Abstract
Recently, research into the development of new targeted therapies has focused on specific genetic alterations to create advanced, more personalized treatment. One of the target genes, fibroblast growth factor receptor‐1 (FGFR1), has been reported to be amplified in estrogen receptor (ER)‐positive subtype breast cancer, and is considered one possible mechanism of endocrine resistance through cross‐talk between ER and growth factor receptor signaling. We performed a comprehensive analysis of FGFR1 at the levels of gene copy number, transcript and protein expression, and examined the relationships between FGFR1 status and clinicopathological parameters, including prognosis in 307 ER‐positive/HER2‐negative primary breast cancer patients treated with standard care at our institute. Most notably, a high level of FGFR1 protein expression was observed in 85 patients (27.7%), and was positively associated with invasive tumor size (P = 0.039). Furthermore, univariate analysis revealed that high FGFR1 protein expression was significantly correlated with poor relapse‐free survival rate (P = 0.0019, HR: 2.63, 95% confidence interval: 1.17–5.98), and showed a tendency towards an increase in recurrent events if the observation period extended beyond the 5 years of the standard endocrine treatment term. FGFR1 gain/amplification was found in 43 (14.0%) patients, which was only associated with higher nuclear grade (P = 0.010). No correlation was found between FGFR1 mRNA expression levels and any clinicopathological factors. Overall, the level of FGFR1 protein expression may be a biomarker of ER‐positive/HER2‐negative primary breast cancer with possible resistance to standard treatment, and may be a useful tool to identify more specific patients who would benefit from FGFR‐1 targeted therapy.
Keywords: Biomarker, breast cancer, ER‐positive/HER2‐negative, fibroblast growth factor receptor‐1, protein expression
Recent increases in genomic information related to cancer has expanded the opportunities both to reveal the character of cancer and to guide cancer therapy. Biomarkers are the set of measurable parameters that provide information directly applicable to the clinical course of cancer, and which are associated with drug sensitivity and resistance. For example, amplification of human epidermal growth factor receptor‐2 (HER2), found in 15–20% of breast cancers, has been regarded as an important predictive marker of the therapeutic effect of anti‐HER2 targeted therapy and breast cancer prognosis.1, 2, 3, 4, 5 While progress in local and systemic treatment has clearly improved the prognosis of breast cancer patients, many patients still die from this disease. The development of new targeted therapy is focused on the specific genetic alterations in individual breast cancers, resulting in more advanced and more personalized treatment.6, 7
Fibroblast growth factor receptor‐1 (FGFR1) is a member of the receptor tyrosine kinase family that plays an important role in mediating fibroblast growth factor (FGF) signaling. Upon activation, FGFR1 promotes cancer cell proliferation, migration, angiogenesis and survival. This gene is also related to activation of the phosphatidylinositol 3‐kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling pathway and the mitogen‐activated protein kinase (MAPK) pathway. There have been a number of reports of FGFR1 gene amplification in breast cancer, and, consequently, FGFR1 has attracted attention as a target for personalized therapy. Previous studies have suggested that FGFR1 amplification is present in around 10% of breast cancer patients, and is associated with a poor outcome.8, 9 The Cancer Genome Atlas (TCGA) showed that copy number gain/amplification of FGFR1 was around 10% in breast cancer (TCGA Network: data are available online at the cBio Cancer Genomics Portal; http://cbioportal.org).10 Another study indicates that FGFR1 amplification is implicated in 15% of cancers of the estrogen receptor (ER)‐positive/HER2‐negative subtype.11 In particular, amplification and subsequent overexpression of FGFR1 contributes to poor prognosis in luminal‐type breast cancers and drives resistance to endocrine therapy.12
In spite of these findings there are no reports comparing copy number aberration (CNA), mRNA expression and protein expression, and the correlation of each parameter with breast cancer characteristics. In this study, we analyzed gene amplification and mRNA and protein overexpression of FGFR1, and their potential association with cilinicopathological factors and prognoses in ER‐positive/HER2‐negative primary breast cancer.
Materials and Methods
Patient characteristics and tumor material
We studied a consecutive series of 307 invasive breast cancer specimens from women treated at Kumamoto University Hospital between June 2000 and January 2011. The median duration of patient follow‐up was 65 months. Informed consent was obtained from all patients. No exclusion criteria were applied. The ethics committee of Kumamoto University Graduate School of Medical Sciences approved the study protocol. All patients had undergone biopsy before neoadjuvant therapy or surgical treatment. Samples were snap‐frozen in liquid nitrogen and stored at −80°C until being used for simultaneous total RNA and genomic DNA (gDNA) extraction. Neoadjuvant and adjuvant treatment were administered depending on the risk evaluation according to tumor biology, such as ER, progesterone receptor (PR), and HER2 expression except Ki67 status, and clinical staging in accordance with the recommendations of the St Gallen international expert consensus on the primary therapy of early breast cancer.13, 14, 15, 16, 17 We based our evaluation on the REporting recommendations for tumor MARKer prognostic studies (REMARK).18
Gene copy number assays
Each patient's gDNA was extracted by using the AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN, Venlo, the Netherlands) following the manufacturer's protocol. The concentration and purity of the prepared gDNA were measured by the A260/A280 absorbance ratios (Nano‐Drop Technologies, Wilmington, DE, USA). FGFR1 gene amplification was analyzed with copy number assay by real‐time quantitative PCR (qPCR) on an ABI 7900HT Fast System (Applied Biosystems, Foster City, USA). RNase P was chosen as a reference for gene dosage because of its single copy number. Each reaction was performed in a reaction mixture containing 5.0 μL of 2 × TaqMan Genotyping Master Mix (Applied Biosystems), 0.5 μL of TaqMan Copy Number Assay (FGFR1: Hs02882334_cn; Applied Biosystems), 0.5 μL of TaqMan Copy Number Reference Assay (RNase P 20X Primer‐Probe VIC; Applied Biosystems), 2.0 μL of nuclease‐free water and 2.0 μL (10 ng) of gDNA sample in a final volume of 10 μL. Thermal cycling conditions included an initialization step at 95°C and 60 s at 60°C. Calculation of the gene copy number was carried out using the absolute quantification method. FGFR1 gene status was defined by the ratio of FGFR1 versus RNase P gene. In total, a ratio from 1.5 to <2.0 was defined as a gain, a ratio larger than or equal to 2.0 as an amplification, and a ratio less than 1.5 as normal range.
