Key Points
Early postnatal loss of Pten protein in mice with Nf1 haploinsufficiency causes a fatal juvenile myeloproliferative neoplasm.
Akt and MAPK activities are elevated in juvenile mice with Nf1 haploinsufficiency and Pten protein loss.
Abstract
Juvenile myelomonocytic leukemia (JMML) is an aggressive pediatric mixed myelodysplastic/myeloproliferative neoplasm (MDS/MPN). JMML leukemogenesis is linked to a hyperactivated RAS pathway, with driver mutations in the KRAS, NRAS, NF1, PTPN11, or CBL genes. Previous murine models demonstrated how those genes contributed to the selective hypersensitivity of JMML cells to granulocyte macrophage–colony-stimulating factor (GM-CSF), a unifying characteristic in the disease. However, it is unclear what causes the early death in children with JMML, because transformation to acute leukemia is rare. Here, we demonstrate that loss of Pten (phosphatase and tensin homolog) protein at postnatal day 8 in mice harboring Nf1 haploinsufficiency results in an aggressive MPN with death at a murine prepubertal age of 20 to 35 days (equivalent to an early juvenile age in JMML patients). The death in the mice was due to organ infiltration with monocytes/macrophages. There were elevated activities of protein kinase B (Akt) and mitogen-activated protein kinase (MAPK) in cells at physiological concentrations of GM-CSF. These were more pronounced in mice with Nf1 haploinsufficiency than in littermates with wild-type Nf1, but this model is insufficient to cause cells to be GM-CSF hypersensitive. This new model represents a murine MPN model with features of a pediatric unclassifiable mixed MDS/MPN and mimics many clinical manifestations of JMML in terms of age of onset, aggressiveness, and organ infiltration with monocytes/macrophages. Our data suggest that the timing of the loss of PTEN protein plays a critical role in determining the disease severity in myeloid malignancies. This model may be useful for studying the pathogenesis of pediatric diseases with alterations in the Ras pathway.
Introduction
Juvenile myelomonocytic leukemia (JMML) is an aggressive, mixed myelodysplastic/myeloproliferative neoplasm (MDS/MPN) with a median age at presentation of 2 years. Left untreated, the majority of patients die within a year after diagnosis, primarily due to organ failure and bleeding resulting from infiltration with monocytes/macrophages.1 Hematopoietic stem cell (HSC) transplant is the only cure for JMML, but the relapse rate remains high at 30% to 40%.2,3 Although many investigators have shown that JMML is caused by hyperactivity of the RAS pathway due to driver mutations in KRAS, NRAS, NF1, PTPN11, and CBL genes,4-11 clinical outcomes are independent of these driver mutations.12 Two recent publications of whole-exome sequencing in JMML arrived at a similar conclusion; that is, the absolute number of somatic alterations at diagnosis is a major determinant of survival.13,14
Although the majority of JMML patients experience an aggressive disease course, small subsets have more indolent disease and/or even spontaneous remission,15 leading to difficult clinical decisions. JMML is also a very age-restricted disease. It rarely presents congenitally, but often develops within the first year after birth, with 96% of patients presenting by age 5 years. Children with clinical neurofibromatosis have a 200- to 500-fold increased risk of developing JMML. Other pediatric diseases with a deregulated RAS/mitogen-activated protein kinase (MAPK) pathway, such as neuro-cardio-facial-cutaneous syndromes and Noonan syndrome, also have a markedly increased risk for JMML.3 Of interest, in nearly all of these inherited genetic syndromes, the children “outgrow” their risk for JMML by approximately age 6 years.16 The mechanism underlying this dramatic shift in risk for JMML development remains unknown.
Previous murine models of JMML were developed based on single-gene disruptions and demonstrated how mutant driver genes contribute to selective granulocyte macrophage–colony-stimulating factor (GM-CSF) hypersensitivity, a unifying characteristic of JMML. However, none of them mimic the JMML clinical phenotype of a very age-restricted disease onset coupled with significant aggressiveness. Most of them were absent in demonstrating hyperactivities of protein kinase B (Akt) and MAPK,4,8,17-21 which are found in 55% and 73% of JMML patients, respectively.22 Nevertheless, they have tremendously advanced our understanding underlying JMML leukemogenesis. It is unfortunate that it remains unclear what causes the early death in JMML patients, given that very few transform to acute leukemia.
PTEN (phosphatase and tensin homolog) negatively regulates phosphatidylinositol-3-kinase (PI3K) and MAPK signaling, which are downstream of RAS. Loss of PTEN protein is frequently found in solid tumors, especially breast, lung, prostate, and ovarian cancers, as well as in acute lymphoid leukemia (ALL).23-28 We reported that 67% of JMML patients were PTEN protein deficient.22 Other groups reported that myeloid-specific somatic deletions of Pten in fetal or adult mice produced a transient MPN, with eventual death in adulthood from acute myeloid leukemia (AML) or ALL.29-31 It was also reported that fetal and adult mouse HSCs display marked differences in their self-renewal, differentiated cell output, and gene expression properties, with persistence of a fetal phenotype until 3 weeks after birth.32 The role of Pten in regulating mouse hematopoiesis has been reported to be age dependent.33 We hypothesized that PTEN protein loss at a specific stage in early human childhood might contribute to the early death in JMML patients. In this study, we demonstrate that by inducing Pten protein deficiency at postnatal day (PND) 8 in mice (equivalent to the age of a full-term newborn in humans34), along with haploinsufficiency of Nf1, results in a lethal murine MPN that mimics human pediatric MPNs, including JMML, in terms of age of onset, aggressiveness, and organ infiltration with monocytes/macrophages. Our data suggest that the timing of PTEN protein loss plays a critical role in determining the disease severity in myeloid malignancies.
Materials and methods
For more information on materials and methods, see supplemental Methods, available on the Blood Web site.
Study design and mice
Because mechanisms of PTEN protein loss in human cancers are complex and not well classified, we chose to induce Pten gene deletion at a specific time point to simulate Pten protein loss in mice, even though PTEN mutations are not found in JMML. By crossbreeding the founder mice bearing PtenfloxP/floxP (Ptenfl/fl; B6.129S4-Ptentm1Hwu/J), Nf1Fcr/+ (B6.129S6-Nf1tm1Frc/J), and Mx1-Cre (B6.Cg-Tg[Mx1-Cre]1Cgn/J) (The Jackson Laboratory), we generated experimental mice with Ptenfl/fl with or without Nf1+/ Fcr (Nf1+/−) and Mx1-Cre. Pten deletion was induced by intraperitoneal injection of 30 μL of polyinosinic polycytidylic acid (poly[I:C]) at a concentration of 1 μg/μL in normal saline (InvivoGen) on days 8 and 10 after birth (hereafter referred collectively as PND8), or at the age of 6 weeks (180 μL of poly[I:C] twice; supplemental Figure 1). Mouse tails were clipped at age 2 weeks, and genotyping was performed according to the instructions provided by the vendor. The experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences.
