Abstract
Human chromosome 21 is known to be associated with the high risk of hematological malignancy but with resistance to breast cancer in the study of Down syndrome. In human cancers, we previously observed the significant alterations of the protein expression encoded by the ganp/MCM3AP gene on human chromosome 21q22.3. Here, we investigated GANP protein alterations in human breast cancer samples (416 cases) at various stages by immunohistochemical analysis. This cohort study clearly showed that expression of GANP is significantly decreased in human breast cancer cases with poor prognosis as an independent risk factor (relapse‐free survival, hazard ratio = 2.37, 95% confidence interval, 1.27–4.42, P = 0.007 [univariate analysis]; hazard ratio = 2.70, 95% confidence interval, 1.42–5.13, P = 0.002 [multivariate analysis]). To investigate whether the altered GANP expression is associated with mammary tumorigenesis, we created mutant mice that were conditionally deficient in the ganp/MCM3AP gene using wap‐cre recombinase transgenic mice. Mammary gland tumors occurred at a very high incidence in female mammary gland‐specific GANP‐deficient mice after severe impairment of mammary gland development during pregnancy. Moreover, tumor development also occurred in female post parous GANP‐heterodeficient mice. GANP has a significant role in the suppression of DNA damage caused by estrogen in human breast cancer cell lines. These results indicated that the GANP protein is associated with breast cancer resistance.
Keywords: Breast cancer resistance, chromosome 21, DNA damage, down syndrome, ganp/MCM3AP, prognostic factor
Individuals with trisomy 21 have increased risks of acute lymphoblastic leukemia, acute myeloid leukemia, and testicular tumors. Interestingly, they have decreased risks of developing solid tumors, such as colon and breast cancers, and this phenomenon has been termed “the divine grace”.1 Many studies have sought to identify the genes involved in this cancer resistance trait of trisomy 21; however, the mouse counterpart of human chromosome 21q is distributed among three different chromosomes, located from 17 to 17.4 cM (30.6–31.7 Mb) on chromosome 17, from 41 to 42 cM (78.4–80.6‐Mb region) on chromosome 10, and from 47 to 70 cM (76.7–100 Mb) on chromosome 16.2 Segmental trisomy in a mouse model results in abnormalities in brain size and structural changes and learning and behavior deficits.3, 4, 5, 6 Several phenotypes have been further indicated in single gene mutants, including learning defects, a slowed ability to habituate novel stimuli, cognitive and behavioral deficits, learning and memory deficits, and behavioral defects.7, 8, 9, 10, 11 Nevertheless, the role of trisomy 21 in tumor resistance remains unclear due to difficulties in narrowing down the chromosomal segments and the complex nature of cancer development, which usually occurs with advancing age or due to environmental insult.
We observed altered expression of the ganp/MCM3AP gene, 12 which is located on human chromosome 21q22.3, in the clinical sample of cancer patients who are at different stages of cancer; observations were seen in various tissues from this clinical sample. Our previous work indicated that increased expression of GANP was observed in B‐cell lymphoma cells of the activation‐induced cytidine deaminase (AID)‐expressing germinal center stages and tumor cells from hematopoietic lineages.13 However, for solid tumors, such as glioblastomas, GANP expression was decreased,14 indicating the reverse pattern between solid tumors and hematological malignancies. Because a tumor‐suppressive effect of ganp/MCM3AP has been previously implicated in solid tumors, we addressed whether the protein that is encoded by this gene is expressed differently in clinical breast cancer cases. Our cohort study of breast cancer patients showed that a significant decrease in GANP occurs in invasive ductal carcinoma compared with normal mammary glands, according to clinical and pathological criteria. Thus, we focused on the decreased expression of this protein in tumors as a candidate molecule for the suppression of cancer development. We observed the spontaneous development of mammary gland tumors with a high incidence in heterodeficient female mice after pregnancy. Furthermore, conditional targeting of the ganp/MCM3AP gene showed lineage‐specific tumor development in the mammary glands, which presumably was independent of environmental factors or a bystander effect due to damage to other organs and tissues associated with tumor development. Our study suggests that the ganp/MCM3AP gene that is located on mouse chromosome 10 and in the human chromosome 21q region has a tumor‐suppressive effect in breast cancer development.