Real‐time quantitative reverse transcription polymerase chain reaction analysis
Total RNA was isolated from tissue specimens using the AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN). Total RNA (0.5 μg) was reverse transcribed to complementary DNA (cDNA) using the PrimeScript RT Master Mix (TaKaRa Bio, Otsu, Japan), according to the manufacturer's protocol. Real‐time quantitative reverse transcription PCR (RT‐qPCR) was used to assess FGFR1 mRNA expression. Real‐time RT‐qPCR was carried out in a solution containing 5.0 μL of 2 × TaqMan Fast Advanced Master Mix (Applied Biosystems), 0.5 μL of TaqMan Gene Expression Assay (FGFR1: Hs00915142_m1, β‐Actin: Hs01060665_g1, PUM1: Hs_00982775_m1, TAF‐10: Hs00359540_g1, Applied Biosystems), 3.5 μL of nuclease‐free water and 1.0 μL of cDNA sample (10 ng/μL) in a final volume of 10 μL. Thermal cycling was performed in an ABI 7900HT Fast System (Applied Biosystems). Negative controls were included in each run. Relative mRNA levels were determined from the threshold cycle for amplification using the ΔΔCt method. Determination of Ct values was performed in duplicate and normalized to the Ct values of simultaneous duplicate measurements of the expression of three housekeeping genes, β‐Actin, PUM1 and TAF‐10, from the same samples. These housekeeping genes were selected based on our previous study.19
Immunohistochemistry
Histological sections (4 μm) were deparaffiinized and then rehydrated. The sections were incubated for 10 min in methanol containing 0.3% hydrogen peroxide to block endogenous peroxidase. Antigen retrieval was performed in a microwave for 60 min in a pH 7.0 antigen retrieval solution. After nonspecific staining had been blocked using a blocking agent, sections were incubated overnight with the primary antibody (1:200, ab10646; Abcam, Cambridge, UK) at 4°C. We used Histofine Simple Stain MAX‐PO (MULTI) as the secondary antibody and Histofine DAB Substrate Kit as the chromogenic substrate (NICHIREI BIOSCIENCES, Tokyo, Japan).
Mouse monoclonal antibodies were used for detection of ER (1D5, 1:50; Dako, Tokyo, Japan), PgR (PgR636, 1:800; Dako), human epidermal growth factor receptor 2 (HER2) (1:200; Dako) and Ki67 (MIB‐1, 1:50; Dako), in accordance with the manufacturer's instructions.
The cytoplasmic expression of FGFR1 was semi‐quantitatively evaluated using a microscope and scored by Histo‐score (H‐score). When an immunostaining was present, it was evaluated using a four value intensity score: 0, negative staining; 1, weak, 2, moderate; 3, strong (Fig. 1). Immunostained slides were scored separately for cytoplasmic staining by intensity score and percentage of invasive tumor cells stained at each of the four intensities. H‐score was calculated using the formula: (3 × percentage strong staining) + (2 × weak moderate staining) + (1 × percentage weak staining). As the cut‐off value we selected an H‐score of 180 or more. This cut‐off value was selected to evaluate our study cohort by almost complete low expression and high expression of FGFR1.
Figure 1.
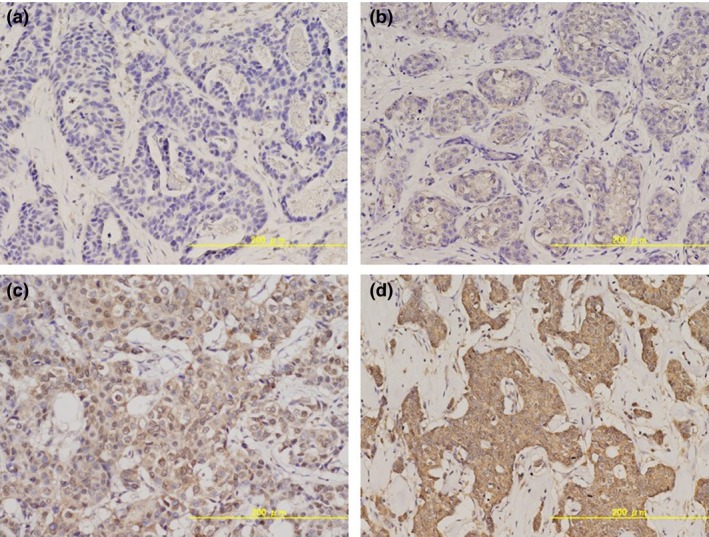
Intensity score of immunohistochemical staining of fibroblast growth factor receptor‐1 (FGFR1). Score 0: (a) negative staining; (b) score 1: weak staining; (c) score 2: moderate staining; and (d) score 3: strong staining.
