Abstract
Long Terminal Repeat (LTR) Retrotransposons are an abundant class of genomic parasites that replicate by insertion of new copies into the host genome. Fungal LTR retrotransposons prevent mutagenic insertions through diverse targeting mechanisms that avoid coding sequences, but conserved principles guiding their target site selection have not been established. Here we show that insertion of the fission yeast LTR retrotransposon Tf1 is guided by the DNA binding protein Sap1, and that the efficiency and location of the targeting depend on the activity of Sap1 as a replication fork barrier. We propose that Sap1 and the fork arrest it causes guide insertion of Tf1 by tethering the integration complex to target sites.
Retrotransposons are mobile genetic elements that replicate through an RNA intermediate that is reverse transcribed into a cDNA capable of insertion elsewhere in the genome. By virtue of this amplifying mechanism, retrotransposons comprise large portions of many eukaryotic genomes, and have a critical influence on their evolution(1). Fungal LTR retrotransposons minimize their mutagenic potential by carefully selecting integration sites away from protein coding sequences(2). The different families of LTR retrotransposons employ a variety of strategies for this selective target site selection, but current models posit tethering interactions between retrotransposon proteins and host DNA binding factors.
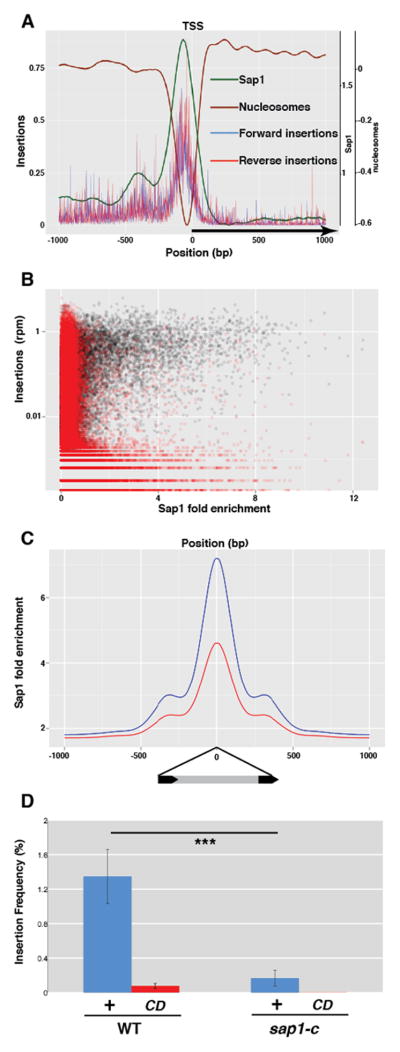
The fission yeast genome shows signs of ancient and persistent colonization by the LTR retrotransposons Tf1 and Tf2, members of the Metaviridae/Ty3-gypsy like group of transposable elements(3). Both Tf1 and Tf2 exhibit a preference for insertion into promoters of RNA polymerase II transcribed genes(4, 5) coinciding with the nucleosome free region (NFR) that is usually present preceding the transcription start site. The main determinant of NFR presence in fission yeast promoters is Sap1(6), which binds DNA as homopolymers to clusters of a 5-bp sequence motif(7, 8). To test whether Sap1 binding coincided with transposition hotspots we performed high-throughput sequencing of transposon-host genome junctions in cultures overexpressing a genetically marked Tf1 transposon(4). Genome-wide correlation analysis shows a strong association of Sap1 enrichment(9) with insertion sites (Fig. 1A,B, fig. S1a). Sap1 is strongly enriched at the previously described Tf1 hotspots, such as the promoters of class II genes (Fig 1A, fig. S1b). Peaks of significant Sap1 enrichment (MACS(10)) account for 63.1% of transposition points, while covering only 5.1% of the host genome, and contained more efficient insertion points than the rest of the genome (fig. S1c). Logistic regression analysis revealed that Sap1 binding is a strong predictor of insertion position (AUC-0.5WT=0.217, fig. S2a,b). However, correlation between Sap1 fold enrichment and number of insertion points, while significant (Spearman’s rho=0.70, p=1e-10), shows a wide variability beyond the threshold of significant enrichment (fig. S1a,b), suggesting that Sap1 binding is not the only factor affecting target site competence. Insertion points coincide precisely with a maximum of Sap1 enrichment(9), strongly indicating that Sap1 determines Tf1 target site selection (Fig. 1C). To test the involvement of Sap1 in Tf1 transposition, we performed high-throughput insertion analysis in a sap1 mutant with a lower affinity for DNA (sap1-c)(9). sap1-c mutants exhibited a drastically reduced transposition frequency (t-test, p<0.001, n=21, Fig. 1D). Additionally, the strong association of insertion points with Sap1 was decreased (Fig. 1C), the portion of insertions in Sap1-enriched regions fell to 49.9%, and the accuracy of Sap1 binding as a predictor of insertion dropped (AUC-0.5sap1-c=0.097, fig. S2a), indicating that transpositions are dispersed away from Sap1 binding peaks. Importantly, the sap1-c background showed no defects in cDNA processing or significantly altered levels of integrase, suggesting that the transposition defect is due to impaired integration (fig. S3). Together, these data show that Sap1 is a major determinant of Tf1 insertion target site selection.
