Abstract
Besides its classical mode of action through activation of specific receptors at the cell surface, fibroblast growth factor 1 (FGF1) can also cross the cellular membrane and translocate into the cytosol and further to the nucleus. The mechanism of this translocation is described partially, but the role of FGF1 inside the cell remains unknown. The aim of our work was to identify novel binding partners of FGF1 to predict its intracellular functions. We combined three methods of identification of such partners based on different principles: yeast two‐hybrid screen and mass spectrometry (MS) analysis of complexes obtained by Tandem Affinity Purification (TAP) or by co‐precipitation from cell lysate using recombinant FGF1. Altogether, we identified twenty novel intracellular proteins interacting with FGF1. For selected proteins, their direct interaction with FGF1 was confirmed by pull‐down assays and SPR measurements. Interestingly, half of the proteins found are involved in processes related to cell viability, such as apoptosis, cell proliferation, and cell cycle regulation. Thus, our study indicates that the role of intracellular FGF1 is to protect the cell against stress conditions by providing an additional signal for cell survival, independently of receptor‐activated signaling cascades. © 2016 IUBMB Life, 68(3):242–251, 2016
Keywords: FGF1, binding partners, Tandem Affinity Purification, intracellular function, translocation, cell survival, apoptosis
Introduction
Fibroblast growth factor 1 (FGF1) is a canonical member of fibroblast growth factor family involved in diverse biological processes including proliferation, cell growth, cell survival, and morphogenesis 1. It induces the biological response by binding to and activating four specific cell‐surface tyrosine kinase receptors (FGFRs). Activation of the FGFRs leads to stimulation of intracellular signaling pathways such as the PKC/phospholipase Cγ, PI3K/Akt, and RAS/mitogen‐activated protein kinase (MAPK) cascades 2, 3. In addition to the classical extracellular activity of FGF1, its unique feature among other growth factors is the ability to cross the cell membrane and to translocate into the cytosol and nucleus 4. However, despite many years of research its intracellular function remains unclear 5.
Several studies aimed at elucidating the role of FGF1 inside the cell by identifying its binding partners. It has been shown that FGF1 interacts directly with: FGF‐BP1 6, 7, protein kinase CK2 8, FGF1 intracellular binding protein (FIBP) 9, mortalin (GRP75/hthsp70/PB74) 10, and p34 11. All these proteins are involved in different biological processes unrelated to each other. Therefore, their identification failed to indicate conclusively an intracellular role of FGF1. We assumed that a comprehensive analysis of the binding partners of FGF1 should allow their grouping into functional classes and in this way shed light on the FGF1 functions inside the cell. Since using any single proteomic approach could bias against certain classes of proteins, we combined three independent methods based on different principles: yeast two‐hybrid screening and mass spectrometry (MS) analysis of complexes obtained after Tandem Affinity Purification (TAP) or after co‐precipitation with recombinant FGF1. Ten of the twenty newly identified FGF1 partners turned out to be related to apoptosis, cell cycle, and proliferation, suggesting that intracellular FGF1 plays a role in cell survival. This hypothesis is supported with our result showing anti‐apoptotic activity of FGF1 in the presence of a potent FGFR inhibitor.
Materials and Methods
Antibodies, Recombinant Proteins, and Chromatographic Resins
The following primary antibodies were used with the catalogue numbers indicated in parentheses: goat anti‐FGF1 (sc‐1884), mouse anti‐nucleophosmin (sc‐53175), mouse anti‐nucleolin (sc‐8031), mouse anti‐MVP (sc‐23917), mouse anti‐p53 (sc‐55476), rabbit anti‐UACA (sc‐135511), mouse anti‐TRIP11 (sc‐135928) from Santa Cruz Biotechnology (TX, USA) and mouse anti‐HSP90 (610419) from BD Transduction Laboratories (CA, USA). Secondary antibodies (mouse, goat, or rabbit) conjugated to HRP were from Santa Cruz Biotechnology (TX, USA) or Jackson ImmunoResearch (PA, USA). Recombinant proteins: UACA (residues 899–1403) with His‐tag was from Abcam (UK), p53 with His‐tag from Enzo Life Sciences (NY, USA), and HSP90α from StressMarq (Canada) or Abcam (UK). Heparin‐Sepharose CL‐6B affinity resin and Glutathione Sepharose™ 4 Fast Flow affinity resin were from GE Healthcare (UK). Anti‐c‐Myc Agarose, IgG Agarose, and Calmodulin Agarose were from Sigma‐Aldrich (MO, USA).
Cell Lines
Human normal foreskin fibroblast cell line (BJ), mouse fibroblast cell line (NIH3T3), and human embryonic kidney cell line (HEK 293) were from ATCC (VA, USA). Flp‐In T‐Rex‐293 cell line was from Invitrogen (CA, USA). Fibroblast cell lines were grown in Quantum 333 medium (PAA laboratories, Austria) supplemented with 2% BS in the case of NIH3T3. HEK 293 cell line was grown in Dulbecco's modified Eagle's medium (DMEM, Life Technologies, CA, USA) supplemented with 10% FBS (PAA Laboratories, Austria). Both the Quantum 333 medium and DMEM medium were supplemented with antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin from Life Technologies, CA, USA) and the cells were grown in a 5% CO2 atmosphere at 37°C.
Plasmids
pET‐3c vector with a DNA fragment encoding Met‐Ala‐FGF122–155 was used for expression of FGF1 without any tag 12. The same plasmid containing additional DNA fragment encoding Streptavidin Binding Peptide (SBP) at N‐terminus (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREPSGGCKLGSGSGSGSGSG) was used for expression of SBP‐FGF1 protein 13. A DNA fragment encoding Met‐Ala‐FGF122–155 was cloned also into pcDNA3 vector (with an N‐terminal myc sequence). pcDNA5FRT/TO/TAPtag (pRS72) with TAP tag sequence (consisting of CBP tag, TEV protease cleavage site and Protein A tag) was a generous gift from Dr. R.J. Szczesny (Institute of Genetics and Biotechnology, Faculty of Biology, University of Warsaw, Warsaw, Poland). A DNA fragment encoding Met‐Ala‐FGF122–155 was cloned into this vector either at N‐terminus or C‐terminus of TAP tag sequence.