Competitive BMT
Competitive bone marrow transplant (BMT) was performed according to the method published by Zhang et al,30 with modifications. Briefly, adult C57BL/6 recipient mice with CD45.1+ background at age 6 to 8 weeks (The Jackson Laboratory) were lethally irradiated with a single dose of 9.5 Gy delivered by a Mark I 137Cs irradiator with a rotating platform (JL Shepherd and Associates, San Fernando, CA) at a rate of 1.08 Gy/min. Unfractionated bone marrow nucleated cells (WBM cells) from 3-week-old wild-type (WT) controls (2 × 105) or PtenΔ/Δ; Nf1+/− mice (1 × 106) with CD45.2+ background were mixed with 2 × 105 rescue cells from WT CD45.1+ mice. Donor cells were suspended in 120 μL of phosphate-buffered saline/5% mouse serum (Fisher Scientific) and injected into the retro-orbital venous sinus of irradiated recipient mice at 6 hours postirradiation. Transplanted mice were maintained with regular mouse chow and sterile water without additives. Four weeks after BMT, peripheral blood (PB) was collected from recipient mice serially at 2-week intervals for complete blood cell (CBC) count or flow cytometric analysis (fluorescence-activated cell sorting [FACS]) until 18 weeks posttransplant, or when recipient mice were euthanized for overall evaluation.
Results
Myeloid-specific deletion of Pten at PND8 results in a lethal juvenile MPN
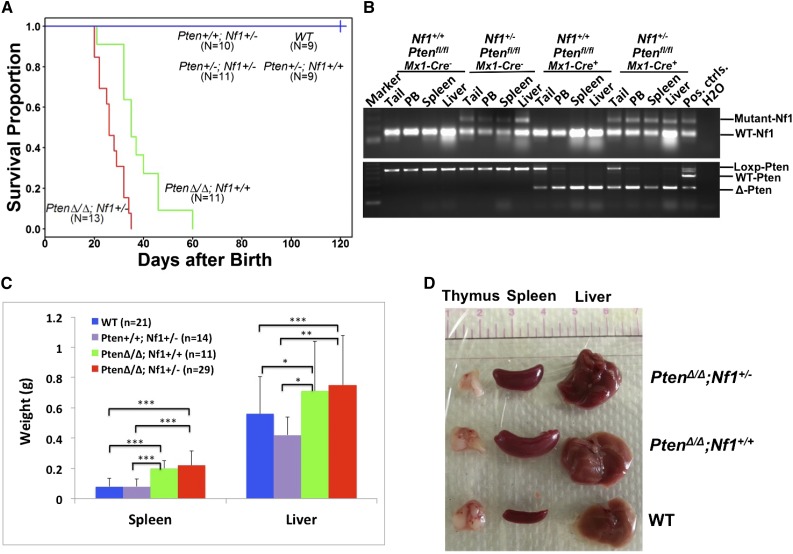
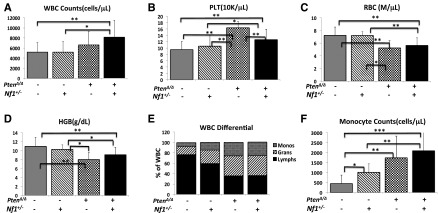
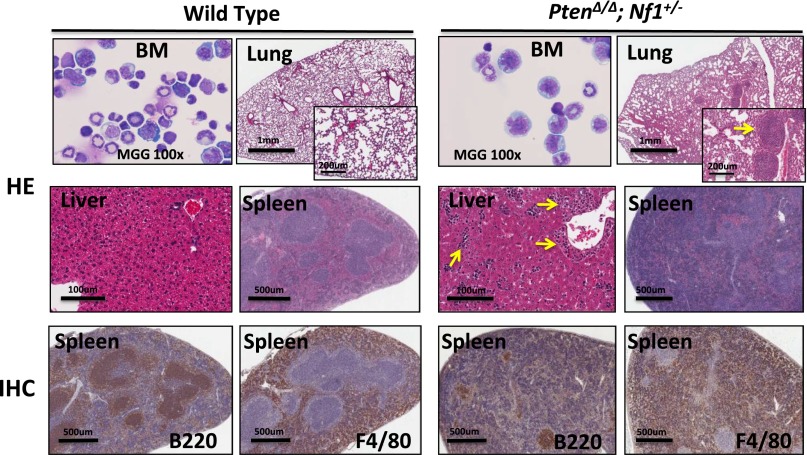
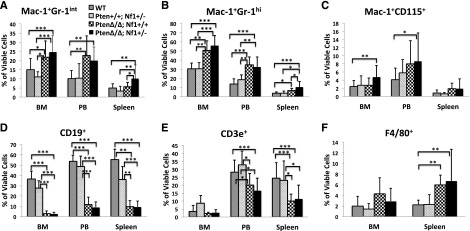
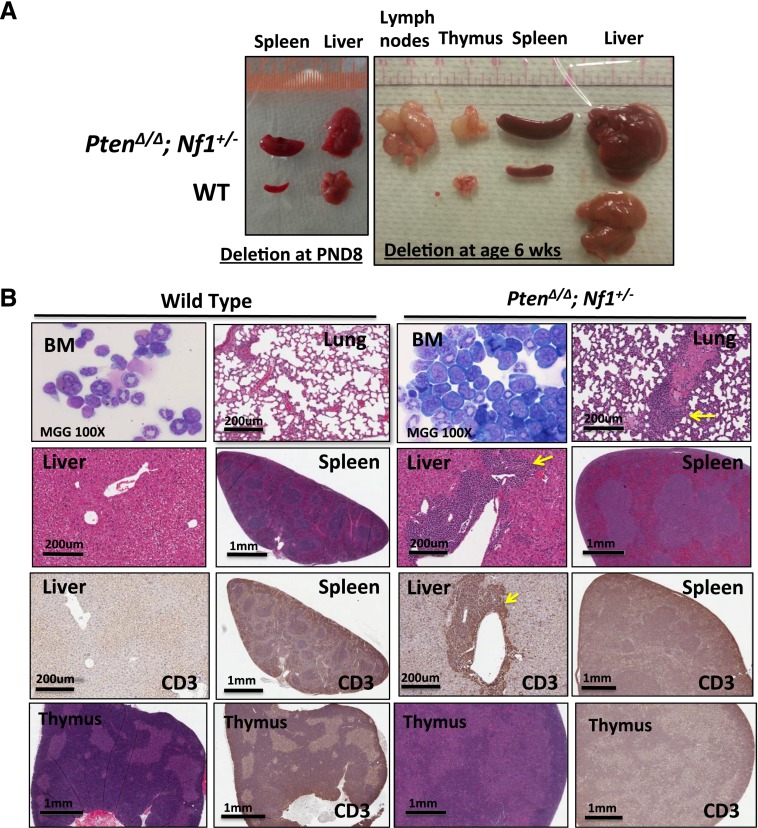
In attempting to test our hypothesis that PTEN protein loss is a second event that contributes to early death in JMML, we designed experimental mice to mimic the putative dynamics of molecular defects in JMML, which is loss of PTEN protein in the first few months after birth following a germ line NF1 mutation. NF1, encoding neurofibromin, negatively regulates Ras activity,35 and was the first well-documented gene that was linked to the selective GM-CSF hypersensitivity in JMML.4,8,17 Mice with germ line homozygous Nf1 inactivation, caused by interrupting the Nf1 exon 31 (Nf1Fcr/Fcr), die during embryonic development. Conversely, heterozygotes do not exhibit any overt disease symptoms until adulthood, with an increased rate of tumorigenesis and a spectrum of tumors similar to WT mice.36,37 Somatic inactivation of Nf1 (Nf1floxP/floxP) or loss of heterozygosity (LOH) caused a JMML-like disease, but with a significant latency (mice did not become sick until age 3 months) (reviewed in Table 1; supplemental Table 4).8,17 In the present study, we generated mice bearing Ptenfl/fl with or without Nf1+/− and Mx1-Cre+, which were born healthy. To simulate an age-specific Pten protein loss in a subject with genetic susceptibility, loss of the Pten gene was induced by intraperitoneal injection with poly(I:C) at PND8, which is equivalent to the age of a full-term newborn in humans.34 Mouse genotypes were identified at an age of 2 weeks, 1 week post–poly(I:C) (supplemental Figure 2). Mice with biallelic Pten deletion and Nf1 haploinsufficiency (PtenΔ/Δ; Nf1+/−) started to display signs of disease (fuller abdomen) in the third week of life (2 weeks post–poly[I:C]). Without intervention, all mice with PtenΔ/Δ; Nf1+/− died at age 20 to 35 days (equivalent to the age of an early juvenile in humans). Mice with PtenΔ/Δ; Nf1+/− had significantly shorter median survival (26 days, n = 13) compared with PtenΔ/Δ; Nf1+/+ mice (35 days, n = 11; P < .001), and with both Pten+/+; Nf1+/− (n = 10) and WT mice (n = 9, 100% survived past the 120-day observation; P < .001) (Figure 1A). Necropsy showed that PtenΔ/Δ mice had substantial hepatosplenomegaly, regardless of Nf1 background, in comparison with both Pten+/+; Nf1+/− (n = 10) and WT littermates (n = 9). No lymphadenopathy or thymus enlargement was found, differing substantially from previous models in which Pten deletion occurred at either the prenatal or the adult stage and in which the mice always died of AML or ALL in adulthood (Table 1; supplemental Table 4).29-31,33 We also noticed that littermates with hemizygous Pten deletion or WT Pten did not show any overt disease symptoms during the 120-day observation, regardless of Nf1 background (Figure 1A). Because our focus was primarily on juvenile leukemia, we studied the mice with only PtenΔ/Δ; Nf1+/−. We euthanized the diseased mice and littermates at age 3 to 4 weeks (2-3 weeks post–poly[I:C]). Tissue genotyping demonstrated that Pten was deleted in PB, spleens, and livers of mice with Ptenfl/fl and Mx1-Cre+, whereas the WT allele of Nf1 remained detectable in mice with all genotypes (Figure 1B), suggesting that Nf1 LOH