Statistical analysis
The associations of FGFR1 gene copy number, mRNA and protein expression levels with clinicopathological factors were analyzed using the χ2‐test. Relapse‐free survival (RFS) and breast cancer‐specific survival (BCSS) curves were calculated according to the Kaplan–Meier method and verified by the log‐rank test. Univariate and multivariate analyses of prognostic values were performed using Cox's proportional hazard model. A statistically significant difference was defined as P < 0.05. We used JMP software version 11 for Windows (SAS Institute Japan, Tokyo, Japan) for all statistical analyses.
Results
Patient characteristics
The median age at diagnosis was 61 years (range 21–93). Among the patients, 227 (74.2%) of these were postmenopausal women. One hundred and six (34.5%) patients had positive nodal status. The median Ki67 labeling index was 17.8 (range 0.5–87.2). Most patients (n = 287, 93.5%) were treated with endocrine therapy, and 78 (25.4%) patients were treated with chemotherapy. A total of 217 (70.7%) patients underwent breast conserving surgery, 90 (29.3%) patients underwent total mastectomy and 169 (55.0%) received radiation therapy (Fig. S1).
Association of fibroblast growth factor receptor‐1 gene copy number, levels of mRNA and protein expression with clinicopathological factors
Among the 307 ER‐positive/HER2‐negative primary breast cancer patients, we analyzed FGFR1 gene copy number, levels of mRNA and protein expression. FGFR1 gain/amplification was found in 43 (14.0%) patients. Relative FGFR1 mRNA expression ranged from 0.00075 to 34.73 (25th percentile 0.14, median 0.42, 75th percentile 1.25). We divided the mRNA expression level into two groups: a high mRNA expression group (upper 25th percentile, n = 76) and a low mRNA expression group (lower 75th percentile, n = 231). In the IHC assessment of FGFR1 protein expression, an H‐score ≥180 was observed in 85 patients (27.7%), who were defined as having a high protein expression level, and an H‐score <180 was observed in 222 patients (72.3%), who were defined as having a low protein expression level.
The detailed results of correlation analysis between amplification, expression of FGFR1 and clinicopathological factors are shown in Table 1. Patients with FGFR1 gain/amplification had higher nuclear grade (P = 0.010), but no other factors had any correlation with FGFR1 gain/amplification. Similarly, there was no correlation between FGFR1 mRNA expression level and any clinicopathological factors. However, higher FGFR1 protein expression level was significantly associated with larger invasive tumor size (P = 0.039). No significant difference was observed in histological subtype20 and these three parameters (FGFR1 gain/amplification; P = 0.89, mRNA expression; P = 0.26, protein expression; P = 0.77).
Table 1.
Association of FGFR1 gene copy number, mRNA expression and protein expression with clinicopathological factors
| Total number of patients | FGFR1 amplification | FGFR1 mRNA expression | FGFR1 protein expression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | Gain/amplification | P‐value | Low | High | P‐value | Low | High | P‐value | ||
| n = 307 | n (%) | n (%) | ||||||||
| Age | ||||||||||
| <50 | 67 | 56 (83.6) | 11 (16.4) | 0.52 | 50 (74.6) | 17 (25.4) | 0.89 | 46 (68.7) | 21 (31.3) | 0.45 |
| ≥50 | 240 | 208 (86.7) | 32 (13.3) | 181 (75.4) | 59 (24.6) | 176 (73.3) | 64 (26.7) | |||
| Menstrual status | ||||||||||
| Premenopause | 79 | 65 (82.3) | 14 (17.7) | 0.28 | 58 (73.4) | 21 (26.6) | 0.62 | 53 (67.1) | 26 (32.9) | 0.24 |