A DNA fragment encoding nucleolin (residues 284–707, nucleolin‐C) 14 was obtained from GenScript (NJ, USA) and cloned into pDEST15 expression vector (with N‐terminal GST). pEX‐N‐GST vectors encoding full‐length nucleophosmin or fragment of major vault protein (113–474) 15 were obtained from Origene Technologies (MD, USA).
Stable Transfection of HEK 293 Cells
Flp‐In T‐Rex 293 cells were seeded on polylysine‐coated culture plate. Cells were co‐transfected with one of TAP tag vectors together with pOG44 (with Flp recombinase gene) using Escort IV reagent (Sigma‐Aldrich, MO, USA). After 8 h, DMEM supplemented with 10% FBS was added. Selection was conducted in the presence of 15 μg/mL blasticidin and 600 μg/mL hygromycin B (Invitrogen, CA, USA). Selective medium was replaced every 2–3 days for at least 30 days. Finally, we obtained three variants of stably transfected cell lines named: TAP, N‐TAP‐FGF1, FGF1‐C‐TAP.
Protein Expression and Purification
Recombinant FGF1, SBP‐FGF1, and C‐terminal fragment of nucleolin (residues 284–707) with GST tag were produced in E. coli Bl21(DE3)pLysS or Bl21(DE3)‐RIL strains (from Merck, Germany). SBP‐FGF1 was purified on Heparin‐Sepharose CL‐6B column. GST‐nucleolin was expressed and purified using Glutathione Sepharose 4 Fast Flow column as described previously 5. Nucleophosmin and major vault protein (residues 113–474) were expressed as fusion proteins with N‐terminal GST in E. coli Rosetta 2(DE3)pLacI strain (from Merck, Germany). Expression of proteins was induced for 16 h by addition of 1 mM isopropyl‐β‐d‐thiogalactopyranoside at 37°C. Afterwards proteins were purified from bacterial lysates using Glutathione Sepharose 4 Fast Flow column.
Protein homogeneity was verified by Sodium Dodecyl Sulfate‐Polyacrylamide Gel Electrophoresis (SDS‐PAGE) and protein identity was confirmed by mass spectrometry (4800 MALDI TOF/TOF Analyzer, Applied Biosystems/MDS Sciex, Canada). To verify native conformation of purified proteins circular dichroism (Jasco J‐715 spectropolarimeter) and fluorescence (Jasco FP‐750 or FP‐8500 spectrofluorimeter) measurements were applied as described previously 12, 16.
Tandem Affinity Purification
Cells were starved (DMEM with 1% FBS) for 24 h, then stimulated with tetracycline (200 μg/mL) for another 24 h. Cells were scraped, pelleted, and lysed in Lysis Buffer (25 mM Tris pH 8.0, 10 mM MgCl2, 100 mM NaCl, 0.5% Igepal, 10% glycerol, protease, and phosphate inhibitors). Next, lysates were sonicated and centrifuged for 10 min to pellet cellular debris. IgG Agarose beads were washed three times with Lysis Buffer and then incubated with cleared cell lysates overnight at 4°C. Subsequently, the beads were washed three times in Lysis Buffer and three times in TEV Cleavage Buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% Igepal, 0.5 mM EDTA, 1 mM DTT). Next, rTEV protease was added to the beads and incubated overnight at 4°C. Eluates containing protein complexes were diluted 1:1 with Calmodulin Binding Buffer (CBB; 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1% Triton X‐100, 1 mM imidazole, 4 mM CaCl2, 10 mM β‐mercaptoethanol), added to Calmodulin Agarose beads prewashed with CBB, and incubated overnight at 4°C. Then, the beads were washed four times with CBB, transferred to Micro Bio‐Spin Columns (Bio‐Rad) and incubated for 2 min with Calmodulin Elution Buffer (50 mM (NH4)HCO3, 25 mM EGTA). Eluates were mixed 1:4 with cold acetone and incubated overnight at −20°C. Protein complexes were pelleted by centrifugation (10 min, 14,000 rpm) and subjected to mass spectrometry analysis (performed at the Mass Spectrometry LAB, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw using MALDI‐TOF/TOF UltrafleXtreme from Bruker Daltonics, MA, USA).
Co‐Precipitation Assay for Protein Identification
NIH3T3 cells (15–20 × 106) were lysed in Lysis Buffer (20 mM Tris‐HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100 supplemented with protease inhibitor cocktail (Roche, Switzerland)) and sonicated. Cellular debris was pelleted by centrifugation. Cleared cell lysate was incubated with recombinant SBP‐FGF1 for 1 h followed by incubation with 50 μL of Streptavidin‐coated dynabeads for 1 h at room temperature or with dynabeads alone (negative control). Dynabeads were washed four times in PBS with 2% Triton X‐100 (PBST) and protein complexes were eluted by 10‐min boiling in SDS sample buffer. Proteins were subjected to SDS‐PAGE and subsequent mass spectrometry analysis performed by the Core Facility for Proteomics and Mass Spectrometry of Oslo University Hospital‐Rikshospitalet (Oslo, Norway) using Ultraflex II MALDI‐TOF/TOF from Bruker Daltonics (MA, USA).
Yeast Two‐Hybrid Screen
Yeast two‐hybrid screen was performed by Hybrigenics Services, S.A.S., Paris, France. A DNA fragment encoding human FGF1 (Met‐Ala‐FGF122–155) was PCR‐amplified and cloned into pB66 as C‐terminal fusion to Gal4 DNA‐binding domain (N‐Gal4‐FGF1‐C) and used as a bait to screen random‐primed human Fibroblasts_RP1 cDNA library containing ∼10 millions of independent clones in pP6 vector. pB66 and pP6 derived from the original pBTM116 17, pAS2ΔΔ 18, and pGADGH 19 plasmids, respectively. For the Gal4 construct, 78.6 millions clones (8‐fold the complexity of the library) were screened using a mating approach with HGX13 (Y187 ade2‐101::loxP‐kanMX‐loxP, matα) and CG1945 (mata) yeast strains. A total of 4 His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions and selected interacting domain (SID) was identified. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (NCBI) using a fully automated procedure. A confidence score (PBS, for Predicted Biological Score) was attributed to each interaction as described by Formstecher et al. 20.
