji Z, Gao H, Yu H. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 2015; 348:1260-4; PMID:26068854; http://dx.doi.org/ 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- 45.Hiruma Y, Sacristan C, Pachis ST, Adamopoulos A, Kuijt T, Ubbink M, von Castelmur E, Perrakis A, Kops GJ. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 2015; 348:1264-7; PMID:26068855; http://dx.doi.org/ 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]
- 46.Aravamudhan P, Goldfarb AA, Joglekar AP. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol 2015; 17:868-79; PMID:26053220; http://dx.doi.org/ 10.1038/ncb3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol 2011; 13:475-82; PMID:21394085; http://dx.doi.org/ 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nijenhuis W, Vallardi G, Teixeira A, Kops GJ, Saurin AT. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol 2014; 16:1257-64; PMID:25402682; http://dx.doi.org/ 10.1038/ncb3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espert A, Uluocak P, Bastos RN, Mangat D, Graab P, Gruneberg U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J Cell Biol 2014; 206:833-42; PMID:25246613; http://dx.doi.org/ 10.1083/jcb.201406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol 2011; 21:942-7; PMID:21640906; http://dx.doi.org/ 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol 2012; 22:900-6; PMID:22521787; http://dx.doi.org/ 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol 2010; 188:809-20; PMID:20231380; http://dx.doi.org/ 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barisic M, Geley S. Spindly switch controls anaphase: spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle 2011; 10:449-56; PMID:21252629; http://dx.doi.org/ 10.4161/cc.10.3.14759 [DOI] [PubMed] [Google Scholar]
- 54.Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Gen Dev 2010; 24:957-71; PMID:20439434; http://dx.doi.org/ 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 2001; 155:1159-72; PMID:11756470; http://dx.doi.org/ 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 2005; 5:773-85; PMID:16195750; http://dx.doi.org/ 10.1038/nrc1714 [DOI] [PubMed] [Google Scholar]
- 57.Hori T, Fukagawa T. Establishment of the vertebrate kinetochores. Chromosome Res 2012; 20:547-61; PMID:22733403; http://dx.doi.org/ 10.1007/s10577-012-9289-9 [DOI] [PubMed] [Google Scholar]
- 58.Takeuchi K, Fukagawa T. Molecular architecture of vertebrate kinetochores. Exp Cell Res 2012; 318:1367-74; PMID:22391098; http://dx.doi.org/ 10.1016/j.yexcr.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 59.Matson DR, Stukenberg PT. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J Cell Biol 2014; 205:541-54; PMID:24862574; http://dx.doi.org/ 10.1083/jcb.201307137 [DOI] [PMC free article] [PubMed] [Google Scholar]