| Postmenopause | 227 | 198 (87.2) | 29 (12.8) | 173 (76.2) | 54 (23.8) | 168 (74.0) | 59 (26.0) | |||
| BMI | ||||||||||
| <23 | 156 | 137 (87.8) | 19 (12.2) | 0.30 | 119 (76.3) | 37 (23.7) | 0.57 | 113 (72.4) | 43 (27.6) | 0.95 |
| ≥23 | 147 | 123 (83.7) | 24 (16.3) | 108 (73.5) | 39 (26.5) | 106 (72.1) | 41 (27.9) | |||
| Nuclear grade | ||||||||||
| 1 | 183 | 165 (90.2) | 18 (9.8) | 0.010* | 138 (75.4) | 45 (24.6) | 0.90 | 129 (70.5) | 54 (29.5) | 0.33 |
| 2, 3 | 123 | 98 (79.7) | 25 (20.3) | 92 (74.8) | 31 (25.2) | 93 (75.6) | 30 (24.4) | |||
| Tumor invasion size | ||||||||||
| ≤20 mm | 170 | 150 (88.2) | 20 (11.8) | 0.21 | 128 (75.3) | 42 (24.7) | 0.98 | 131 (77.1) | 39 (22.9) | 0.039* |
| >20 mm | 137 | 114 (83.2) | 23 (16.8) | 103 (75.2) | 34 (24.8) | 91 (66.4) | 46 (33.6) | |||
| Nodal status | ||||||||||
| − | 201 | 175 (87.1) | 26 (12.9) | 0.46 | 154 (76.6) | 47 (23.4) | 0.45 | 145 (72.1) | 56 (27.9) | 0.93 |
| + | 106 | 89 (84.0) | 17 (16.0) | 77 (72.6) | 29 (27.4) | 77 (72.6) | 29 (27.4) | |||
| Ki67 | ||||||||||
| <15% | 115 | 104 (90.4) | 11 (9.6) | 0.092 | 91 (79.1) | 24 (20.9) | 0.18 | 85 (73.9) | 30 (26.1) | 0.45 |
| ≥15% | 169 | 141 (83.4) | 28 (16.6) | 122 (72.2) | 47 (27.8) | 118 (69.8) | 51 (30.2) | |||
| Histological subtype | ||||||||||
| Pap‐tub ca. | 201 | 175 (87.1) | 26 (12.9) | 154 (76.6) | 47 (23.4) | 146 (72.6) | 55 (27.4) | |||
| Sol‐tub ca. | 28 | 23 (82.1) | 5 (17.9) | 0.44 | 17 (60.7) | 11 (39.3) | 0.12 | 22 (78.6) | 6 (21.4) | 0.12 |
| Scirrhous ca. | 31 | 24 (77.4) | 7 (22.6) | 22 (71.0) | 9 (29.0) | 17 (54.8) | 14 (45.2) | |||
| Other type | 45 | 40 (15.3) | 5 (11.1) | 38 (84.4) | 7 (15.6) | 35 (77.8) | 10 (22.2) | |||
*χ2‐test: P < 0.05. BMI, body mass index; FGFR1, fibroblast growth factor receptor‐1; pap‐tub ca., papillo tubular carcinoma; scirrhous ca., scirrhous carcinoma; sol‐tub ca., solid tubular carcinoma.
Prognostic relevance of fibroblast growth factor receptor‐1 gene copy number, mRNA and protein expression
In the analysis of RFS, both local recurrence and distant metastases were considered as events. Among 21 recurrent cases, distant metastases occurred in 13 cases and local recurrence in 18 cases (10 of 18 cases had both local recurrences and distant metastases). Eight patients died as a result of breast cancer, and these were regarded as events when analyzing BCSS.
Survival analysis using the Kaplan–Meier method revealed that higher FGFR1 protein expression was associated with shorter RFS (P = 0.015). No correlation was found with BCSS (P = 0.70) (Fig. 2). FGFR1 gene copy number and mRNA expression were not significantly associated with either RFS or BCSS using the Cox proportional hazards model and could not be verified by the Kaplan–Meier curve (data not shown).
Figure 2.
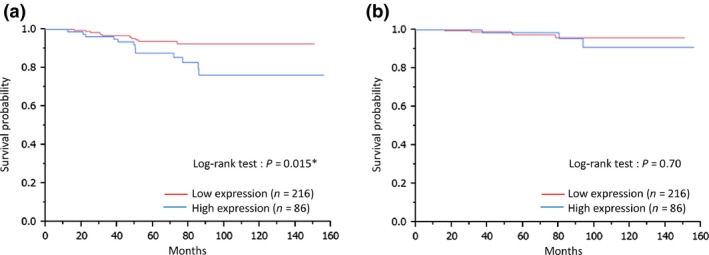
Protein expression of fibroblast growth factor receptor‐1 (FGFR1) and survival. Kaplan–Meier plots of the association of FGFR1 protein expression with relapse free survival (RFS) (a) and breast cancer specific survival (BCSS) (b) in estrogen receptor (ER)‐positive/human epidermal growth factor receptor‐2 (HER2)‐negative primary breast cancer. RFS verified by the log‐rank test.
The prognostic relevance of FGFR1 gene copy number, mRNA and protein expression are summarized in Tables 2 and 3. According to univariate analysis for RFS, FGFR1 protein expression (low level versus high level, P = 0.019, hazard ratio [HR]: 2.63, 95% confidence interval [CI: 1.17–5.98]) was statistically significant.
Table 2.