Materials and Methods
Patients and samples
Primary invasive breast carcinoma specimens were obtained by surgical excision from 92 female patients (1995–2000) at the Department of Breast and Endocrine Surgery, Nagoya City University Hospital (NCU) (Nagoya, Japan) and from 376 female patients (2001–2008) at the Department of Breast and Endocrine Surgery, Kumamoto University Hospital (KUH) (Kumamoto, Japan). Detailed information of the clinical parameters for these two cohorts is summarized in Table S1. Informed consent was obtained from all patients before surgery. Adjuvant and neoadjuvant treatments were carried out after risk evaluation according to clinical staging (Union for International Cancer Control classification) and analysis of tumor biology, including estrogen receptor (ER) status, progesterone receptor (PgR) status, and beginning in 2006, human epidermal growth factor receptor 2 (Her2) status, in accordance with the recommendation of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. The patients underwent postoperative follow‐up every 3 months. The median follow‐up period was 66 months (range, 15–144 months) for the KUH cohort.
For ductal carcinoma in situ, specimens were obtained by surgical excision from 23 female patients at NCU and from 40 female patients at KUH. Normal samples (31 cases) were derived from adjacent histologically normal tissues (18 cases from ductal carcinomas in situ and 13 cases from invasive ductal carcinomas) that we used for immunohistochemical staining in Figure 1(A). The background information about the clinicopathological factors is shown in Table S2. The ethics committees of NCU and KUH independently approved this study protocol for breast cancer.
Figure 1.
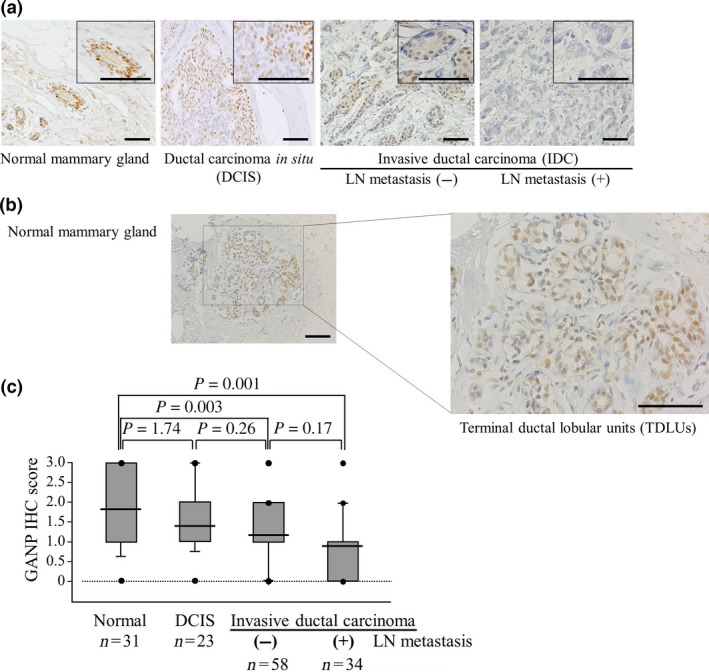
Reduced expression of GANP is significantly associated with the degree of malignancy in breast cancers. (A) Representative images of GANP immunostaining of a normal mammary gland, ductal carcinoma in situ and invasive ductal carcinoma. Images at higher magnification are shown in the insets. Scale bar = 100 μm. (B) GANP expression in terminal duct lobular units. Representative images of GANP immunostaining are shown. Scale bar = 200 μm. (C) A comparison of GANP expression among the four groups is shown by immunohistochemistry (IHC) scores using box plots with whiskers defining the range of scores. LN, lymph node.
Mice
Mammary gland‐specific ganp‐deficient mice were obtained by crossing ganp‐floxed mice and wap‐cre transgenic mice (Jackson Laboratory, Bar Harbor, Maine).15, 16 GANP‐heterodeficient (ganp +/d) mice were established previously.17 Three types of mutant strains (ganp‐floxed, wap‐cre transgenic, and ganp +/d mice) were backcrossed to C57BL/6J mice at least 15 times. All animal protocols were carried out following the approval of the Animal Care and Use Committee of the Graduate School of Medical Sciences of Kumamoto University.