Fig. 1.
Tf1 transposition into Sap1 binding regions. (A) Sap1, nucleosome positioning and average insertion number in reads per million (rpm) at type II genes aligned at the Transcription Start Site (TSS)(B) Genome-wide correlation between transposition (insertion number, rpm) and Sap1 binding in 500bp windows. Black: genomic windows; Red: randomized value pairs. (C) WT Sap1 enrichment around WT insertions (blue) and sap1-c insertions (red). (D) Transposition frequency in WT and sap1-c mutant of Integrase + (+) and Catalytic Dead (CD) Tf1. Error bars depict s.d. and asterisks depict statistically significant differences.
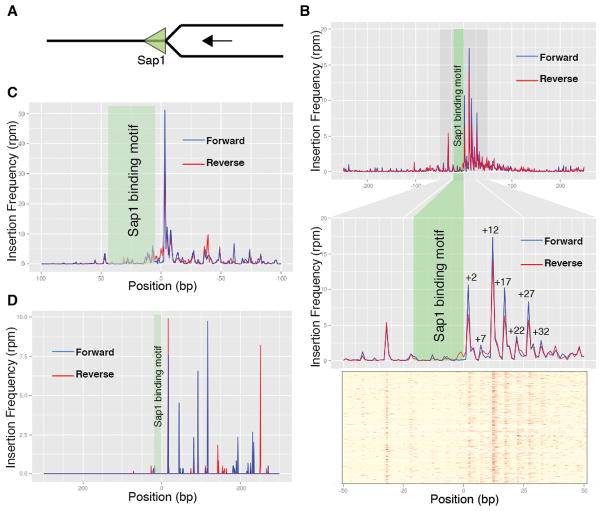
Sap1 is an essential factor with functions affecting genome integrity during DNA replication(11). It has a demonstrated role in forming directional replication fork barriers (RFB)(12, 13). We plotted Tf1 insertion density around Sap1 binding motifs, taking into account their orientation (Fig. 2A). Sap1 binding motifs exhibit enrichment of insertions around them (Fig. 2B), indicating that Sap1 binding directs transposition but protects its footprint. Strikingly, most insertion events occurred 3′ of the Sap1 binding motif (Wilcoxon signed rank test [5,99<mu<7,99] 95%CI, p<2e-16, n=888), displaying a prominent periodicity of peaks (Fig. 2B and fig. S4). This is the side of the motif where confirmed instances of Sap1 dependent RFB cause fork arrest(8, 9, 12). Moreover, in both confirmed Sap1 dependent RFB, the replication terminator Ter1 located at rDNA(12, 13) and the solo LTR interspersed in the genome(9), most insertions occurred on the blocking side of the Sap1 barrier, suggesting that the RFB influences site selection (Fig. 2, C and D). Consistently, Tf1 insertion hotspots and Sap1 binding regions coincide with domains of γH2A deposition and with DNA Pol ε (Cdc20) maxima in undisturbed S-phase, both markers of replication fork arrest(14, 15) (fig. S5). Since Sap1 fork barrier activity is not a function of binding affinity but of binding site structure(8), this observation could potentially explain the variability in transposition competence of Sap1 binding sites (fig. S1a,b), and why the sap1-c allele, which only modestly lowers DNA binding but severely affects RFB activity(9), so dramatically decreases Tf1 transposition (Fig. 1D).