Univariate and multivariate analysis for relapse free survival in ER+/HER2− breast cancer patients
| Univariate | Mutivariate | ||||||
|---|---|---|---|---|---|---|---|
| P‐value | HR | 95% CI | P‐value | HR | 95% CI | ||
| FGFR1 amplification | Normal | ||||||
| Normal | vs | 0.92 | 0.94 | 0.22–2.73 | 0.90 | 1.10 | 0.21–4.26 |
| Gain/amplification | Gain/amplification | ||||||
| FGFR1 mRNA expression | |||||||
| Low | Low versus high | 0.40 | 0.68 | 0.25–1.64 | 0.39 | 0.62 | 0.19–1.76 |
| High | |||||||
| FGFR1 protein expression | |||||||
| Low | Low versus high | 0.019* | 2.63 | 1.17–5.98 | 0.0070* | 3.63 | 1.42–9.95 |
| High | |||||||
| Age | |||||||
| <50 | <50 vs ≥50 | 0.062 | 0.45 | 0.20–1.04 | 0.032* | 0.11 | 0.017–0.83 |
| ≥50 | |||||||
| Menstrual status | |||||||
| Premenopause | Pre versus post | 0.38 | 0.69 | 0.31–1.64 | 0.095 | 5.82 | 0.74–39.53 |
| Postmenopause | |||||||
| BMI | |||||||
| <23 | <23 vs ≥23 | 0.96 | 1.02 | 0.43–2.37 | 0.64 | 1.25 | 0.48–3.29 |
| ≥23 | |||||||
| Nuclear grade | |||||||
| 1 | 1 vs 2, 3 | 0.71 | 1.17 | 0.50–2.62 | 0.50 | 0.70 | 0.23–1.96 |
| 2, 3 | |||||||
| Tumor invasion size | ≤20 mm | ||||||
| ≤20 mm | vs | 0.16 | 1.78 | 0.80–4.13 | 0.48 | 1.43 | 0.53–4.01 |
| >20 mm | >20 mm | ||||||
| Nodal status | |||||||
| + | + vs − | 0.078 | 2.07 | 0.92‐4.70 | 0.43 | 1.48 | 0.51–4.08 |
| − | |||||||
| Ki67 | |||||||
| <15% | <15% vs ≥15% | 0.092 | 2.23 | 0.88–6.79 | 0.45 | 1.51 | 0.53–4.95 |
| ≥15% | |||||||
*χ2‐test: P < 0.05. BMI, body mass index; CI, confidence interval; ER+/HER2−, estrogen receptor‐positive/human epidermal growth factor receptor‐2‐negative; FGFR1, fibroblast growth factor receptor‐1; HR, hazard ratio.
Table 3.
Univariate and multivariate analysis for breast cancer specific survival in ER+/HER2− breast cancer patients
| Univariate | Mutivariate | ||||||
|---|---|---|---|---|---|---|---|
| P‐value | HR | 95% CI | P‐value | HR | 95% CI | ||
| FGFR1 amplification | Normal | ||||||
| a: normal | versus | 0.92 | 0.90 | 0.048–5.09 | 0.74 | 0.65 | 0.026–6.02 |
| b: Gain/amplification | Gain/amplification | ||||||
| FGFR1 mRNA expression | |||||||
| a: low | Low versus high | 0.51 | 0.59 | 0.086–2.63 | 0.46 | 0.49 | 0.045–3.01 |
| b: high | |||||||
| FGFR1 protein expression | |||||||
| a: low | Low versus high | 0.70 | 1.33 | 0.27–5.43 | 0.31 | 2.50 | 0.40–14.63 |
| b: high | |||||||
| Age | |||||||
| a: <50 | <50 vs ≥50 | 0.93 | 0.93 | 0.21–6.34 | 0.77 | 0.59 | 0.022–14.32 |
| b: ≥50 | |||||||
| Menstrual status | |||||||
| a: premenopause | Pre versus post | 0.83 | 1.19 | 0.27–8.13 | 0.69 | 2.09 | 0.10–75.83 |
| b: postmenopause | |||||||
| BMI | |||||||
| a: <23 | <23 vs ≥23 | 0.27 | 2.20 | 0.54–10.76 | 0.082 | 4.65 | 0.82–40.62 |
| b: ≥23 | |||||||
| Nuclear grade | |||||||
| a: 1 | 1 vs 2, 3 | 0.16 | 2.72 | 0.67–13.30 | 0.86 | 1.16 | 0.22–6.72 |
| b: 2, 3 | |||||||
| Tumor invasion size | ≤20 mm | ||||||
| a: ≤20 mm | vs | 0.32 | 2.04 | 0.50–9.93 | 0.72 | 1.38 | 0.26–10.53 |
| b: >20 mm | >20 mm | ||||||
| Nodal status | |||||||
| a: + | + vs − | 0.032* | 4.97 | 1.14–33.95 | 0.018* | 9.87 | 1.43–201.32 |
| b: − | |||||||
| Ki67 | |||||||
| a: <15% | <15% vs ≥15% | 0.14 | 3.91 | 0.67–73.92 | 0.42 | 2.37 | 0.33–47.96 |
| b: ≥15% | |||||||
*χ2 test: P < 0.05; CI, confidence interval; ER+/HER2−, estrogen receptor‐positive/human epidermal growth factor receptor‐2‐negative; FGFR1, fibroblast growth factor receptor‐1; HR, hazard ratio.
In Cox's proportional hazards model, our data indicate that FGFR1 protein expression (low versus high, P = 0.0070, HR: 3.63, 95% CI: 1.42–9.95) and age (<50 vs ≥50, P = 0.032, HR: 0.11, 95% CI: 0.017–0.83) was an independent prognostic factor of a poor prognosis in terms of RFS for ER positive HER2 negative primary breast cancer. As for BCSS, nodal status was a significant univariate parameter (−versus +, P = 0.032, HR: 4.97, 95% CI: 1.14–33.95), while FGFR1 gene amplification, mRNA and protein expression showed no significant difference.