Transient Transfection of HEK 293 Cells
Transient expression of myc‐FGF1 was performed by transfecting HEK 293 cells with plasmid DNA using Escort IV Transfection Reagent (Sigma‐Aldrich, MO, USA) in Minimum Essential Medium (MEM, Gibco, NY, USA) according to the manufacturer's protocol. After 7 h, the medium was changed for DMEM supplemented with 10% FBS and the cells were grown for 24 h 5.
Co‐Precipitation Assays for Verification of Protein–Protein Interactions
Pull‐down assays were performed as described above for protein identification, using recombinant SBP‐FGF1 and either BJ cell lysate (0.5 × 106 cells per sample) or recombinant partner proteins. Protein complexes were eluted from the Streptavidin‐coated dynabeads resin by 10 min boiling in SDS sample buffer, and subjected to SDS‐PAGE and Western blotting using specific antibodies.
Additionally, HEK 293 cells transfected with myc‐FGF1 were treated as described previously 5. Briefly, cells were lysed in lysis buffer (20 mM Tris‐HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100 supplemented with a protease inhibitor cocktail), sonicated and centrifuged to remove cellular debris. Cleared lysate was incubated with 20 μL of anti‐c‐Myc Agarose for 1 h at room temperature. The resins were washed four times in PBS with 1% Triton X‐100 before the protein complexes were eluted by 10 min boiling in SDS sample buffer. Protein complexes were subjected to SDS‐PAGE and Western blotting.
Surface Plasmon Resonance
Interaction between putative partner proteins and FGF1 was investigated by surface plasmon resonance (SPR) analysis using Biacore® 3000 (GE Healthcare, UK). Recombinant binding proteins were covalently immobilized on carboxymethylated dextran sensor chip (CM5, GE Healthcare, UK) using Amine Coupling Kit (GE Healthcare). Hepes‐Buffered Saline‐P (10 mM Hepes, 0.15 M NaCl, 3 mM EDTA, 0.005% P20, pH 7.4) was used as Immobilization Running Buffer. Aliquots (60 μL) of FGF1 in Running Buffer (PBS with 0.005% P20, pH 7.3) were injected at a flow rate of 30 μL/min and specific protein–protein interactions were measured. Dissociation of FGF1 from proteins of interest was monitored over 4‐min period. Regeneration of sensor chip surface was performed with 1 M NaCl or 2 mM NaOH. Data were analyzed using the BIAevaluation 3.1 software (GE Healthcare, UK). Final sensorgrams were generated by subtracting response in reference flow cell from responses in sample flow cell.
Caspase‐3/7 Activity and Viability Assay
NIH3T3 cells were subjected to serum starvation for 24 h to induce apoptosis. Then the cells were treated for 6 h with FGF1 (200 ng/mL) and heparin (10 U/mL) in the presence of the specific FGFR tyrosine kinase inhibitor, 100 nM PD173084, used to prevent receptor autophosphorylation. Next, caspase‐3/7 activity, reflecting the apoptosis progression, along with cell viability was measured using ApoLive‐Glo Multiplex Assay (Promega, WI, USA), according to the manufacturer's protocol. Caspase‐3/7 activity data were divided by cell viability values, then normalized toward the cells untreated with FGF1 (control), and denoted as relative caspase‐3/7 activity. For statistical analysis one‐way ANOVA with Tukey's post test was applied using SigmaPlot (Systat Software).
Results
Proteins Interacting With FGF1 Identified Using Tandem Affinity Purification
To identify intracellular protein partners of FGF1, we used three unrelated methods: Tandem Affinity Purification followed by MS analysis, co‐precipitation followed by MS analysis, and yeast two‐hybrid screen (Fig. 1). First, we performed five independent series of TAP experiments identifying each time proteins that bound to N‐TAP‐FGF1, FGF1‐C‐TAP, and TAP‐tag as a negative control. Proteins present in any of the five negative‐control samples were excluded from further considerations. Among 78 specific FGF1 interactors identified, only those with Mascot score above 30 were considered, as such value indicated identity or extensive homology (P < 0.05). Altogether, we obtained seven intracellular proteins including two chaperons (HSP90α and HSC70), two proteins associated with cell cytoskeleton (MAPRE1 and ACTBL2), sideroflexin‐1 that acts as a channel in the mitochondria membrane 21, vacuolar protein V‐type proton ATPase, and tumor suppressor p53 (Table 1).
Figure 1.
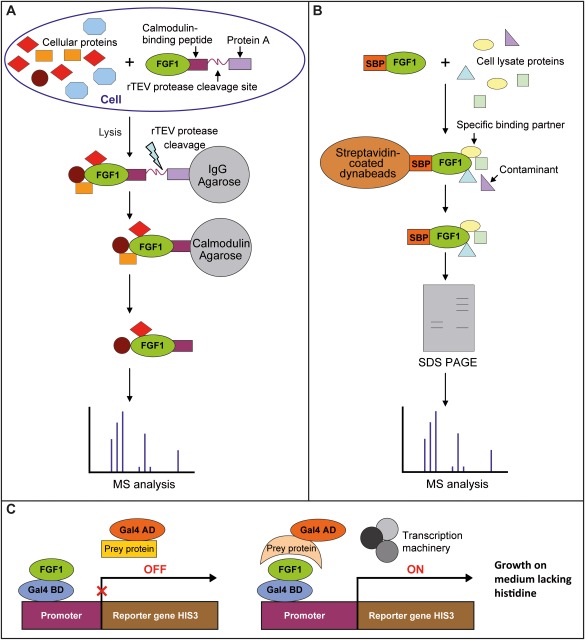
Three methods for identification of partner proteins: Tandem Affinity Purification (TAP) (A), co‐precipitation using recombinant FGF1 as bait and cell lysate (B), and yeast two‐hybrid screen (C).