had not occurred in diseased mice. The size and the weight of spleens and livers were significantly increased in mice with PtenΔ/Δ, regardless of Nf1 background, in comparison with WT and Pten+/+; Nf1+/− mice, but there was no difference between Pten+/+; Nf1+/− and WT mice (Figure 1C-D). CBC counts showed modestly elevated white blood cells only in PtenΔ/Δ; Nf1+/− mice, which was significantly different from littermates with WT Pten with or without Nf1 haploinsufficiency (Figure 2A). Significantly increased platelets and anemia were also found in PtenΔ/Δ mice, regardless of Nf1 background, but mice with PtenΔ/Δ; Nf1+/− had significantly lower platelets than those with PtenΔ/Δ; Nf1+/+ (Figure 2B-D). Blood smear evaluation showed markedly elevated monocytes and granulocytes with normal morphology, but decreased lymphocytes in all PtenΔ/Δ mice (Figure 2E-F; supplemental Table 2). Tissue sections of formalin-fixed organs showed substantial monocyte/macrophage infiltration in the spleens, livers, and lungs from all PtenΔ/Δ mice (Figure 3). Of interest, total cell numbers in bone marrow (BM) were significantly reduced in PtenΔ/Δ; Nf1+/− mice in comparison with WT mice (48.88 × 106 ± 6.49 × 106 vs 82.80 × 106 ± 24.38 × 106; P < .05). BM cytospins demonstrated increases in monocytes and granulocytes with normal morphology in PtenΔ/Δ; Nf1+/− mice (Figure 3). FACS analysis confirmed that PtenΔ/Δ mice with or without Nf1 haploinsufficiency had significant increases of monocytes/macrophages (Mac-1+Gr-1int) and granulocytes (Mac-1+Gr-1high) in BM, blood, and spleen (Figure 4A-B; supplemental Figure 3). Importantly there was a significant difference in monocytes/macrophages and granulocytes in spleens from mice with PtenΔ/Δ; Nf1+/− and those with PtenΔ/Δ; Nf1+/+ (Figure 4A-B). We also found significantly decreased B-cell (CD19+) and T-cell (CD3e+) populations in PB and spleen in PtenΔ/Δ mice, regardless of Nf1 background (Figure 4D-E; supplemental Figure 3). Mature monocytes/macrophages (Mac-1+CD115+) were elevated significantly in BM and PB only in PtenΔ/Δ; Nf1+/− mice, but not in PtenΔ/Δ; Nf1+/+ mice, compared with WT mice (Figure 4C; supplemental Figure 3A). Macrophage (F4/80+) infiltration was confirmed in spleens from PtenΔ/Δ mice regardless of Nf1 background (Figures 3 and 4F; supplemental Figure 3B). FACS profiles showed no difference in mice with WT Pten with or without Nf1 haploinsufficiency. Chromosome stability analysis of BM cells showed no significant difference between mice with PtenΔ/Δ; Nf1+/− (n = 5) and WT littermates (n = 5) (P = .107) (data not shown).
Table 1.
Comparison of JMML patients and the Ptenfl/flNf1+/− mouse model with previously reported mouse models
| JMML patients | Mx1-Cre | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ptenfl/fl Nf1+/− | Ptenfl/fl | Ptenfl/fl | Nf1fl/fl or Nf1-/fl | Lox-Stop-Lox (LSL) | ||||||
| p55−/− | p55+/− | KrasG12D | NrasG12D | Ptpn11D61Y/+ | ||||||
| Age mutation introduced | NApp | PND8 | 6 wk | PND2 | 6 wk | 3 or 6 wk | PND3-PND5 | 3 wk | 3 wk | 3-4 wk |
| Age MPN occurs (postinduction) | 2 y, often <1 y | 3 wk (2 wk) | 9 wk (3 wk) | T-ALL (7 wk) | T-ALL (3 wk) | 7 wk (1-4 wk) | >3 mo (3 mo) | 6 wk (3 wk) | >7 mo (6 mo) | 5 mo (4 mo) |
| Survival | <1 y | 3.5 wk* | 11 wk | 6-8 wk | 7-12 wk | 7.5 mo | >3 mo | >12 mo | >10 mo | |
| Penetrance, % | NApp | 100 | 100 | 100 | 90 | 100 | 100 | 100 | 100 | |
| Leukocytosis | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | |
| Monocytosis | Yes | Yes | Yes | N/A | Yes | Yes | Yes | N/A | Yes | |
| Lymphocytes | N/A | Decreased | Yes | Yes | N/A | Increased | N/A | Increased | N/A | |
| Hemoglobin (hematocrit) | Decreased | Decreased | No | N/A | N/A | Normal | Decreased | Decreased | Decreased | |
| Platelets counts | Normal/decreased | Elevated | No | N/A | N/A | Normal | Normal | Normal | Normal | |
| Hypersensitivity to GM-CSF | Yes | No | N/A | N/A | N/A | Selective | Independent† | Independent† | Independent† | |
| Hepatosplenomegaly | Yes | Yes | Yes | N/A | Yes | Yes | Yes | Yes | Yes | |
| Cell infiltration in Lung | Yes | Yes | Yes‡ | N/A | N/A | No | N/A | Yes§ | No | |
| AML or ALL | No (blasts <20%) | No | T-ALL | T-ALL | Yes | No | No | No | No | |
| Other organs | Skin, lymph nodes, colon | Not found | Thymus, lymph nodes | Thymus, lymph nodes | Lymphoma | No | N/A | Lymph nodes, histiocytoma | No | |
| Elevated Akt activity | 55% patients | Constitutive | N/A | No | Yes | N/A | No | Yes | N/A | Yes |
| Elevated MAPK activity | 73% patients | Yes‖ | N/A | N/A | N/A | Yes‡ | Yes | No | Yes | |
| Reference | 1, 22 | The present study | 33 | 29, 30 | 17 | 20, 21 | 19 | 18 | ||
NApp, Not applicable; N/A, not available.