Fig. 2.
Insertion profile around Sap1 binding motifs. (A) Sap1 binding motifs are oriented as blocking replication forks advancing in the right-to-left orientation. (B) Averaged transposition frequency around Sap1 binding motifs (n=888). Upper panel: 500bp window; middle panel: 100bp zoom-in window; lower panel: 100bp heat-map of individual motifs. (C) Averaged transposition frequency around Tf2 LTR (n=152). (D) Averaged transposition frequency around Ter1 (n=3).
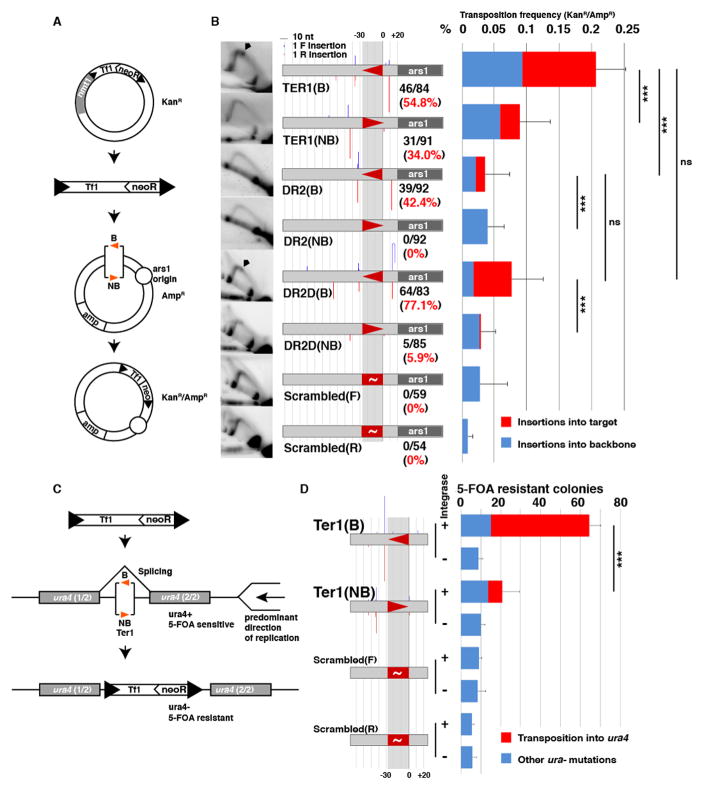
To test the hypothesis that Sap1-dependent fork arrest guides Tf1 insertion, we assessed whether the transposition competence of Sap1 binding sites correlates with the intensity of Sap1 binding or with their RFB activity. We tested the influence of Sap1 binding site orientation with respect to fork progression on transposition efficiency in wild type cells, using three well characterized Sap1 binding sites: (i) the rDNA replication terminator Ter1(12, 13), a very efficient RFB; (ii) the synthetic sequence DR2, derived from in vitro Sap1 binding selection(16) but an inefficient RFB; and (iii) DR2D, a mutation of DR2 that enhances its RFB activity (8). We introduced these Sap1 binding sites in one of the two orientations (Blocking: B; or Non-Blocking: NB) into autonomously replicating plasmids, in close proximity to a replication origin so as to control the predominant direction of fork progression over the motif. We then used these plasmids as transposition acceptors in a targeting assay (17) (Fig. 3A). The results are summarized in Fig. 3B. 2D native-native gel electrophoresis of replication intermediates confirmed that Ter1 and DR2D, but not DR2, are efficient RFB in their blocking orientation (Fig. 3B). ChIP analysis showed little difference in Sap1 enrichment between the two orientations of each motif but revealed that DR2 is the strongest Sap1 binder, with DR2D as the weakest and Ter1 showing intermediate enrichment (fig. S6A). The results of the transposition trap experiment show that RFB competency (Ter1 and DR2D, but not DR2) as well as blocking ability (B orientation) determined higher transposition frequency into the target site (n=3 biological replicates per condition, Tukey Range test, p=0.0006). All insertions displayed 5bp target site duplications (TSD, not shown), indicating that they were integrase-mediated transpositions. These results indicate that transposition into Sap1 binding regions depends not on their Sap1 binding affinity but on their efficiency as RFB.