Table 1.
FGF1 binding partners identified using TAP system
| Protein | Coverage (%) | Mascot score | Matched peptides | TAP‐tag position |
|---|---|---|---|---|
| Heat shock protein 90‐alpha, HSP90α | 23 | 359 | 11 | C‐TAP, N‐TAP |
| Microtubule‐associated protein RP/EB family member 1, MAPRE1 | 17 | 314 | 4 | C‐TAP, N‐TAP |
| V‐type proton ATPase subunit B, V‐ATPase | 3 | 225 | 4 | C‐TAP, N‐TAP |
| Heat shock cognate 71 kDa protein, HSC70 | 4 | 126 | 2 | C‐TAP, N‐TAP |
| Beta‐actin‐like protein 2, ACTBL2 | 10 | 115 | 3 | C‐TAP, N‐TAP |
| Sideroflexin 1, SFN1 | 4 | 71 | 1 | N‐TAP |
| Cellular tumor antigen p53 | 3 | 31 | 1 | N‐TAP |
Proteins Interacting With FGF1 Determined by Co‐precipitation Method
In the second approach, we used FGF1 fused with SBP‐tag to co‐precipitate interacting proteins from cell lysates. Proteins were eluted from Streptavidin‐coated dynabeads using biotin, which provided mild conditions, and resolved by SDS‐PAGE (Supporting Information Fig. 1). Bands, present in the SBP‐FGF1 precipitate but not in the negative control, were cut out from the gel and subjected to MS analysis. In twenty independent experiments, we found 26 proteins as potential FGF1 binding partners. Among them, eleven were identified with a Mascot score above 30 and were considered to be positive hits (Table 2). The most abundant result was nucleolin, a multifunctional nucleolar protein, already described by us in the context of FGF1 intracellular trafficking 5. Other proteins found using this approach were: the component of nuclear membrane—lamin‐A/C, three proteins involved in maintaining cell structure (γ‐actin, α‐actinin‐4, gelsolin), three RNA interacting proteins (heterogeneous nuclear ribonucleoproteins C1/C2 and A2/B1 and IF2A protein), Ca2+‐dependent, anionic phospholipid‐binding protein—annexin A2 22, nucleophosmin—multifunctional protein, involved in many different cellular processes such as: ribosome biogenesis, centrosome duplication, protein chaperoning, histone assembly, and cell proliferation 23, and major vault protein that functions as a scaffold protein for both SHP‐2 and ERK, thus playing an important role in cell survival signaling 24.
Table 2.
Proteins identified as FGF1 partners by co‐precipitation with recombinant SBP‐FGF1
| Protein | Coverage (%) | Mascot score | Matched peptides |
|---|---|---|---|
| Lamin‐A/C | 29 | 257 | 20 |
| Nucleolin (Protein C23), NCL | 24 | 219 | 19 |
| Actin, cytoplasmic 2,γ‐actin | 46 | 213 | 15 |
| Major vault protein, MVP | 13 | 145 | 10 |
| Nucleophosmin (nucleolar phosphoprotein B23), NPM | 31 | 95 | 9 |
| Heterogeneous nuclear ribonucleoproteins A2/B1, hnRNP A2/B1 | 24 | 88 | 7 |
| α‐Actinin‐4 | 9 | 77 | 6 |
| Gelsolin | 10 | 75 | 5 |
| Annexin A2 | 17 | 54 | 5 |
| Heterogeneous nuclear ribonucleoproteins C1/C2, hnRNP C1/C2 | 14 | 50 | 4 |
| Eukaryotic translation initiation factor 2 alpha subunit, IF2A | 8 | 30 | 3 |
Proteins Interacting With FGF1 Found by Yeast Two‐Hybrid Screen
The Y2H experiment in which human Fibroblasts_RP1 was used as prey library and pB66 (N‐GAL4‐FGF1‐C) as a bait vector was performed by Hybrigenics. Among four positive clones detected, two clones were identical and one clone was out of frame (Table 3). The Y2H screening revealed two proteins interacting with FGF1: thyroid receptor‐interacting protein 11 (TRIP11) with a confidence score D (moderate confidence) and uveal autoantigen with coiled‐coil domains and ankyrin repeats (UACA) with a confidence score C (good confidence) (Fig. 2). TRIP11 binds the thyroid receptor (THRB) and enhances THRB‐modulated transcription 25. It has also been associated with Golgi apparatus structure 26. UACA interacts with Apaf‐1 and induces its translocation into the nucleus upon pro‐apoptotic stress 27. Moreover, UACA regulates the sensitivity of cancer cells to extracellular inducers of apoptosis 28.
Table 3.
Proteins identified by yeast two‐hybrid screen using human FGF1 (Met‐Ala‐FGF122–155) as a bait
| Clone name | Gene name | Frame | PBS |
|---|---|---|---|
| pB66_A‐1 | Homo sapiens—TGFB1 | OOF | N/A |
| pB66_A‐3 | Homo sapiens—TRIP11 | IF | D |
| pB66_A‐2 | Homo sapiens—UACA | IF | C |
| pB66_A‐4 | Homo sapiens—UACA | IF | C |
PBS shows confidence of interaction: C: good confidence, D: moderate confidence, and N/A: not applicable. In frame (IF) denotes fragments in the same frame as Gal4AD, OOF denotes fragment out of this frame.
Figure 2.
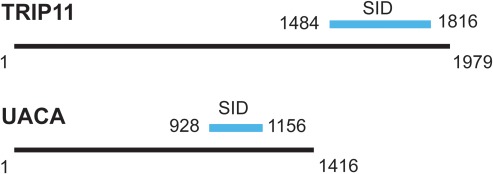
Schematic presentation of TRIP11 and UACA identified as FGF1 partners using yeast two‐hybrid screen. Blue lines indicate selected interaction domains (SID) identified within these proteins.