Equivalent to 1 to 3 y old in humans.
Colony growth in the absence of GM-CSF.
Only observed in Mac-1+ cells.
Lymphocytes.
Preference to GM-CSF over IL-3.
Figure 1.
Tissue genotype, phenotype, and survival of mice with or without Pten deletion and Nf1 mutation. (A) Kaplan-Meier analysis demonstrated the high penetration (100%) of disease and early lethal effect of Pten deletion that was induced at PND8 in mice with or without Nf1 mutation. WT mice were euthanized after age 120 days. Survival in mice with PtenΔ/Δ; Nf1+/− was significantly shorter, with a median survival of 26 days (95% confidence interval [CI], 22-32) (n = 13), and differed from PtenΔ/Δ; Nf1+/+ (35 days [95% CI, 32-46]; P = .001), Pten+/−; Nf1+/−, Pten+/+; Nf1+/−, and WT mice (P < .001). Survival in mice with PtenΔ/Δ; Nf1+/+ significantly differed from littermates with Pten+/−; Nf1+/− or Pten+/+; Nf1+/− and with WT (P < .001). (B) Genotype analysis by polymerase chain reaction on tissues that were collected at age 3 to 4 weeks from the littermate mice received poly(I:C) at PND8, demonstrating that Pten was deleted in the PB, spleen, and liver of mice with Ptenfl/fl and Mx1-Cre+, whereas the WT Nf1 allele was detectable in mice with all genotypes, suggesting that the Nf1 LOH had not occurred in diseased mice. In agreement with prior studies regarding Nf1, the intensity of signals of the mutant and WT Nf1 alleles combine to roughly equal totals. Thus, when a mutant Nf1 allele is present, the density of the WT allele is proportionately decreased (haploinsufficient). (C) Average spleen and liver weights demonstrate hepatosplenomegaly in PtenΔ/Δ mice. Data are presented as mean ± SD, *P < .05; **P < .01; ***P < .001. (D) Photographs of organs from representative littermate mice at age 4 weeks demonstrating hepatosplenomegaly; however, the thymus was spared in PtenΔ/Δ mice regardless of Nf1 background, in comparison with WT littermates (additional relevant data are presented in supplemental Figures 1 and 2). SD, standard deviation.
Figure 2.
CBC counts and white blood cell (WBC) differential analysis in mice at age 3 to 4 weeks (2-3 weeks post–poly[I:C]). CBC counts were performed by a Vet ABC Hematology Analyzer. WBCs (A), platelets (PLT) (B), red blood cells (RBC) (C), hemoglobin (HGB) (D), and differentials (E) were manually counted from blood smears stained with May-Grünwald-Giemsa (MGG). (F) Absolute numbers of monocytes from 4 groups of mice were compared. Data are presented as mean ± SD (n = 10-14 per group). *P < .05; **P < .01; ***P < .001. Additional details are presented in supplemental Table 2. Gran, granulocytes; Lymphs, lymphocytes; Monos, monocytes.
Figure 3.
Morphologic analysis of organ tissues from PtenΔ/Δ; Nf1+/− mice induced at PND8 and from control littermates at age 3 weeks (2 weeks post–poly[I:C]). Cytospins from BM show increased granulocytes and monocytes and substantial monocytic infiltration was found in the lungs, liver, and spleen of mice with PtenΔ/Δ; Nf1+/−. The lungs show nodular monocytic infiltration (indicated by an arrow). Monocytic infiltration was observed in the sinuses of the liver (indicated by arrows). Monocytic infiltration involved both red and white pulps of the spleen, which is accompanied by decreased lymphoid tissues. Images were acquired using a Nikon Eclipse NV with a 100× lens (BM cytospins) and an Aperio CS2 scanner (HE and IHC). BM, bone marrow; HE, hematoxylin and eosin staining, IHC, immunohistochemistry staining with indicated specific antibodies.
Figure 4.
FACS analysis of cell subpopulations from the BM, PB, and spleens of PtenΔ/Δ; Nf1+/− mice induced at PND8 and of control littermates at age 3 to 4 weeks (2-3 weeks post–poly[I:C]). Mice with PtenΔ/Δ, regardless of Nf1 background, had significantly increased monocytes/macrophages and granulocytes (A-C) in BM, PB, and spleen, but elevated mature monocytes were seen in BM and PB from mice with PtenΔ/Δ; Nf1+/− but not PtenΔ/Δ; Nf1+/+ (C). B cells (D) and T cells (E) were both markedly decreased in PB and spleen from mice with PtenΔ/Δ, regardless of Nf1 background; macrophage infiltration was confirmed in spleens from mice with PtenΔ/Δ, regardless of Nf1 background (F). Data are presented as mean ± SD (n = 7-21 per group). *P < .05; **P < .01; ***P < .001. Additional supportive data are presented in supplemental Table 3 and supplemental Figure 4.
In summary, mice with PtenΔ/Δ; Nf1+/− induced at PND8 had decreased hemoglobin; markedly elevated normal-appearing monocytes/macrophages infiltrating BM, spleen, liver, and lungs; and the mice died rapidly at an age equivalent to that of an early juvenile in humans. On the basis of the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice,38 we believe that PtenΔ/Δ; Nf1+/− mice represent a juvenile mouse MPN model that has features of an unclassifiable juvenile mixed MDS/MPN and mimics many JMML clinical manifestations in terms of age of onset, aggressiveness, and organ infiltration with monocytes/macrophages. Our data also suggest that Nf1 haploinsufficiency worsens the accumulation of monocytes/macrophages in the BM and blood (Figure 4C) and the infiltration of monocytes/macrophages and granulocytes in the spleens of mice with PtenΔ/Δ; Nf1+/− compared with littermates with PtenΔ/Δ; Nf1+/+ (Figure 4A-B), which results in a substantially shorter survival (Figure 1A).
Loss of Pten protein promotes HSC migration but does not alter cell sensitivities to GM-CSF and IL-3
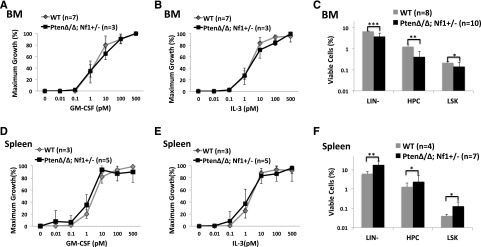
We previously reported that selective GM-CSF hypersensitivity is a unifying characteristic in JMML.1,39 Other groups reported that circulating CD34+ cells were significantly increased, with a lower apoptosis rate in JMML.40 WBM and spleen cells from PtenΔ/Δ; Nf1+/− or WT mice were serum starved for 4 hours after overnight depletion of adherent cells and seeded in semisolid medium with varying concentrations of recombinant mouse GM-CSF or interleukin-3 (IL-3). We found a slight leftward shift in GM-CSF and IL-3 growth curves in spleen cells, but not in WBM cells (Figure 5A-B,D-E). We performed FACS analysis on BM and spleen cells, and found that BM hematopoietic progenitor cells (HPCs), including LIN−, LIN−Scal-1−cKit+, and LIN−Scal-1+cKit+, were significantly decreased, with less apoptosis, whereas HPCs were increased in the spleen (Figure 5C,F; supplemental Figure 4). These findings, along with markedly decreased total cell numbers in the BM of PtenΔ/Δ; Nf1+/− mice, suggest an increased migration of HPCs to the spleen, which was also observed in mice with Pten deletion induced at the other ages.29,30
Figure 5.