Fig. 3.
Transposition competence of Sap1 binding sites depends on RFB activity. (A) Plasmid transposition trap strategy. (B) Transposition into Ter1, DR2, DR2D and Scrambled binding motifs in plasmid transposition trap assay. Left column: 2D gel electrophoresis; RFB signals are marked with an arrowhead. Middle column: diagram of target site with insertion sites as columns (blue in forward, red in reverse orientation) at the insertion position of height proportional to number of insertions. Sap1 binding sites depicted by triangles, in blocking (B, pointing left) and non-blocking (NB, pointing right) orientations. Number of insertions into the target site (150bp window around the Sap1 binding motif) over the total number of insertions in black and percentage in red numbers. Right column: Frequency of transposition into the plasmid (KanR/AmpR plasmids over AmpR total plasmids), with insertions into the target site in red, and insertions into the plasmid backbone in blue. Error bars depict s.d. (C) Intron transposition trap strategy. (D) Insertion into Ter1 and Scrambled binding motifs in intron transposition trap assay. Diagram of motif arrangement and insertions as in (B). Proportion of 5-FOA resistant colonies due to transposition into ura4 in red, due to other mutations in blue.
We next tested if the effect of target site orientation extended to genomic positions. We set up a transposon trap system in which the target site is placed inside an artificial intron in the reporter gene ura4, allowing selection of insertions by treatment with the counterselection drug 5-Fluoroorotic acid (5-FOA). ura4 is passively replicated by forks approaching from two nearby replication origins on its centromeric side (18) allowing us to correlate the target site efficiency with its competence as a RFB (Fig. 3C). Blocking (B) or non-blocking (NB) orientations of Ter1 showed equal binding of Sap1 (fig. S6b). However, insertion frequency was 10 fold larger in the Ter1 motif placed in the blocking orientation (t-test, p<0.001, n=4 biological replicates per condition). Once again, all insertions exhibited TSD (not shown). We conclude that the efficiency of insertion near a Sap1 binding motif depends on its ability to cause fork arrest.
These observations prompted us to examine how Sap1, the Tf1 intasome (integration complex), and the replication fork interact. We split Sap1 into functional domains and tested for interactions with the full length Tf1 integrase with a yeast two-hybrid assay, which revealed that the Tf1 integrase interacts directly with the C-terminal dimerization domain of Sap1(16) (fig. S7). To evaluate the role of this interaction and the arrested fork in Tf1 transposition, we turned to chromosome conformation capture (3C) to measure tethering of mature cDNA at Sap1 dependent and independent RFB (Fig 4A). Sap1 bound to Ter1 in the blocking orientation led to prominent recruitment of Tf1 cDNA, while the non-blocking orientation was unable to recruit (Fig. 4B Ter1 B/NB). Tethering to Ter1 was also dependent on WT Sap1 and the presence of Tf1 integrase in the intasome (Fig. 4B, sap1-c, Δint panels). This suggests that the direct interaction of Sap1 with Integrase (fig. S7) participates in intasome recruitment, and that Sap1 bound to cDNA (fig. S8) through its cognate binding sequences in the LTR(9) is not sufficient to localize the intasome by multimerization with genome-bound Sap1. A Sap1-independent RFB (Ter2, dependent on the DNA binding factor Reb1 (19)) did not tether the cDNA (Fig. 4B, Ter2B/Ter2NB). This is consistent with our genome-wide observations that class III genes and other Sap1 independent RFB are not hotspots for Tf1 transposition despite causing polar fork arrest (not shown) (15, 19). Combined, these results suggest that integrase, Sap1, and fork barrier activity must be present to tether Tf1 cDNA to the target site and guide insertion.
Fig. 4.