Verification of Identified Interactions by Co‐precipitation
In the next step, we performed co‐precipitation experiments using BJ cells and recombinant FGF1, followed by immunoblotting. First, SBP‐FGF1 was incubated with cell lysate and then the whole mixture was incubated with Streptavidin‐coated dynabeads. After washing with PBST, retained protein complexes were eluted from the beads with SDS sample buffer followed by electrophoresis and Western blotting with antibodies against selected proteins identified in the earlier steps as potential interactors of FGF1. As shown in Fig. 3A, HSP90, nucleolin, nucleophosmin, MVP, p53, UACA, and TRIP11 were present among the protein complexes interacting with FGF1 and absent in negative control (beads alone), indicating direct or indirect binding. To substantiate the physiological significance of the identified interactions, we also tested whether FGF1 could interact with endogenous proteins in mammalian cells. Similarly to pull‐down experiments with recombinant FGF1 and our previous results obtained for nucleolin 5, we were able to pull‐down nucleolin, nucleophosmin, p53, and UACA from HEK 293 cells by precipitating transiently overexpressed myc‐FGF1 with myc‐agorose (Fig. 3B). These proteins were not detected in untransfected cells upon incubation with anti‐c‐Myc Agarose. This result confirms that tested proteins are interaction partners of FGF1 in mammalian cells.
Figure 3.
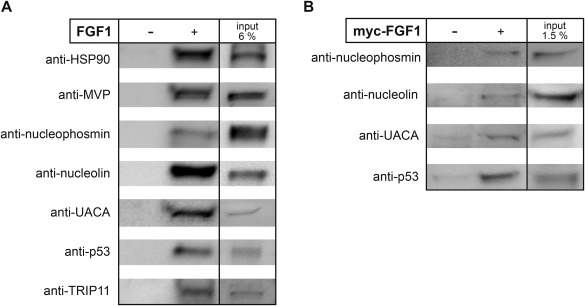
Verification of intracellular FGF1 partners by co‐precipitation. A: BJ cell lysate was incubated with SBP‐FGF1 and then with dynabeads, or with dynabeads alone. Protein complexes were eluted from the beads with sample buffer, resolved by SDS‐PAGE and subjected to Western blotting using specific antibodies as indicated. B: HEK 293 cells transiently transfected with myc‐FGF1 were lysed and then subjected to immunoprecipitation followed by SDS‐PAGE and Western blotting with an anti‐nucleolin, anti‐p53, anti‐nucleophosmin, or anti‐UACA antibody. As controls non‐transfected cells were used.
Unfortunately, we could not validate any of FGF1 interactions directly in cells, due to the limitations of immunofluorescence technique. We are able to visualize exogenous FGF1 exclusively in endosomes as only in this small compartment the concentration of the growth factor is high enough to allow detection using fluorescent labeling or antibody staining.
Validation of Direct Interaction of FGF1 With Selected Proteins
In order to analyze whether the interaction between FGF1 and the identified proteins is direct or not, we performed a pull‐down assay using recombinant SBP‐FGF1 and selected pure recombinant proteins identified earlier as its potential binding partners. We tested the following proteins (intact or fragments as indicated in parentheses): HSP90α, MVP (113–474), nucleolin‐C (284–707), UACA (899–1403), p53, and nucleophosmin. TRIP11 was not commercially available in active form and we were unable to express it. As shown in Fig. 4A, all the potential partners tested bound to SBP‐FGF1 immobilized on Streptavidin‐coated dynabeads but not to dynabeads alone, indicating a direct interaction with FGF1. In the case of HSP90α, nucleolin‐C (284–707) and p53 the results were confirmed further by surface plasmon resonance experiments, in which FGF1 without SBP tag was used (Fig. 4B).
Figure 4.
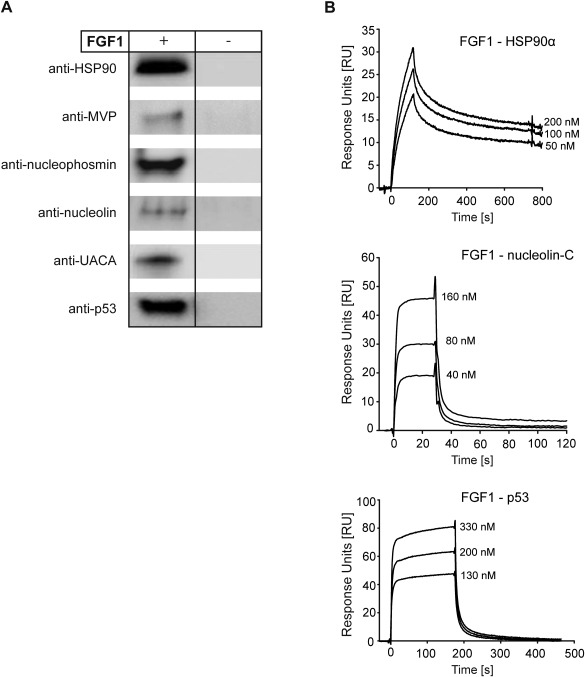
Confirmation of direct interaction of FGF1 with HSP90α, MVP (113–474), nucleolin‐C (284–707), UACA (899–1403), p53, and nucleophosmin. A: Recombinant potential FGF1 binding partners were incubated with SBP‐FGF1 and then Streptavidin‐coated dynabeads or with dynabeads alone. Protein complexes were eluted from the beads with sample buffer, resolved by SDS‐PAGE and subjected to Western blotting using specific antibodies. B: Recombinant HSP90α, nucleolin‐C, or p53 were immobilized on carboxymethylated dextran sensor chip CM5 at the level of 500–1000 RU (response units). Recombinant FGF1 was injected at increasing concentrations at a flow rate of 30 μL/min and specific protein–protein interactions were measured.