Analysis of progenitor cells from the BM and spleens of PtenΔ/Δ; Nf1+/− mice induced at PND8 and of control littermates at age 3 weeks (2 weeks post–poly[I:C]). Colony-forming unit–granulocyte-macrophage (CFU-GM) assays: after serum starvation for 4 hours, 5 × 104 BM cells (A-C) or 2 × 105 spleen cells (D-F) were seeded in triplicate on 1-mL semisolid cultures in 35-mm plates with 0.3% agar and McCoy’s 5A Medium supplemented with nutrients and 15% fetal calf serum containing various concentrations of recombinant mouse GM-CSF or IL-3. (A-B,D-E) Data show there was no significant aberrant curve shift for GM-CSF and IL-3 sensitivities. (C,F) FACS analysis suggests hematopoietic migration from the BM to the spleen in mice with PtenΔ/Δ; Nf1+/−. LIN−, lineage markers negative; HPC, LIN−Scal-1−c-Kit+; LSK, LIN−Scal-1+c-Kit+. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001. Additional supportive data are presented in supplemental Figure 3.
MPN with induced Pten protein loss is transplantable
It was reported that Pten deletion in adult mice caused either a transient or lethal MPN, but mice in those models lived much longer (51-61 days) than that in this reported model, with a significant potential to transform into acute leukemia (reviewed in Table 1).29,30 We questioned whether the MPN phenotype in mice with induced Pten protein loss at PND8 and Nf1 haploinsufficiency could be transplantable without transforming into acute leukemia. WBM cells from PtenΔ/Δ; Nf1+/− or WT littermates with CD45.2+ background mixed with WBM cells from WT mice with CD45.1+ were transplanted into lethally irradiated WT mice (CD45.1+). Recipient mice transplanted with cells from PtenΔ/Δ; Nf1+/− mice developed an indolent myelodysplastic syndrome (MDS) (1/8) or MPN (7/8) by 8 weeks posttransplantation (supplemental Figure 5A), whereas all mice transplanted with control WT BM (n = 5) lived without signs of disease through the 20-week post-BMT observation period. By 12 weeks posttransplantation, PtenΔ/Δ; Nf1+/− mice (3/8) started to die, so we euthanized 5 diseased recipients at 13 to 14 weeks posttransplantation and analyzed grafted tissues. We found that >50% of cells in recipient mice were positive for CD45.2. One recipient mouse was identified with donor-derived T-cell ALL (T-ALL) (CD45.2+); the other 4 mice had signs of MPN after 13 weeks posttransplantation without any signs of acute leukemia (supplemental Figure 5). This finding suggests that the MPN phenotype in Nf1+/− mice with Pten protein loss induced at PND8 is transplantable and that PtenΔ/Δ; Nf1+/− cells possess a proliferative advantage over their WT counterparts. However, the disease latency and severity is significantly different from mice with PtenΔ/Δ induced after 3 weeks of age (adulthood), in which recipients always died of acute leukemia after 12 weeks posttransplantation.29-31,33
Timing of Pten protein loss plays a critical role in determining whether the disease transforms to acute leukemia
Other groups previously reported that mice with Pten deletion at age 6 weeks (equivalent to human adulthood) developed a transient MPN and eventually died of acute leukemia,29,30,33 whereas mice with PtenΔ/Δ and p53−/− died of T-ALL at a similar time, irrespective of whether PtenΔ/Δ was induced at PND2 (equivalent to the third trimester of pregnancy in humans34) or 6 weeks after birth33 (Table 1; supplemental Table 4). To investigate whether the timing of Pten deletion plays a role in determining phenotype and disease aggressiveness, we induced PtenΔ/Δ in mice with or without Nf1+/− at age 6 weeks. As expected, all 14 mice with PtenΔ/Δ developed a transient MPN after 3 weeks post–poly(I:C), and 6 of them subsequently transformed into T-ALL (Figure 6; supplemental Figure 6). Of interest, T-ALL occurred more frequently in mice with PtenΔ/Δ; Nf1+/+ (5/7) than with PtenΔ/Δ; Nf1+/− (1/7), further supporting that Pten protein loss induced at PND8 determines disease severity and that the timing causes a more aggressive MPN in mice than when it occurs in adulthood.29,30,33 This finding also suggests that Nf1 haploinsufficiency contributes to the hematopoietic bias favoring the myeloid lineage.
Figure 6.
Significantly different phenotype in mice with Pten deletion induced at 6 weeks after birth in comparison with mice induced at PND8. (A) Photographs of organs from representative littermate mice with Pten deletion induced at PND8 (euthanized at age 3 weeks) or at age 6 weeks (euthanized at age 9 weeks). Hepatosplenomegaly and significant enlargement of lymph nodes and thymus are demonstrated in mice with PtenΔ/Δ; Nf1+/− induced at age 6 weeks. (B) Micrographs demonstrate BM with increased lymphoblasts and associated lymphocyte infiltration in the lung and liver (indicated by arrows), spleen, and thymus in mice with PtenΔ/Δ; Nf1+/−. BM cytospins were stained by MGG; lung with HE; liver, spleen, and thymus with HE and IHC with antibody against CD3. Images were acquired by an Aperio CS2 scanner (additional relevant FACS data are presented in supplemental Figure 6).
Pten loss at PND8 causes preferential MAPK activation in response to GM-CSF
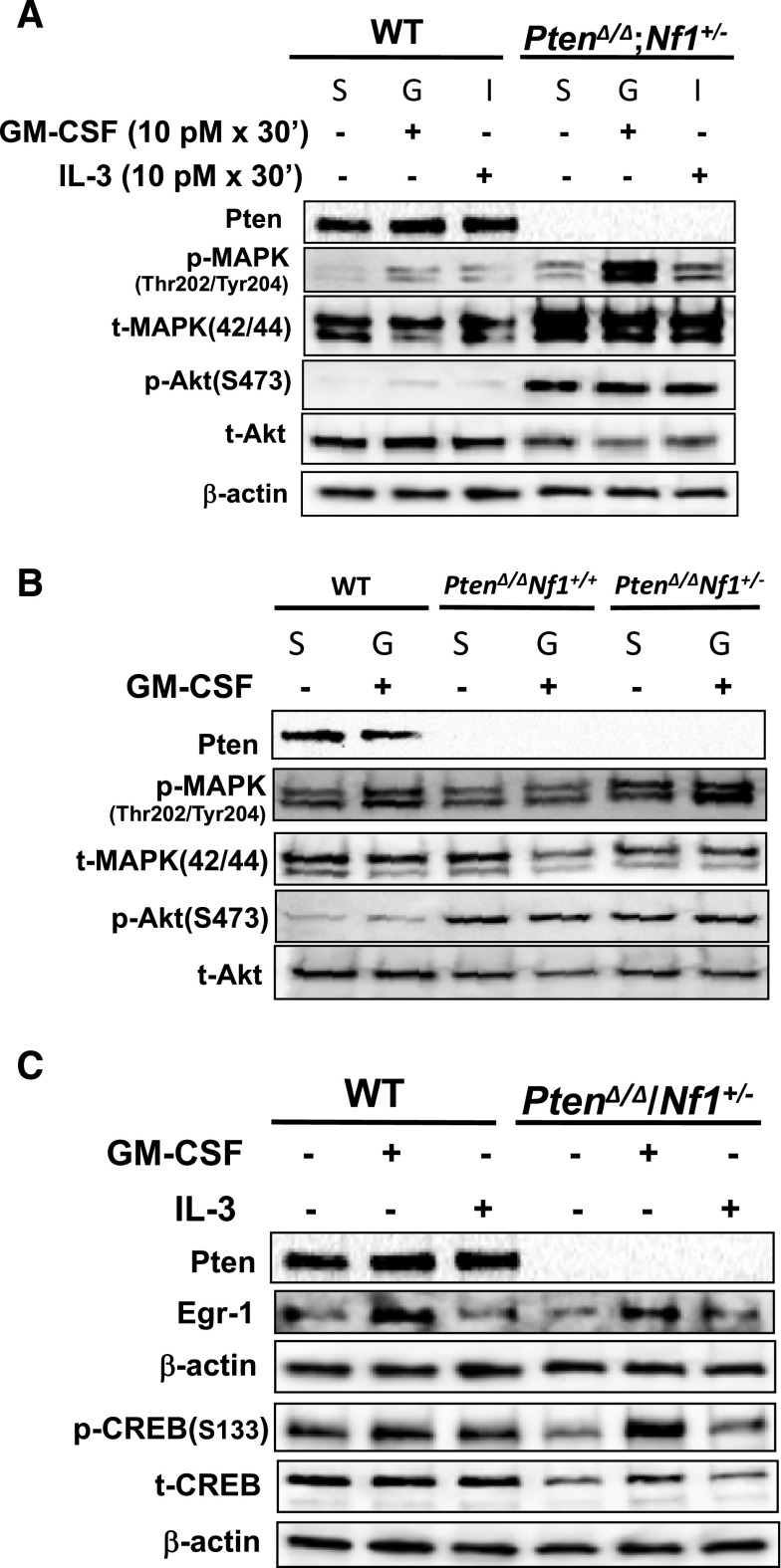