Statistical analysis
For quantitative comparisons across groups, we applied one‐way anova when equal variance in the results was identified by Bartlett's test. We applied the Kruskal–Wallis test or Mann–Whitney U‐test for the data that are not normally distributed. We applied Fisher's exact test or χ2‐test for categorical variables as appropriate. When the results of overall comparisons were statistically significant, we carried out further pairwise multiple comparisons (Dunnett's or Tukey's tests as appropriate). The survival data were analyzed using the Kaplan–Meier method and the log–rank test was used to compare statistical significances. Cox's proportional hazards model or the competing risk regression model18 were used for univariate and multivariate analyses of GANP to detect other potential confounders. Differences were considered significant when P < 0.05 (two‐sided).
Detailed methods are described in Document S1.
Results
Deficient GANP expression in human breast cancer
We investigated the expression of the GANP protein in mammary gland tumors from clinical cases at NCU (115 patients in comparison with 31 normal mammary glands). GANP was prominently detected in the nuclei of normal human mammary gland epithelial cells but was absent in cells from the surrounding region (Fig. 1A). GANP expression was upregulated in normal mammary glands, particularly at the terminal duct lobular units, which are known to be associated with breast cancer development (Fig. 1B). However, GANP expression decreased gradually with advancing stages of breast cancer (Fig. 1A). There were marked decreases in invasive ductal carcinoma (IDC) without lymph node (LN) metastasis and IDC with LN metastasis. We classified the clinical samples from the NCU by the degree of GANP expression as grades 0, 1, 2, and 3 because most tumor cells (more than 90%) in the individual samples showed levels almost similar to those detected in a homogeneous population (Fig. S1). Analysis of immunohistochemical (IHC) scores revealed that GANP expression was statistically low in carcinoma with or without LN metastasis compared with normal tissues (Fig. 1C). The reduction of GANP expression in IDC with LN metastasis was not statistically significant compared with in IDC without LN metastasis (P = 0.17), suggesting there is differential expression of GANP in IDC. We also analyzed the GANP expression in IDC after TNM classification; however, a weak trend was only observed between stages I and II (P = 0.108) or stages I and III (P = 0.120), which is perhaps due to the limited number of analyzed patients in the NCU cohort (Fig. S2).
The clinical samples for this study were collected from 1995 to 2000; thus, the standard treatment regimen of the patients at that time was different from the latest guidelines for breast cancer medical treatment. To confirm the value of GANP expression as a parameter of breast cancer malignancy grade, we investigated its expression compared with other known parameters in another retrospective cohort analysis of patients at KUH (416 patients; Table 1). Low tumor GANP expression was not significantly associated with certain clinical parameters, such as menopause, tumor size, nodal status, nuclear grade (grades 1–3), ERα expression, PgR expression, Her2 expression, Ki67 positivity, subtypes of ERα+Her2 positivity, or triple negativity of ERα, PgR, and Her2 (Table S3). However, univariate analysis of relapse‐free survival (RFS) showed a significant (P = 0.007) association of reduced GANP expression with worse survival, similar to what was detected for other parameters, including tumor size, node positivity, nuclear grade, ERα positivity, PgR positivity, and Ki67 positivity. These findings were further confirmed by multivariate analysis (P = 0.002), which revealed that nuclear grade and ERα expression were significant independent prognostic factors (Table 1). A comparison of RFS with breast cancer‐specific survival (BCSS) also showed that GANP expression was a significant independent prognostic factor as determined by univariate (P = 0.005) and multivariate (P = 0.004) analyses. This highly reliable marker was comparable to the well‐established parameter of ERα expression (univariate, P < 0.001; multivariate, P = 0.007) compared with the other parameters used in clinical therapeutic regimens (Table S4). We further applied competing risk regression models and confirmed our findings of the associations between GANP IHC, the other covariates and BCSS (Table S5). In support of these findings, low GANP expression cases (n = 170) showed a significantly poorer prognosis compared with those with high GANP expression (n = 206) in terms of both BCSS (P = 0.0027) and RFS (P = 0.0054) over a period of 100 months (Fig. 2). Based on human cohort studies using GANP IHC, low GANP expression was presumably associated with malignant advancement of breast cancers rather than breast carcinogenesis.
Table 1.