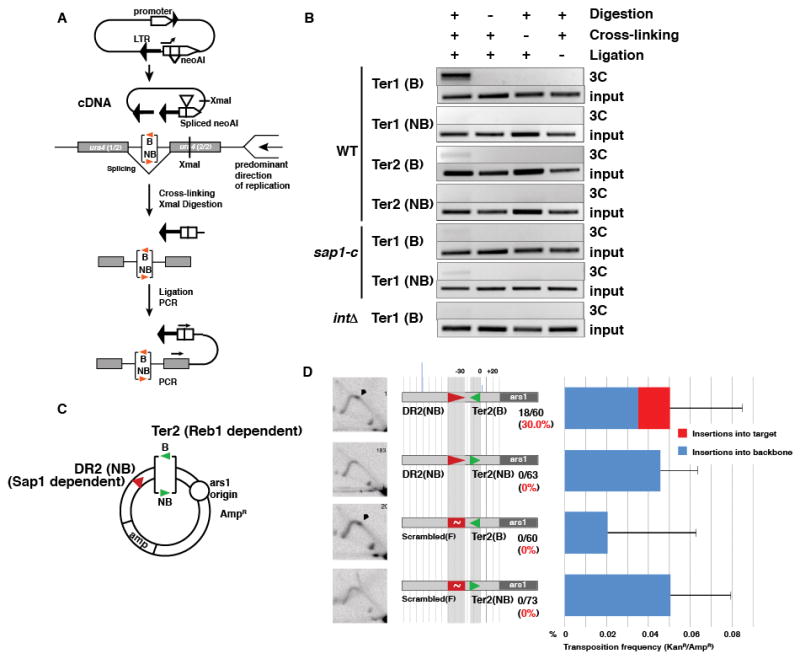
Sap1 binding and RFB activity collaborate to tether the intasome. (A) 3C analysis of cDNA tethering. (B) cDNA tethering at Sap1 dependent (Ter1) and independent (Ter2) RFB in WT, sap1-c mutant and integrase frameshift Tf1 mutant. (C) Separation of Sap1 binding and RFB activities. (D) Results of plasmid trap assay. Left column: 2D gel electrophoresis. Middle column: Diagram of arrangement of Sap1 (DR2 and Scrambled) and Reb1 (Ter2) binding motifs, insertion points and frequency depicted as in Fig. 3B.
We next tested if the RFB and Sap1 binding requirements are separable. If so, we could rescue insertion into a non-RFB Sap1 binding site by providing an independent RFB in cis. We cloned Ter2 next to the DR2 binding site placed in the non-blocking orientation (Fig. 4C and fig. S9). Ter2 rescued the targeting efficiency of DR2 only when placed in the blocking orientation (Mann-Whitney U test, p<0.001, n=3 biological replicates per condition, Fig. 4D). Part of the increase in targeting to DR2 could be caused by replication forks converging onto the Ter2 blocked fork to complete S phase, approaching DR2 in the blocking orientation. Accordingly, insertions are detectable on the blocking side of DR2 (Fig. 4D). However, transposition also occurred near Ter2 into the side of the motif where Reb1 stops the fork, suggesting that features of the arrested fork, and not the location of binding sites, are the major determinants of target site choice. Together, these results reveal that Tf1 transposition targeting requires two separable conditions, both necessary but neither sufficient: (1) Sap1 binding and (2) an active RFB.
Insertion of Tf1 into an arrested replication fork could be a result of strand-transfer reactions between the cDNA and replication intermediates, like the insertion of the bacterial Tn7 transposon into the lagging-strand template at replication terminators(20). Alternatively, fork arrest or restart may fire DNA damage checkpoint and repair mechanisms that participate in insertion(21), or leave epigenetic marks to guide it(22, 23) (fig. S5 and S10). Other LTR retrotransposons may display similar preference for RFB. The S cerevisiae LTR retrotransposons Ty1 (Copia group) and Ty3 (Gypsy group) insert upstream of RNA Pol III transcribed genes like tRNA and 5S(22–25) which are confirmed RFB(26) (fig. S10). Similarly, other LTR retrotransposons with insertion preference for heterochromatin might use fork stalling at satellite repeats in pericentromeric DNA(27).
Supplementary Material
Acknowledgments
The authors are indebted to Henry Levin for strains, reagents, and technical advice as well as critical discussions. This work was supported by a Searle Scholar Award and NIH grant 1R01GM105831 (to MZ) and a Rutgers Biotechnology Training Program Grant 5T32GM008339 (to JZJ). The high throughput sequencing data reported in this paper are archived at the GEO repository (http://www.ncbi.nlm.nih.gov/geo), under accession GSE67692.