Exogenous FGF1 Inhibits Apoptosis Induced by Serum Starvation in the Presence of FGFR Kinase Inhibitors
Since translocation of FGF1 is stimulated by stress conditions 29 and many of the novel FGF1‐intreracting proteins identified by us are associated with cell survival processes, we examined the effect of translocated FGF1 on NIH3T3 cells upon apoptosis induction.
In order to observe only the effect of translocated FGF1, we blocked the FGFR‐dependent signaling using highly specific FGFR kinase activity inhibitor (PD173074) 30. We analyzed apoptosis progression by tracking two separate signals, the first corresponding to cell viability and the second proportional to caspase‐3/7 activity. Upon apoptosis induction in NIH3T3 cells by 24‐h serum starvation and subsequent 6‐h treatment with FGF1 in the presence of 100 nM PD173074, we observed significant decrease in caspase‐3/7, as compared to control cells (untreated with FGF1) (Fig. 5). Relative caspase‐3/7 activity lowered from 1 to 0.5, suggesting that the translocated FGF1 exhibits anti‐apoptotic activity independently of FGF receptor stimulation.
Figure 5.
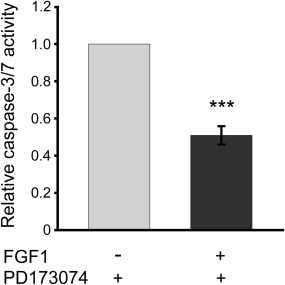
FGFR‐independent anti‐apoptotic activity of exogenous FGF1. NIH3T3 cells were serum starved for 24 h and then treated for 6 h with FGF1 (200 ng/mL) and heparin (10 U/mL) in the presence of the specific FGFR tyrosine kinase inhibitor, 100 nM PD173084. Apoptosis was measured using ApoLive‐Glo Multiplex Assay (Promega) Next, caspase‐3/7 activity, reflecting the apoptosis progression, along with cell viability were measured using ApoLive‐Glo Multiplex Assay (Promega). Caspase‐3/7 activity data were divided by cell viability values, then normalized toward the cells untreated with FGF1 (control), and denoted as relative caspase‐3/7 activity. Graphs represent the mean ± SD of four independent experiments (***P < 0.001).
Discussion
We identified twenty intracellular proteins as potential binding partners for FGF1 using three different methods: Tandem Affinity Purification (TAP) and standard co‐precipitation, both followed by MS analysis, and yeast two‐hybrid screen. Independent N‐TAP and C‐TAP experiments identified seven and five proteins, respectively, altogether revealing seven novel binding partners of FGF1. We find this method highly reliable since all the C‐TAP hits were also found in N‐TAP assay. Co‐precipitation identified the largest number of FGF1 interactors, eleven proteins, and the yeast two‐hybrid test provided only two hits. There were no overlaps between any of the three sets of FGF1 interactors identified using those three approaches. For selected proteins (HSP90α, nucleolin, nucleophosmin, MVP, p53, UACA, and TRIP11), we confirmed their ability to form protein complexes with FGF1 using in vitro techniques. In the case of nucleolin, nucleophosmin, p53, and UACA, physiological significance of the interactions was confirmed by co‐precipitation of FGF1 intracellular binders from HEK cells transfected with FGF1 construct.
For two out of the twenty proteins identified here, HSP90α and p53, there were earlier literature data indicating their involvement in the intracellular activity of FGF1. Wesche et al. 31 demonstrated that when HSP90 was inhibited by geldanamycin, the process of translocation of FGF1 from endosomes to cytosol and further to the nucleus was blocked. However, those authors did not show a direct interaction between the two proteins. Here we have documented the direct interaction between FGF1 and HSP90α using two independent techniques, co‐precipitation of pure proteins and SPR measurement. The second protein mentioned in the literature as a putative FGF1 partner was p53. Renaud and coworkers showed that FGF1 was immunoprecipitated together with p53 from PC12 cell lysate but their direct interaction was not investigated either 32, 33. They showed that endogenous FGF1 inhibited p53‐dependent apoptosis upstream of mitochondrial and nuclear events. FGF1 prevented phosphorylation, stabilization, and nuclear translocation of p53 and its transcriptional activity 33. In this study, we confirmed not only the presence of p53 within FGF1 complex, but also direct binding of these two proteins. Interestingly, both HSP90α and p53 were identified here by the TAP approach.
The co‐precipitation experiment showed eleven FGF1‐binding proteins among which nucleolin seemed the most interesting one due to its direct involvement in the cytosol‐nucleus trafficking. We have studied the role of nucleolin in FGF1 intracellular transport earlier and found that it regulates the phosphorylation and thereby the export of FGF1 from the nucleus 5.
For the remaining seventeen proteins identified here, no literature indications or even suggestions on their involvement in the intracellular fate of FGF1 could be found. Therefore, we decided to explore the biological processes our FGF1 binding partners are involved in. An analysis of the Gene Ontology (GO) annotations for “biological processes” from the UniProtKB database (www.uniprot.org) allowed us to classify these proteins into ten groups (Fig. 6). As we discussed above, the role of HSP90α belonging to ATP binding and signal transduction proteins and nucleolin (involved, among other functions, in nucleo‐cytoplasmic transport) is related to the mechanism of FGF1 translocation itself rather than its intracellular function. In this classification, the most abundant are proteins involved in cell survival (50% of identified proteins): apoptosis: 6; cell cycle: 5; and cell proliferation: 3 proteins, suggesting that the intracellular functions of FGF1 can be related to stimulation of the cell to overcome unfavorable conditions. In agreement, translocation of exogenous FGF1 to the cytosol and nucleus occurs in stress environment such as deprivation of serum 29, 34 and is augmented in the presence of toxic compounds such as: H2O2, thapsigargin, or mannitol 35. Moreover, it has been shown that the presence of FGF1 (due to transfection process) in the nucleus of PC12 cells prevents apoptosis induced by serum starvation or etoposide 33.
Figure 6.