Univariate and multivariate analyses for relapse‐free survival of 376 female patients (2001–2008) treated at Kumamoto University Hospital
| Variables | Categories | n | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value† | HR | 95% CI | P‐value† | |||
| GANP | IHC 0–1 vs. IHC 2–3 | 170/206 | 2.37 | 1.27–4.42 | 0.007 | 2.70 | 1.42–5.13 | 0.002 |
| Other covariates | ||||||||
| Menopause | Post. vs. pre | 123/253 | 0.62 | 0.34–1.13 | 0.120 | Excluded | – | – |
| Tumor size | ≥2 cm vs. <2 cm | 162/214 | 2.66 | 1.43–4.97 | 0.002 | 1.61 | 0.79–3.30 | 0.19 |
| Nodal status | Positive vs. negative | 168/204‡ | 2.47 | 1.36–4.49 | 0.003 | 1.84 | 0.96–3.54 | 0.07 |
| Nuclear grade | 2–3 vs. 1 | 168/204§ | 3.28 | 1.68–6.41 | 0.001 | 1.26 | 1.06–1.49 | 0.01 |
| ERα | Positive vs. negative | 298/78 | 0.24 | 0.13–0.44 | <0.001 | 0.32 | 0.12–0.81 | 0.02 |
| PgR | Positive vs. negative | 255/121 | 0.33 | 0.18–0.60 | <0.001 | 0.82 | 0.32–2.09 | 0.68 |
| Her2 | Positive vs. negative | 55/321 | 1.54 | 0.74–3.21 | 0.250 | 0.59 | 0.26–1.35 | 0.21 |
| Ki67 | Positive vs. negative | 179/181¶ | 3.45 | 1.70–7.00 | 0.001 | 2.13 | 1.00–4.54 | 0.05 |
Hazard ratios (HR) and 95% confidence intervals (CI) were estimated by Cox proportional hazard model. † P‐values were based on two‐sided test. ‡Four cases were of unknown nodal status. §Four cases were of unknown nuclear grade. ¶Sixteen cases were of unknown Ki67 status. Cut‐off values for estrogen receptor α (ERα), progesterone receptor (PgR), human epidermal growth factor receptor 2 (Her2), and Ki67 were determined based on the American Society of Clinical Oncology/College of American Pathologists guideline (2013). IHC, immunohistochemistry.
Figure 2.
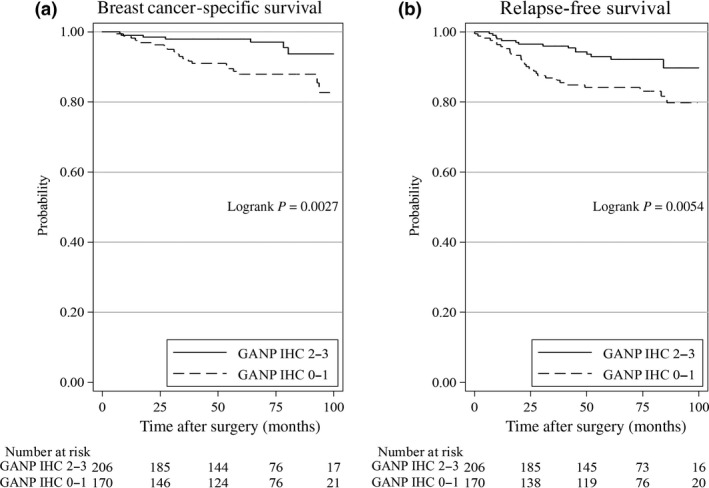
Statistical analysis of GANP expression levels and breast cancer‐specific survival (A) or relapse‐free survival (B) using the Kaplan–Meier method for 376 female patients (2001–2008) treated at Kumamoto University Hospital. Patients with high GANP‐expressing tumors had a better prognosis than those with lower GANP expression.