Magee et al reported that Pten deletion induced in adult mice (6 weeks old) increased phosphorylated Akt levels in HSCs and multipotential progenitor cells, whereas Pten deletion at PND2 (equivalent to the third trimester of pregnancy in humans34) did not manifest until 3 to 4 weeks after birth.33 However, MAPK was not impaired in either group.33 To investigate the impact of Pten loss at PND8 on the Akt and MAPK pathways in response to stimulation by GM-CSF or IL-3, WBM cells were harvested from littermates who were WT or PtenΔ/Δ with or without Nf1+/− at age 3 to 4 weeks and stimulated with 10 pM GM-CSF after serum starvation for 4 hours, which is in the physiological concentration range that promotes a “survival-only” response in blood cells41 and in the serum concentration range in AML and CML patients.42,43 We found that Akt was constitutively active in WBM cells with PtenΔ/Δ, regardless of Nf1 background (Figure 7A-B). Importantly, the MAPK pathway was hyperactivated, in a cytokine-dependent manner, with a preferential response to stimulation by GM-CSF over IL-3 in cells with PtenΔ/Δ; Nf1+/− (Figure 7A; supplemental Figure 7A). This finding suggests that disruption of the Akt signaling pathway was undifferentiated in GM-CSF and IL-3 pathways, whereas the MAPK pathway was preferentially upregulated in response to GM-CSF stimulation at physiological concentrations in mice with Pten loss. This observation has not been reported in other murine models (Table 1; supplemental Table 4). We also found that mice with PtenΔ/Δ; Nf1+/− had significantly higher MAPK activities compared with littermates with PtenΔ/Δ; Nf1+/+ (Figure 7B; supplemental Figure 7B), suggesting that elevated MAPK activity may play a critical role in the disease severity in Nf1-deficient mice with PtenΔ/Δ.
Figure 7.
Western blot analysis of the elements in GM-CSF signal transduction pathway in PtenΔ/Δ; Nf1+/− mice induced at PND8 and in control littermates at age 3 weeks (2 weeks post–poly[I:C]). After serum starvation (S) and stimulation with GM-CSF (G) or IL-3 (I), mouse BM cells were lysed in a density of 2 × 107/mL in 2× Laemmli Sample Buffer (Bio-Rad Laboratories). One aliquot with 20 μL per well was loaded onto a 4% to 20% Mini-PROTEAN TGX gel (Bio-Rad). (A) Comparison of the activities of Akt and MAPK pathways in mice with WT and PtenΔ/Δ; Nf1+/− induced at PND8 in response to stimulation by GM-CSF and IL-3 at a physical concentration. Representative data from 8 experiments in littermate mice (age 3-4 weeks) are shown. (B) Comparison the activities of Akt and MAPK pathways in mice with PtenΔ/Δ with or without Nf1+/− in response to stimulation by GM-CSF. Representative data from 3 experiments are shown. (C) Comparison the activation of CREB and expression of Egr-1 in mice with WT and PtenΔ/Δ; Nf1+/−. Representative data from 4 experiments are shown. CREB and Egr-1 were detected on 2 separate blots. p, phosphorylated protein; t, total protein. Additional relevant semiquantitative data are presented in supplemental Figure 7.
Pten protein loss at PND8 significantly decreases total CREB protein but does not alter Egr-1 expression
We reported that JMML patients were deficient in cyclic adenosine monophosphate response element–binding protein (CREB) and early growth response protein 1 (EGR1) levels (85% and 87%, respectively).44 CREB has a dual function in regulating c-JUN, being a repressor in the absence of phosphorylation and an activator when phosphorylated.45 c-JUN is elevated in JMML.46 CREB phosphorylated at Ser133 differentially responds to stimulations by GM-CSF and IL-347 and upregulates Egr-1 transcription. Egr-1 plays a deterministic role in governing hematopoietic development along the macrophage lineage.48 We hypothesized that deficiency of both CREB and Egr-1 was necessary for JMML cells to be selectively GM-CSF hypersensitive. To understand why the selective GM-CSF hypersensitivity was absent in PtenΔ/Δ; Nf1+/− mice, we investigated protein expression of CREB and Egr-1. We stimulated WBM cells with 10 pM GM-CSF or IL-3.41 We found that although total CREB protein was consistently decreased in PtenΔ/Δ; Nf1+/− mice, the pattern of CREB phosphorylation was not altered in response to GM-CSF or IL-3 stimulation, nor was Egr-1 expression (Figure 7C). This finding supports the notion that the regulation of CREB activation and Egr-1 expression is intact in PtenΔ/Δ; Nf1+/− mice. It further suggests that without deregulated CREB activation or altered Egr-1 expression, Pten protein loss along with Nf1 haploinsufficiency is insufficient to alter the cell sensitivity to GM-CSF (proliferation related), although it can cause the lethal monocyte/macrophage accumulation in organs by promoting the mechanism for “survival only.”41
Discussion
Although JMML does not readily transform to acute leukemia, these very young patients still face an early death. We were therefore motivated to identify molecular defects responsible for the aggressiveness and early demise. Prior murine models of JMML have not reproduced the clinical phenotype typified by a very age-restricted and aggressive disease that rarely transforms to acute leukemia (reviewed in Table 1; supplemental Table 4). The discoveries of differences in properties and transcriptional programs of HSCs during development and aging32,49 prompted us to focus on the unique physical and pathological conditions of hematopoiesis in early development. The new murine model we report here simulates PTEN protein loss, a condition observed in many human malignancies. Importantly, we designed our model to occur in the setting of germ line Nf1 haploinsufficiency. Although we recognize that PTEN mutations are not found in JMML, we chose to introduce Pten gene loss merely as a means to simulate the consequence of Pten protein loss. More important, the protein loss was induced at a specific time in juvenile mice at an age equivalent to that of human newborns. This timing resulted in a deadly murine MPN at a juvenile age equivalent to the age of JMML patients. This murine MPN is very aggressive, features monocyte/macrophage accumulation, and does not readily transform to acute leukemia. Our data support our hypothesis that PTEN protein loss in the first few months of life is a significant contributor to early death in JMML.