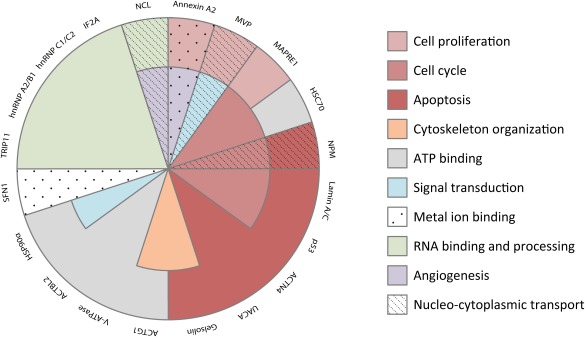
Gene ontology analysis of proteins identified as FGF1 binding partners. The classification was performed according to the GO annotation for “biological process” using UniProtKB database. Note that some proteins belong to more than one class.
To test our hypothesis on pro‐survival and anti‐apoptotic role of translocated FGF1, we performed functional experiment in NIH3T3 cells in which apoptosis was induced by 24‐h starvation. Our pilot experiment revealed that even when the FGFR downstream signaling cascades were completely blocked due to the presence of potent inhibitor, 6‐h treatment with FGF1 reduced significantly the caspase‐3/7 activity and protected NIH3T3 cells against apoptosis. These data were in keeping with previous studies on FGF1 translocation demonstrating the maximal translocation efficiency 6 h after the addition of FGF1 to the cell medium 36. Altogether, our results suggest that the presence of exogenously added FGF1 inside the cell may provide an additional signal for cell survival that is independent of the classical signaling pathways initiated by FGF binding to its cell‐membrane receptors.
Supporting information
Supporting Information Figure 1.
Acknowledgements
This work was supported by the Polish‐Norwegian Research Programme operated by the National Centre for Research and Development under the Norwegian Financial Mechanism 2009–2014 in the frame of Project Contract No. Pol‐Nor/197969/50/2013, and the Polish‐Norwegian Research Fund (PNRF‐87‐AI‐1/07). The cost of publication was covered by the by the Polish Ministry of Science and Higher Education under the Leading National Research Centre (KNOW) Programme (2014‐2018). The skilful technical assistance of Mandana Yadollahi, Anne Engen and Agnieszka Kubiak is gratefully acknowledged.
References
- 1. Powers, C. J. , McLeskey, S. W. , and Wellstein, A. (2000) Fibroblast growth factors, their receptors and signaling. Endocr. Relat. Cancer 7, 165–197. [DOI] [PubMed] [Google Scholar]
- 2. Schlessinger, J. (2000) Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 3. Eswarakumar, V. P. , Lax, I. , and Schlessinger, J. (2005) Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139–146. [DOI] [PubMed] [Google Scholar]
- 4. Wiedlocha, A. and Sorensen, V. (2004) Signaling, internalization, and intracellular activity of fibroblast growth factor. Curr. Top. Microbiol. Immunol. 286, 45–79. [DOI] [PubMed] [Google Scholar]
- 5. Sletten, T. , Kostas, M. , Bober, J. , Sorensen, V. , Yadollahi, M. , et al. (2014) Nucleolin regulates phosphorylation and nuclear export of fibroblast growth factor 1 (FGF1). PLoS One 9, e90687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu, D. , Kan, M. , Sato, G. H. , Okamoto, T. , and Sato, J. D. (1991) Characterization and molecular cloning of a putative binding protein for heparin‐binding growth factors. J. Biol. Chem. 266, 16778–16785. [PubMed] [Google Scholar]
- 7. Tassi, E. , Al‐Attar, A. , Aigner, A. , Swift, M. R. , McDonnell, K. , et al. (2001) Enhancement of fibroblast growth factor (FGF) activity by an FGF‐binding protein. J. Biol. Chem. 276, 40247–40253. [DOI] [PubMed] [Google Scholar]
- 8. Skjerpen, C. S. , Nilsen, T. , Wesche, J. , and Olsnes, S. (2002) Binding of FGF‐1 variants to protein kinase CK2 correlates with mitogenicty. EMBO J. 27, 4058–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolpakova, E. , Wiedlocha, A. , Stenmark, H. , Klingenberg, O. , Falnes, P. , et al. (1998) Cloning of an intracellular protein that binds selectively to mitogenic acidic fibroblast growth factor. Biochem. J. 336, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mizukoshi, E. , Suzuki, M. , Loupatov, A. , Uruno, T. , Hayashi, H. , et al. (1999) Fibroblast growth factor‐1 interacts with the glucose‐regulated protein GRP75/mortalin. Biochem. J. 343, 461–466. [PMC free article] [PubMed] [Google Scholar]
- 11. Skjerpen, C. S. , Wesche, J. , and Olsnes, S. (2002) Identification of ribosome‐binding protein p34 as an intracellular protein that binds acidic fibroblast growth factor. J. Biol. Chem. 277, 23864–23871. [DOI] [PubMed] [Google Scholar]
- 12. Zakrzewska, M. , Krowarsch, D. , Wielocha, A. , Olsnes, S. , and Otlewski, J. (2005) Highly stable mutants of human fibroblast growth factor‐1 exhibit prolonged biological action. J. Mol. Biol. 352, 860–875. [DOI] [PubMed] [Google Scholar]
- 13. Zakrzewska, M. , Zhen, Y. , Wielocha, A. , Olsnes, S. , and Wesche, J. (2009) Size limitation in translocation of fibroblast growth factor 1 fusion proteins across the endosomal membrane. Biochemistry 48, 7209–7218. [DOI] [PubMed] [Google Scholar]
- 14. Yang, C. , Kim, M. S. , Chakravarty, D. , Indig, F. E. , and Carrier, F. (2009) Nucleolin binds to the proliferating cell nuclear antigen and inhibits nucleotide excision repair. Mol. Cell. Pharmacol. 1, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu, Z. , Fotouhi‐Ardakani, N. , Wu, L. , Maori, M. , Wang, S. , et al. (2002) PTEN associates with the vault particles in Hela cells. J. Biol. Chem. 277, 40247–40252. [DOI] [PubMed] [Google Scholar]