Role of GANP in estrogen‐induced DNA damage
We investigated the association of the GANP protein level and cell damage in ERα‐positive MCF7 cells, which have been commonly used to examine estrogen responsiveness and ER target genes. Ganp‐knockdown by representative ganp siRNA of three sequence regions14, 19 reduced the GANP protein level to 25% of the level observed with control siRNA (siCtrl) treatment (Fig. 3A; left). As a complementary approach, we prepared stable MCF7 transfectants with a retroviral vector carrying human ganp (h‐ganp)–internal ribosome entry site (IRES)–green fluorescent protein (gfp) (Fig. 3A; right). To investigate the effect of GANP on estrogen‐responding cells, DNA damage was measured in vitro in MCF7 cells after estrogen stimulation using the Comet assay, which detects single‐stranded and double‐stranded DNA damage with high sensitivity (Fig. 3B). While estradiol (E2) caused DNA damage in the MCF7 cells, resulting in a 17% average tail moment, siGanp markedly augmented the E2‐induced DNA damage in these cells (to a 35% average tail moment) (Fig. 3C; P = 0.009). This finding indicated that GANP insufficiency exacerbated the DNA damage induced by estrogen. Ganp‐transfected MCF7 cells had lower E2‐induced DNA damage (12% average tail moment) than mock IRES‐gfp‐transfected control cells (21% average tail moment) (Fig. 3C; P = 0.006). Cellular changes after E2‐induced DNA damage were examined with a cell proliferation assay. siGanp caused a decrease in proliferation compared with the control, whereas ganp transfection had the opposite effect on proliferation (Fig. 3D). In addition, the TUNEL assay also revealed that siGanp induces cell apoptosis (Fig. 3E; right and representative pictures on the left), which was confirmed by the increase in sub‐G1 cells according to flow cytometric analysis (Fig. 3F; right and representative profiles on the left). Although the cell apoptotic change was not marked in GANP overexpression, loss of GANP causes marked change in apoptosis. Taken together, these results suggest that GANP is necessary to prevent estrogen‐induced DNA damage in breast cancer cells.
Figure 3.
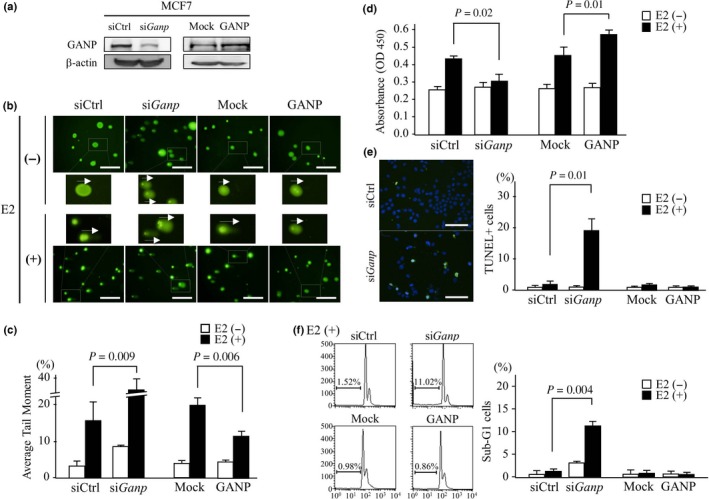
Effect of ganp‐knockdown or ganp transfection in an E2‐stimulated breast cancer cell line. (A) MCF7 cells were treated with siGanp or control siRNA (siCtrl) and transfected with h‐ganp cDNA (GANP) or a mock control (Mock). GANP expression was detected by Western blot analysis and compared with a β‐actin control. (B,C) E2‐induced DNA damage was measured by the Comet assay. Cells were transfected with siGanp or h‐ganp cDNA and then examined after stimulation with E2 with siCtrl‐treated and mock‐transfected cells used as controls, respectively. The tail of degraded DNA was measured as the tail moment by CometScore. Representative tail lengths are indicated by arrows. Scale bar = 100 μm (B). Error bars represent SD (C). P‐values calculated by Student's t‐test. The data are representative of three independent experiments. (D) Cell proliferation was measured using MTT assay. MCF7 cell treatment was similar to that in (A). Cells were stimulated with E2 for 4 days and then subjected to MTT assay using a Cell Counting Kit‐8. (E) Measurement of apoptotic cells was carried out by the TUNEL assay. MCF7 cells transfected with siGanp or ganp were stimulated with E2 for 4 days then TUNEL positivity per DAPI‐stained cell was counted by fluorescence microscopy. Representative images of E2‐stimulated MCF7 cells are shown (left). Scale bar = 100 μm. (F) Measurement of apoptotic cells with sub‐G1 gating as determined by propidium iodide staining. On day 4 after E2 stimulation, the cells were harvested, stained with propidium iodide solution and analyzed by flow cytometry. Representative cell‐cycle profiles of E2‐stimulated MCF7 cells are shown (left). (D–F) Each column represents the mean and SD. P‐values calculated by Student's t‐test. The experiments were carried out more than three times.