We previously reported that JMML PTEN protein deficiency was not due to PTEN mutation. If one considers that (1) although somatic deletion of Nf1 or mutant Ras in mice can mimic aberrant GM-CSF hypersensitivity, these mice lack juvenile lethality; and (2) somatic loss of Pten without detectable mutation occurs in T-ALL murine cell lines with K-RasD12/G6450; then taken together, it suggests that PTEN protein loss is an event that follows driver mutations that cause GM-CSF hypersensitivity in JMML and results in lethal disease progression.
Our data demonstrated that MAPK pathway disruption was more pronounced in GM-CSF signaling in mice with Nf1 haploinsufficiency than in WT Nf1 littermates when Pten protein loss was induced at PND8. Others reported that the MAPK pathway was not impaired in mice with Pten deletion induced on either PND2 or at age 6 weeks, and the phosphorylated Akt did not manifest until 3 to 4 weeks after birth in HSCs/multipotential progenitor cells in mice with PtenΔ/Δ induced on PND2.33 Taken together, these results suggest that the upregulated MAPK pathway is a critical player responsible for the early lethal monocyte/macrophage infiltration in vital organs, in addition to the deregulated Akt pathway in mice with Nf1 haploinsufficiency and Pten protein loss induced at PND8. This reasoning may explain why Nf1-deficient mice with Pten loss developed a more severe MPN at a juvenile age, whereas other mice with PtenΔ/Δ induced in the prenatal or adulthood stages developed a latent and transient MPN but eventually transformed into acute leukemia in adulthood.31,33 It is possible that the cells were able to accumulate more molecular defects during expansion, given a less aggressive course. Furthermore, it was reported that in vitro, somatic loss of Pten expression in cells with K-RasD12/G64 is strongly correlated with resistance to MEK inhibition.50 We also found that cells with Pten deletion are resistant to a MAPK inhibitor, PD0325901 (Y.L.L., unpublished observations). Therefore, using MAPK inhibitors as monotherapy should be approached cautiously in JMML patients with PTEN deficiency. Alternatively, because rapamycin, a mTOR inhibitor, can selectively inhibit colony formation of CFU-GM of JMML cells22 and deplete leukemia-initiating cells while restoring normal HSC function,29 mTOR inhibitors could be potential therapeutics for JMML and other pediatric MPNs with PTEN deficiency.
Our finding that the phenotypes in Nf1 haploinsufficient mice with Pten protein loss induced at PND8 were dramatically different from mice induced at age 6 weeks provides a proof-of-concept that the timing of Pten loss determines the disease severity in myeloid malignancy. Furthermore, the notion that Pten disruption caused the differential MAPK activities in juvenile mice with or without Nf1+/− (Figure 7B; supplemental Figure 7B) suggests that the role of Pten in the HSC switch during development and aging may have a different impact on hematopoiesis in hosts with Nf1 haploinsufficiency compared with WT. This idea enriches the observation made by Magee et al,33 wherein they demonstrated that Pten was not required for neonatal hematopoiesis in WT mice. Therefore, this murine model may provide insight for studying other molecules, such as LIN28B, HMGA2, SOX17, BMI1, and CEBPA, which are key players in regulating the HSC switch,49 in patients with Nf1 haploinsufficiency or other diseases with genetic defects.
Given the knowledge that mice with somatic inactivation of Nf1 (Nf1floxP/floxP) or LOH, which causes a JMML-like disease but with significant latency (reviewed in Table 1; supplemental Table 4),8,17 we propose that driver mutations found in NF1 and other genes in the Ras pathway in JMML initialize the cells to be GM-CSF hypersensitive but are insufficient to cause the full juvenile clinical phenotype with lethal results. This premise echoes 2 recent reports that demonstrate that the absolute number of mutations, in addition to the drivers, are critical in predicting the prognosis of JMML.13,14
Acknowledgments
The authors would like to thank to Yuanyan Xu at the University of Alabama at Birmingham for her invaluable help in experimental design and in troubleshooting the mouse breeding.
This work was supported by an Envoys Seeds of Science Award and the Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences (Y.L.L.); the Dr F. E. Joyce Molecular Pathology Laboratory Endowment (P.D.E.); the Arkansas Space Grant Consortium through the National Aeronautics and Space Administration grant NNX13AB29A (R.P.); an Edward P. Evan’s Foundation grant and the National Institutes of Health National Cancer Institute grant R01 CA122023 and National Institute for General Medical Sciences grant P20 GM109005 (M.H.-J. and D.Z.); and the University of Arkansas for Medical Sciences Winthrop P. Rockefeller Endowment for Leukemia Research (D.Z.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.L.L. conceived the project, designed and performed the experiments, analyzed the data, prepared the figures, and wrote the manuscript; Y.Y. designed and performed the experiments and analyzed the data; C.W. performed the experiments; L.S. designed and performed the FACS analyses and BMTs; S.Y.L. designed the experiments, performed the statistical analyses, prepared the figures, and wrote the manuscript; H.N. analyzed the morphologic data from blood, BM, and organ tissues and edited the manuscript; W.F. assisted with the FACS analyses; N.C. performed the blood profiling experiments and analyzed the data; R.P. performed the chromosome stability analysis; Z.X. designed the experiments, analyzed the data, prepared the figures, and edited the manuscript; M.H.-J. supervised an experiment and edited the manuscript; S.L. designed the experiments and edited the manuscript; D.Z. designed and supervised the experiments, analyzed the data, and wrote the manuscript; P.D.E. helped conceive the project, designed and supervised the experiments, analyzed the data, and wrote the manuscript. All authors reviewed the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Y. Lucy Liu, Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences, 4301 W Markham St, #753, Little Rock, AR 72205; e-mail: ylucyliu@uams.edu; and Peter D. Emanuel, Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences, 4301 W Markham St, #623, Little Rock, AR 72205; e-mail: pdemanuel@uams.edu.