Footnotes
The authors declare no financial conflicts of interest.
References and Notes
- 1.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 3.Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Research. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y, Levin HL. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010;20:239–248. doi: 10.1101/gr.099648.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee AG, et al. Serial number tagging reveals a prominent sequence preference of retrotransposon integration. Nucleic Acids Res. 2014;42:8449–8460. doi: 10.1093/nar/gku534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsankov A, Yanagisawa Y, Rhind N, Regev A, Rando OJ. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res. 2011;21:1851–1862. doi: 10.1101/gr.122267.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arcangioli B, Ghazvini M, Ribes V. Identification of the DNA-binding domains of the switch-activating-protein Sap1 from S.pombe by random point mutations screening in E.coli. Nucleic Acids Res. 1994;22:2930–2937. doi: 10.1093/nar/22.15.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krings G, Bastia D. Molecular architecture of a eukaryotic DNA replication terminus-terminator protein complex. Mol Cell Biol. 2006;26:8061–8074. doi: 10.1128/MCB.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaratiegui M, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lahondes R, Ribes V, Arcangioli B. Fission Yeast Sap1 Protein Is Essential for Chromosome Stability. Eukaryotic Cell. 2003;2:910. doi: 10.1128/EC.2.5.910-921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mejía-Ramírez E, Sánchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernández P. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol. 2005;25:8755–8761. doi: 10.1128/MCB.25.19.8755-8761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krings G, Bastia D. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem. 2005;280:39135–39142. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- 14.Rozenzhak S, et al. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 2010;6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014;12:101. doi: 10.1186/s12915-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazvini M, Ribes V, Arcangioli B. The essential DNA-binding protein sap1 of Schizosaccharomyces pombe contains two independent oligomerization interfaces that dictate the relative orientation of the DNA-binding domain. Mol Cell Biol. 1995;15:4939–4946. doi: 10.1128/mcb.15.9.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leem YE, et al. Retrotransposon Tf1 Is Targeted to Pol II Promoters by Transcription Activators. Mol Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Sánchez-Gorostiaga A, López-Estraño C, Krimer DB, Schvartzman JB, Hernández P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol. 2004;24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricker AD, Peters JE. Vulnerabilities on the Lagging-Strand Template: Opportunities for Mobile Elements. Annu Rev Genet. 2014 doi: 10.1146/annurev-genet-120213-092046. [DOI] [PubMed] [Google Scholar]
- 21.Curcio MJ, et al. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol Cell Biol. 2007;27:8874–8885. doi: 10.1128/MCB.01095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mularoni L, et al. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 2012;22:693–703. doi: 10.1101/gr.129460.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Research. 2012;22:704–713. doi: 10.1101/gr.129585.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridier-Nahmias A, et al. An RNA polymerase III subunit determines sites of retrotransposon integration. Science. 2015;348:585–588. doi: 10.1126/science.1259114. [DOI] [PubMed] [Google Scholar]
- 25.Qi X, et al. Retrotransposon profiling of RNA polymerase III initiation sites. Genome Research. 2012;22:681–692. doi: 10.1101/gr.131219.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 27.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Castro E, et al. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. The EMBO Journal. 2012;31:124–137. doi: 10.1038/emboj.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi A, et al. RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc Natl Acad Sci U S A. 2012;109:E2306–15. doi: 10.1073/pnas.1205278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atwood A, Choi J, Levin HL. The Application of a Homologous Recombination Assay Revealed Amino Acid Residues in an LTR-Retrotransposon That Were Critical for Integration. J Virol. 1998;72:1324. doi: 10.1128/jvi.72.2.1324-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee AG, Leem YE, Kelly FD, Levin HL. The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection. J Virol. 2009;83:2675–2685. doi: 10.1128/JVI.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Pidoux A, Mellone B, Allshire R. Analysis of chromatin in fission yeast. Methods. 2004;33:252–259. doi: 10.1016/j.ymeth.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Atwood A, Lin JH, Levin HL. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods. 2009;48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin HL, Weaver DC, Boeke JD. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin HL. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.