Correlation between fibroblast growth factor receptor‐1 amplification, mRNA and protein expression
Modest positive correlations between these three variables (FGFR1 gene gain/amplification, expression levels of mRNA and protein) were found, as shown in Figure 3. The median level of FGFR1 mRNA expression was significantly higher in the FGFR1 gene gain/amplification group (median mRNA 0.95) than in the normal range group (median mRNA 0.37) (P = 0.0004). The median level of FGFR1 protein expression was also higher in the gene gain/amplification group (median H‐score 160) than the normal range group (median H‐score 100) (P = 0.039). We compared the mRNA expression levels between the groups with high and low FGFR1 protein expression. The median levels of mRNA expression were 0.31 in the low protein expression group and 0.84 in the high protein expression group (P = 0.001).
Figure 3.
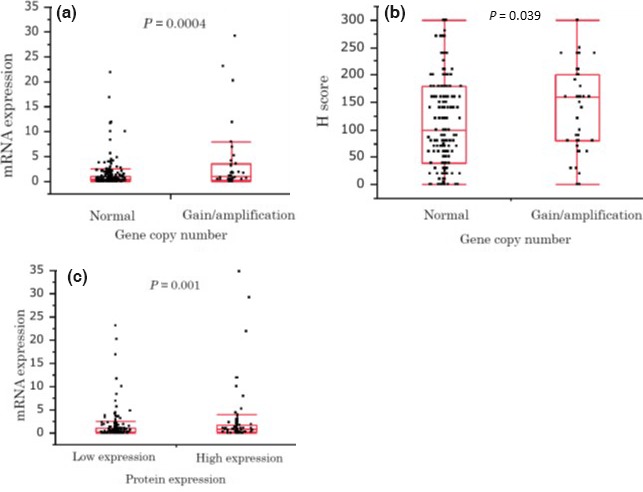
Correlation between fibroblast growth factor receptor‐1 (FGFR1) amplification, mRNA and protein expression. Relationship between FGFR1 gene copy number and mRNA expression level (a), gene copy number and protein expression level (b), and mRNA and protein expression level (c), verified by the Wilcoxon test. The line within each box represents the median value for that group. Whiskers above and below each box show the maximum and minimum values in that group, respectively.
Discussion
We investigated gene copy number and mRNA and protein expression levels of FGFR1 in ER‐positive/HER2‐negative primary breast cancer, and the effects of three expression levels. Among them, protein overexpression of FGFR1 was significantly associated with invasive tumor size as a correlation with clinicopathological factors. One significant finding in this study is the observation that higher FGFR1 protein expression is associated with significantly worse RFS using the log‐rank test by Kaplan–Meier curve and univariate Cox regression analysis. The widening difference in the late observation period which was revealed by the log‐lank test may imply the presence of resistance for present standard therapies resulting in late recurrence in the higher FGFR1 protein expression group.
There is some prior evidence to substantiate our theory, suggesting that higher protein expression of FGFR1 might influence the likelihood of recurrence. First, abnormal FGFR signaling has been shown to contribute to breast cancer progression. In the transgenic mouse model, which uses a mouse mammary tumor virus‐inducible FGFR1, sustained activation of FGFR1 in the mouse mammary epithelium induces alveolar hyperplasia and long‐term activation resulting in stromal invasion, which is associated with extracellular matrix remodeling and vascular branching in the stroma adjacent to these lesions.21 In another study, inducible FGFR1 activation was demonstrated to result in a gain of invasive properties and promotion of the epithelial–mesenchymal transition, which is caused by induction of matrix metalloproteinase‐3.22 Second, FGFR1 makes an important contribution to the resistance to endocrine therapy through activation of the MAPK and PI3K pathways.12 As our results show, most of the patients in this study (93.5%) were treated with standard endocrine therapy for 5 years, after which the recurrent events were comparatively increased in the higher FGFR1 expression group during the late observation period. The management of late recurrences of ER‐positive breast cancer is currently a critical matter of debate. Tumor size and nodal status in particular, as well as several multi gene prognostic tests, have been highlighted as the prognostic factors of late recurrence,23, 24, 25 which is in partial agreement with our findings concerning tumor size. To date, few studies have used IHC to examine expression of FGFR1 in breast cancer. There are only two reports describing effective treatment with FGFR signal inhibitors in triple negative subtype breast cancer.26, 27 We believe that our report provides valuable information regarding a potential target for effective usage of FGFR inhibitors in ER‐positive/HER2‐negative primary breast cancer.
In the metastatic setting, FGFR pathway targeting agents have been developed, such as SU5402, PD173074, lucitanib (E‐3810), dovitinib (TKI258), AZD4547 and BGJ398.28, 29, 30 Some of these agents are currently undergoing phase I or II clinical trials.28 A combination of endocrine therapy and FGFR inhibitors can be adopted as a subsequent therapy if ER‐positive metastatic breast cancer patients have little hormone responsiveness.31 These agents are multiple tyrosine kinase inhibitors targeting receptors including FGFR1‐3, vascular endothelial growth factor receptor (VEGFR)1‐3, c‐KIT, fms‐related tyrosine kinase 3 (FLT3) and platelet‐derived growth factor receptor (PDGFR).