References
- 1.Emanuel PD, Shannon KM, Castleberry RP. Juvenile myelomonocytic leukemia: molecular understanding and prospects for therapy. Mol Med Today. 1996;2(11):468–475. doi: 10.1016/1357-4310(96)10044-7. [DOI] [PubMed] [Google Scholar]
- 2.Locatelli F, Nöllke P, Zecca M, et al. European Working Group on Childhood MDS; European Blood and Marrow Transplantation Group. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105(1):410–419. doi: 10.1182/blood-2004-05-1944. [DOI] [PubMed] [Google Scholar]
- 3.Chang TY, Dvorak CC, Loh ML. Bedside to bench in juvenile myelomonocytic leukemia: insights into leukemogenesis from a rare pediatric leukemia. Blood. 2014;124(16):2487–2497. doi: 10.1182/blood-2014-03-300319. [DOI] [PubMed] [Google Scholar]
- 4.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat Genet. 1996;12(2):137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 5.Bader JL, Miller RW. Neurofibromatosis and childhood leukemia. J Pediatr. 1978;92(6):925–929. doi: 10.1016/s0022-3476(78)80362-x. [DOI] [PubMed] [Google Scholar]
- 6.Sheng XM, Kawamura M, Ohnishi H, et al. Mutations of the RAS genes in childhood acute myeloid leukemia, myelodysplastic syndrome and juvenile chronic myelocytic leukemia. Leuk Res. 1997;21(8):697–701. doi: 10.1016/s0145-2126(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 7.Flotho C, Valcamonica S, Mach-Pascual S, et al. RAS mutations and clonality analysis in children with juvenile myelomonocytic leukemia (JMML). Leukemia. 1999;13(1):32–37. doi: 10.1038/sj.leu.2401240. [DOI] [PubMed] [Google Scholar]
- 8.Bollag G, Clapp DW, Shih S, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12(2):144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 9.Kratz CP, Niemeyer CM, Castleberry RP, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106(6):2183–2185. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh ML, Sakai DS, Flotho C, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114(9):1859–1863. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakaguchi H, Okuno Y, Muramatsu H, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013;45(8):937–941. doi: 10.1038/ng.2698. [DOI] [PubMed] [Google Scholar]
- 12.Bresolin S, Zecca M, Flotho C, et al. Gene expression-based classification as an independent predictor of clinical outcome in juvenile myelomonocytic leukemia. J Clin Oncol. 2010;28(11):1919–1927. doi: 10.1200/JCO.2009.24.4426. [DOI] [PubMed] [Google Scholar]
- 13.Stieglitz E, Taylor-Weiner AN, Chang TY, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47(11):1326–1333. doi: 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caye A, Strullu M, Guidez F, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47(11):1334–1340. doi: 10.1038/ng.3420. [DOI] [PubMed] [Google Scholar]
- 15.Matsuda K, Shimada A, Yoshida N, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109(12):5477–5480. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 16.Niemeyer CM, Arico M, Basso G, et al. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS) Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. Blood. 1997;89(10):3534–3543. [PubMed] [Google Scholar]
- 17.Le DT, Kong N, Zhu Y, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103(11):4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 18.Chan G, Kalaitzidis D, Usenko T, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113(18):4414–4424. doi: 10.1182/blood-2008-10-182626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Haigis KM, McDaniel A, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117(6):2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun BS, Tuveson DA, Kong N, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101(2):597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Meter ME, Díaz-Flores E, Archard JA, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109(9):3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YL, Castleberry RP, Emanuel PD. PTEN deficiency is a common defect in juvenile myelomonocytic leukemia. Leuk Res. 2009;33(5):671–677. doi: 10.1016/j.leukres.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsui S, Inoue H, Yasuda K, et al. Reduced expression of PTEN protein and its prognostic implications in invasive ductal carcinoma of the breast. Oncology. 2005;68(4-6):398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Yang R, Yue D, Zhang Z. Expression of FAK and PTEN in bronchioloalveolar carcinoma and lung adenocarcinoma. Lung. 2009;187(2):104–109. doi: 10.1007/s00408-008-9130-6. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez M, Siwko S, Zeng L, Li J, Yi Z, Liu M. Prostate-specific G-protein-coupled receptor collaborates with loss of PTEN to promote prostate cancer progression [published online ahead of print June 1, 2015]. Oncogene. doi: 10.1038/onc.2015.170. doi:10.1038/onc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schöndorf T, Göhring UJ, Roth G, et al. Time to progression is dependent on the expression of the tumour suppressor PTEN in ovarian cancer patients. Eur J Clin Invest. 2003;33(3):256–260. doi: 10.1046/j.1365-2362.2003.01116.x. [DOI] [PubMed] [Google Scholar]
- 27.Silva A, Yunes JA, Cardoso BA, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762–3774. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuurbier L, Petricoin EF, III, Vuerhard MJ, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica. 2012;97(9):1405–1413. doi: 10.3324/haematol.2011.059030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Grindley JC, Yin T, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441(7092):518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Li Y, Gao C, et al. Relevant mouse model for human monocytic leukemia through Cre/lox-controlled myeloid-specific deletion of PTEN. Leukemia. 2010;24(5):1077–1080. doi: 10.1038/leu.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowie MB, Kent DG, Dykstra B, et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci USA. 2007;104(14):5878–5882. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11(3):415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West JR. Fetal alcohol-induced brain damage and the problem of determining temporal vulnerability: a review. Alcohol Drug Res. 1987;7(5-6):423–441. [PubMed] [Google Scholar]
- 35.Bollag G, McCormick F, Clark R. Characterization of full-length neurofibromin: tubulin inhibits Ras GAP activity. EMBO J. 1993;12(5):1923–1927. doi: 10.1002/j.1460-2075.1993.tb05841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brannan CI, Perkins AS, Vogel KS, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8(9):1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 37.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7(3):353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 38.Kogan SC, Ward JM, Anver MR, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 39.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77(5):925–929. [PubMed] [Google Scholar]
- 40.Timeus F, Crescenzio N, Baldassarre G, et al. Functional evaluation of circulating hematopoietic progenitors in Noonan syndrome. Oncol Rep. 2013;30(2):553–559. doi: 10.3892/or.2013.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas D, Powell JA, Green BD, et al. Protein kinase activity of phosphoinositide 3-kinase regulates cytokine-dependent cell survival. PLoS Biol. 2013;11(3):e1001515. doi: 10.1371/journal.pbio.1001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sallerfors B, Olofsson T. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and granulocyte colony-stimulating factor (G-CSF) in serum during induction treatment of acute leukaemia. Br J Haematol. 1991;78(3):343–351. doi: 10.1111/j.1365-2141.1991.tb04447.x. [DOI] [PubMed] [Google Scholar]
- 43.Balleari E, Bason C, Visani G, Gobbi M, Ottaviani E, Ghio R. Serum levels of granulocyte-macrophage colony-stimulating factor and granulocyte colony-stimulating factor in treated patients with chronic myelogenous leukemia in chronic phase. Haematologica. 1994;79(1):7–12. [PubMed] [Google Scholar]
- 44.Liu YL, Lensing SY, Yan Y, Cooper TM, Loh ML, Emanuel PD. Deficiency of CREB and over expression of miR-183 in juvenile myelomonocytic leukemia. Leukemia. 2013;27(7):1585–1588. doi: 10.1038/leu.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamph WW, Dwarki VJ, Ofir R, Montminy M, Verma IM. Negative and positive regulation by transcription factor cAMP response element-binding protein is modulated by phosphorylation. Proc Natl Acad Sci USA. 1990;87(11):4320–4324. doi: 10.1073/pnas.87.11.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Kondo T, Voorhorst CS, et al. Increased c-Jun expression and reduced GATA2 expression promote aberrant monocytic differentiation induced by activating PTPN11 mutants. Mol Cell Biol. 2009;29(16):4376–4393. doi: 10.1128/MCB.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HJ, Mignacca RC, Sakamoto KM. Transcriptional activation of egr-1 by granulocyte-macrophage colony-stimulating factor but not interleukin 3 requires phosphorylation of cAMP response element-binding protein (CREB) on serine 133. J Biol Chem. 1995;270(27):15979–15983. doi: 10.1074/jbc.270.27.15979. [DOI] [PubMed] [Google Scholar]
- 48.Krishnaraju K, Hoffman B, Liebermann DA. Early growth response gene 1 stimulates development of hematopoietic progenitor cells along the macrophage lineage at the expense of the granulocyte and erythroid lineages. Blood. 2001;97(5):1298–1305. doi: 10.1182/blood.v97.5.1298. [DOI] [PubMed] [Google Scholar]
- 49.Babovic S, Eaves CJ. Hierarchical organization of fetal and adult hematopoietic stem cells. Exp Cell Res. 2014;329(2):185–191. doi: 10.1016/j.yexcr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Shieh A, Ward AF, Donlan KL, et al. Defective K-Ras oncoproteins overcome impaired effector activation to initiate leukemia in vivo. Blood. 2013;121(24):4884–4893. doi: 10.1182/blood-2012-05-432252. [DOI] [PMC free article] [PubMed] [Google Scholar]