- 16. Zakrzewska, M. , Wielocha, A. , Szlachcic, A. , Krowarsch, D. , Otlewski, J. , et al. (2009) Increased protein stability of FGF1 can compensate for its reduced affinity for heparin. J. Biol. Chem. 284, 25388–25403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vojtek, A. B. and Hollenberg, S. M. (1995) Ras‐Raf interaction: two‐hybrid analysis. Methods Enzymol. 255, 331–342. [DOI] [PubMed] [Google Scholar]
- 18. Fromont‐Racine, M. , Rain, J. C. , and Legrain, P. (1997) Toward a functional analysis of the yeast genome through exhaustive two‐hybrid screens. Nat. Genet. 16, 277–282. [DOI] [PubMed] [Google Scholar]
- 19. Bartel, P. , Chien, C. T. , Sternglanz, R. , and Fields, S. (1993) Elimination of false positives that arise in using the two‐hybrid system. Biotechniques 14, 920–924. [PubMed] [Google Scholar]
- 20. Formstecher, E. , Aresta, S. , Collura, V. , Hamburger, A. , Meil, A. , et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fowler, S. L. , Akins, M. , Zhou, H. , Figeys, D. , and Bennett, S. A. L. (2013) The liver connexin32 interactome is a novel plasma membrane‐mitochondrial signaling nexus. J. Proteome Res. 12, 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rescher, U. and Gerke, V. (2004) Annexins–—unique membrane binding proteins with diverse functions. J. Cell Sci. 117, 2631–2639. [DOI] [PubMed] [Google Scholar]
- 23. Maggi, L. B. , Kuchenruether, M. , Dadey, D. Y. A. , Schwope, R. M. , Grisendi, S. , et al. (2008) Nucleophosmin serves as a rate‐limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell Biol. 28, 7050–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolli, S. , Zito, C. I. , Mossink, M. H. , Wiemer, E. A. , and Bennett, A. M. (2004) The major vault protein is a novel substrate for the tyrosine phosphatase SHP‐2 and scaffold protein in epidermal growth factor signaling. J. Biol. Chem. 279, 29374–29385. [DOI] [PubMed] [Google Scholar]
- 25. Chang, K. H. , Chen, Y. , Chen, T. T. , Chou, W. H. , Chen, P. L. , et al. (1997) A thyroid hormone receptor coactivator negatively regulated by the retinoblastoma protein. Proc. Natl. Acad. Sci. USA 94, 9040–9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Infante, C. , Ramos‐Morales, F. , Fedriani, F. , Bornens, M. , and Rios, R. M. (1999) GMAP‐210, a cis‐Golgi network‐associated protein, is a minus end microtubule‐binding protein. J. Cell Biol. 145, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakai, T. , Liu, L. , Teng, X. , Mukai‐Sakai, R. , Shimada, H. , et al. (2004) Nucling recruits Apaf‐1/pro‐caspase‐9 complex for the induction of stress‐induced apoptosis. J. Biol. Chem. 279, 41131–41140. [DOI] [PubMed] [Google Scholar]
- 28. Burikhanov, R. , Shrestha‐Bhattarai, T. , Qiu, S. , Shukla, N. , Hebbar, N. , et al. (2013) Novel mechanism of apoptosis resistance in cancer mediated by extracellular PAR‐4. Cancer Res. 73, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malecki, J. , Wesche, J. , Skjerpen, C. S. , Wiedlocha, A. , and Olsnes, S. (2004) Translocation of FGF‐1 and FGF‐2 across vesicular membranes occurs during G1‐phase by a common mechanism. Mol. Biol. Cell 15, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zakrzewska, M. , Sorensen, V. , Jin, Y. , Wiedlocha, A. , and Olsnes, S. (2011) Translocation of exogenous FGF1 into cytosol and nucleus is a periodic event independent of receptor kinase activity. Exp. Cell Res. 317, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 31. Wesche, J. , Malecki, J. , Wiedlocha, A. , Skjerpen, C. S. , Claus, P. , et al. (2006) FGF‐1 and FGF‐2 require the cytosolic chaperone HSP90 for translocation into the cytosol and the cell nucleus. J. Biol. Chem. 281, 11405–11412. [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez‐Enfedaque, A. , Bouleau, S. , Laurent, M. , Courtois, Y. , Mignotte, B. , et al. FGF1 nuclear translocation is required for both its neurotrophic activity and its p53‐dependent apoptosis protection. Biochim. Biophys. Acta 1793, 1719–1727. [DOI] [PubMed] [Google Scholar]
- 33. Bouleau, S. , Parvu‐Ferecatu, I. , Rodriguez‐Enfedaque, A. , Rincheval, V. , Grimal, H. , et al. (2007) Fibroblast Growth Factor 1 inhibits p53‐dependent apoptosis in PC12 cells. Apoptosis 12, 1377–1387. [DOI] [PubMed] [Google Scholar]
- 34. Imamura, T. , Oka, S. , Tanahashi, T. , and Okita, Y. (1994) Cell cycle‐dependent nuclear localization of exogenously added fibroblast growth factor‐1 in BALB/c 3T3 and human vascular endothelial cells. Exp. Cell Res. 215, 363–372. [DOI] [PubMed] [Google Scholar]
- 35. Sorensen, V. , Zhen, Y. , Zakrzewska, M. , Haugsten, E. M. , Walchli, S. , et al. (2008) Phosphorylation of fibroblast growth factor (FGF) receptor 1 at Ser777 by p38 mitogen‐activated protein kinase regulates translocation of exogenous FGF1 to the cytosol and nucleus. Mol. Cell Biol. 28, 4129–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiedlocha, A. , Nilsen, T. , Wesche, J. , Sorensen, V. , Malecki, J. , et al. (2005) Phosphorylation‐regulated nucleocytoplasmic trafficking of internalized fibroblast growth factor‐1. Mol. Biol. Cell 16, 794–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.