However, there are many reports of genetic aberration of FGFR1 in breast cancer, with gene amplification in particular being most commonly described. Previous studies reported that the percentage of FGFR1 amplification found ranges from 7.5% to 17%,9, 11, 12, 32, 33, 34, 35 and two of these studies reported amplification especially in the ER‐positive subtype.11, 12 In addition, Elbauomy Elsheikh et al.9 showed that FGFR1 amplification is associated with PR status (negative status), age (older than 50) and development of distant metastasis. Other studies have shown a positive association with proliferation,34, 36 ER status35 and HER2 status.33 In our study, FGFR1 gene gain/amplification was found to be equivalent to 14.3% in the ER‐positive/HER2‐negative subtype and was only associated with higher nuclear grade, but was not correlated with survival, which was partially inconsistent with previous studies.
We also found that the level of FGFR1 mRNA expression was not associated with any clinicopathological parameters or with survival in this study. This is in contrast to other recent studies which report that FGFR1 mRNA expression in breast cancer is associated with survival, when assessing the effect of single genes on breast cancer prognosis using tools such as Kaplan–Meier–Plotter (www.kmplot.com)37 and PrognoScan (http://www.abren.net/PrognoScan/).38 To our knowledge, there is no convincing evidence to suggest any association between overexpression of FGFR1 mRNA and prognosis of breast cancer.
In our comprehensive analysis, we found modest correlations among the level of FGFR protein expression, mRNA expression and gain/amplification in this study. André et al.32 report that microarray expression analysis of FGFR1 showed a significant correlation between copy number amplification and mRNA expression levels. They suggest that the occurrence of a DNA gain leads to unregulated overexpression of mRNA, because CNA is able to contribute to an increase in DNA instability and lead to genomic imbalance, and that CNA has an effect on inter‐individual variation in gene expression. Our result on the correlation between gene gain/amplification and mRNA expression level provides further evidence in support of this concept. Moreover, gene amplification is the usual mechanism for higher protein expression, but there have been relatively inconsistent conclusions with regard to this. Reis‐Filho et al.8 report that copy number gains are associated with protein overexpression, while by contrast another study shows the opposite result, with no association between gene amplification and protein overexpression.26 Zhang et al.39 demonstrate that many CNA‐driven mRNA level increases do not translate directly into increased abundance of the corresponding proteins; hence, CNA‐mRNA correlations were significantly higher than CNA‐protein correlations for genes. In contrast, the gene expression level of mRNA is typically correlated with protein, but mRNA expression does not always correlate well with the expression of corresponding proteins because of regulation by post‐transcriptional mechanisms.40 Overall, FGFR1 overexpression is regulated not only by gene amplification, but also by other mechanisms such as transcriptional activation and suppression of protein degradation.41 As just described, various mechanisms have been suggested that correlate with gene amplification, mRNA and protein. The best bioassay method for analyzing FGFR1 is as yet unknown, but different analyses of data from various studies are expected to be informative.
Our study has some limitations. Discrepancies between the results of this study and those of previous studies were found in our qPCR systems for analyzing gene amplification. One previous study involving an FGFR inhibitor29 showed that tumor reduction was greater in patients with FGF pathway amplification identified by qPCR than in those with amplification identified by in situ hybridization. This variability of results could be caused not only by use of different techniques to evaluate alterations in copy number but also by differences between tumor sample backgrounds, which showed differences in certain clinicopathological parameters and prognostic follow‐up periods. In addition, the tumor tissue materials used to extract gDNA and mRNA contain not only tumor cells but also stromal cells. The immunohistochemical approach provides information about the localization of the targeted protein, whereas semi‐quantitative expression analysis will show expression in the total cell population and might, therefore, provide a more subjective value.
In conclusion, our results suggest that the expression level of FGFR1 protein may be an independent prognostic factor in terms of RFS for ER‐positive/HER2‐negative breast cancer patients receiving standard care. Although the technical method of IHC has not been standardized, it can be useful for further investigation of biological models to elucidate their interactions, and to establish possible use of FGFR inhibitors for ER‐positive/HER2‐negative breast cancer patients, providing hope for these patients who currently have limited treatment options. We believe our findings provide an essential basis of the role of FGFR1 in breast cancer.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting information
Table S1. Patient characteristics.
Acknowledgments
We thank Y. Azakami, M. Nagame and Y. Tsurusaki for excellent technical support and A. Okabe, R. Tokimatsu, M. Suematsu and M. Tanabe for excellent clinical data management.
Cancer Sci 107 (2016) 491–498
Funding Information
No sources of funding were declared for this study.
References
- 1. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER‐2/neu oncogene. Science 1987; 235: 177–82. [DOI] [PubMed] [Google Scholar]
- 2. Slamon DJ, Leyland‐Jones B, Shak S et al Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344: 783–92. [DOI] [PubMed] [Google Scholar]
- 3. Romond EH, Perez EA, Bryant J et al Trastuzumab plus adjuvant chemotherapy for operable HER2‐positive breast cancer. N Engl J Med 2005; 353: 1673–84. [DOI] [PubMed] [Google Scholar]
- 4. Valachis A, Mauri D, Polyzos NP, Chlouverakis G, Mavroudis D, Georgoulias V. Trastuzumab combined to neoadjuvant chemotherapy in patients with HER2‐positive breast cancer: a systematic review and meta‐analysis. Breast 2011; 20: 485–90. [DOI] [PubMed] [Google Scholar]
- 5. Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER‐2 receptor and breast cancer: ten years of targeted anti‐HER‐2 therapy and personalized medicine. Oncologist 2009; 14: 320–68. [DOI] [PubMed] [Google Scholar]
- 6. Bild AH, Yao G, Chang JT et al Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–7. [DOI] [PubMed] [Google Scholar]
- 7. Alvarez RH, Valero V, Hortobagyi GN. Emerging targeted therapies for breast cancer. J Clin Oncol 2010; 28: 3366–79. [DOI] [PubMed] [Google Scholar]
- 8. Reis‐Filho JS, Simpson PT, Turner NC et al FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res 2006; 12: 6652–62. [DOI] [PubMed] [Google Scholar]
- 9. Elbauomy Elsheikh S, Green AR, Lambros MB et al FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridization analysis. Breast Cancer Res 2007; 9: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumors. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stephens PJ, Tarpey PS, Davies H et al The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486: 400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turner N, Pearson A, Sharpe R et al FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010; 70: 2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thürlimann B, Senn HJ. Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol 2003; 21: 3357–65. [DOI] [PubMed] [Google Scholar]
- 14. Goldhirsch A, Glick JH, Gelber RD et al Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol 2005; 16: 1569–83. [DOI] [PubMed] [Google Scholar]
- 15. Goldhirsch A, Wood WC, Gelber RD et al Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol 2007; 18: 1133–44. [DOI] [PubMed] [Google Scholar]
- 16. Goldhirsch A, Ingle JN, Gelber RD et al Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol 2009; 20: 1319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldhirsch A, Wood WC, Coates AS et al Strategies for subtypes – dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011; 22: 1736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McShane LM, Altman DG, Sauerbrei W et al REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pract Oncol 2005; 2: 416–22. [PubMed] [Google Scholar]
- 19. Ibusuki M, Fu P, Yamamoto S et al Establishment of a standardized gene‐expression analysis system using formalin‐fixed, paraffin‐embedded, breast cancer specimens. Breast Cancer 2013; 20: 159–66. [DOI] [PubMed] [Google Scholar]
- 20. JBCS . General Rules for Clinical and Pathological Recording of Breast Cancer, The 17th Edition. Tokyo, Japan, KANEHARA&CO.,LTD., June 2012; 21.
- 21. Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: Development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol 2002; 157: 703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xian W, Schwertfeger KL, Vargo‐Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol 2005; 171: 663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sestak I, Dowsett M, Zabaglo L et al Factors predicting late recurrence for estrogen receptor‐positive breast cancer. J Natl Cancer Inst 2013; 105: 1504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bianchini G, Pusztai L, Karn T et al Proliferation and estrogen signaling can distinguish patients at risk for early versus late relapse among estrogen receptor positive breast cancers. Breast Cancer Res 2013; 15: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubsky P, Brase JC, Jakesz R et al The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2− breast cancer patients. Br J Cancer 2013; 109: 2959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee HJ, Seo AN, Park SY et al Low prognostic implication of fibroblast growth factor family activation in triple‐negative breast cancer subsets. Ann Surg Oncol 2014; 21: 1561–8. [DOI] [PubMed] [Google Scholar]
- 27. Cheng CL, Thike AA, Tan SY, Chua PJ, Bay BH, Tan PH. Expression of FGFR1 is an independent prognostic factor in triple‐negative breast cancer. Breast Cancer Res Treat 2015; 151: 99–111. [DOI] [PubMed] [Google Scholar]
- 28. André F, Cortés J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res Treat 2015; 150: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. André F, Bachelot T, Campone M et al Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res 2013; 19: 3693–702. [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto‐Ibusuki M, Arnedos M, André F. Targeted therapies for ER+/HER2− metastatic breast cancer. BMC Med 2015; 13: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwase H. Treatment strategy for metastatic breast cancer with estrogen receptor‐positive tumor. Int J Clin Oncol 2015; 20: 249–52. [DOI] [PubMed] [Google Scholar]
- 32. André F, Job B, Dessen P et al Molecular characterization of breast cancer with high‐resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 2009; 15: 441–51. [DOI] [PubMed] [Google Scholar]
- 33. Kadota M, Sato M, Duncan B et al Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA . Cancer Res 2009; 69: 7357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Letessier A, Sircoulomb F, Ginestier C et al Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12 and 20q13 amplifications in breast cancers. BMC Cancer 2006; 6: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moelans CB, de Weger RA, Monsuur HN, Vijzelaar R, van Diest PJ. Molecular profiling of invasive breast cancer by multiplex ligation‐dependent probe amplification‐based copy number analysis of tumor suppressor and oncogenes. Mod Pathol 2010; 23: 1029–39. [DOI] [PubMed] [Google Scholar]
- 36. Jang MH, Kim EJ, Choi Y et al FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res 2012; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gyöffy B, Lanczky A, Eklund AC et al An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat 2010; 123: 725–31. [DOI] [PubMed] [Google Scholar]
- 38. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta‐analysis of the prognostic value of genes. BMC Med Genomics 2009; 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang B, Wang J, Wang X et al Proteogenomic characterization of human colon and rectal cancer. Nature 2014; 513: 382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 2003; 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet 2010; 77: 201–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient characteristics.