Development of mammary gland tumors in ganp‐deficient mice
To investigate the effect of GANP deficiency on mammary gland cells, GANP expression was conditionally targeted in mammary gland tissues by crossing floxed/floxed (fl/fl) ganp mice with wap‐induced cre‐expressing mice (Fig. 4A). The IHC analysis showed that wap‐cre‐ganp fl/fl mice did not express GANP at a detectable level in mammary glands after pregnancy in comparison with ganp fl/fl mice. Real‐time PCR showed a marked reduction in ganp transcription in the mammary glands of the wap‐cre‐ganp fl/fl mice (Fig. 4B). The wap‐cre‐ganp fl/fl mice showed marked abnormalities in mammary gland cell development during the lactation period after pregnancy. The GANP‐deficient mammary glands showed the impaired formation of glandular architecture and irregular alignment of epithelial cells lining the lumen, with increased fibrosis that was presumably due to the tissue damage response (indicated by asterisks) in the interglandular space (Fig. 4C; a–c, g–i, H&E staining). The mammary glands of the wap‐cre‐ganp fl/+ mice showed intermediate phenotypes between the control and wap‐cre‐ganp fl/fl mice (Fig. 4C; d–f). The ganp‐homodeficient mammary glands showed a reduction in Ki67 staining compared with the control (Fig. 4D). The tissue‐specific targeting of the ganp gene caused the generation of mammary gland tumors in both the homodeficient wap‐cre‐ganp fl/fl (35%, 21/60) and heterodeficient wap‐cre‐ganp fl/+ (9.1%, 3/33) mice, with a latency of 1 year after pregnancy (Fig. 4E; a,c, white columns). Notably, lung metastases of tumor cells were observed at multiple sites with a high incidence in the homodeficient wap‐cre‐ganp fl/fl mice (76%, 16/21; Fig. 4E; b,c, black column), whereas no tumors were observed in the control ganp fl/fl mice.
Figure 4.
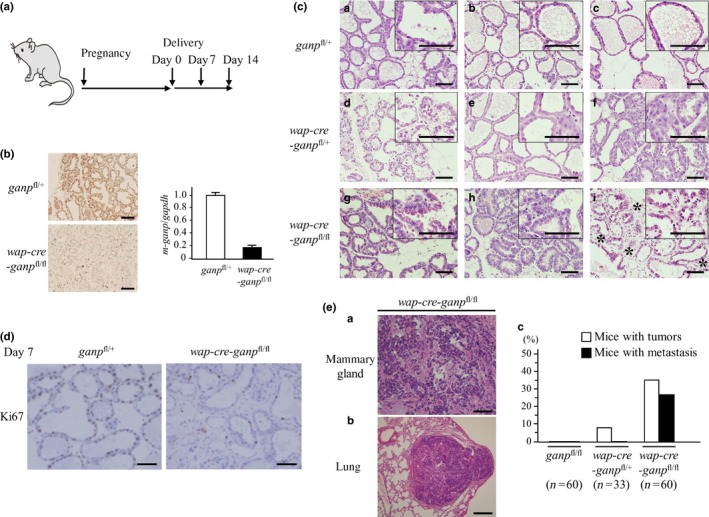
Impaired differentiation of mammary gland cells in ganp‐deficient mice. (A) Conditional targeting of the ganp gene in mammary gland cells from wap‐cre transgenic mice. (B) GANP expression was markedly decreased in wap‐cre‐ganp fl/fl mice compared with control mice after pregnancy on day 14. Scale bar = 100 μm (left). Real‐time PCR showed an 80% reduction in ganp transcripts in mammary glands from wap‐cre‐ganp fl/fl mice compared with control mice (right). Each column represents results from a single mouse analyzed in triplicate with error bars as SD. Results are normalized to the ganp fl/+ mouse. Data are representative of three independent experiments. (C) After pregnancy, the mice showed impaired differentiation of mammary gland cells with minimal lactic acid production, and abnormal gland architecture was observed on days 0 (a–c), 7 (d–f), and 14 (g–i) after delivery. Asterisks indicate fibrotic changes in the interglandular areas of mammary glands. Images at higher magnification are shown in the insets. Scale bar = 100 μm. (D) Comparison of Ki67 positivity in mammary epithelial cells from control and wap‐cre‐ganp fl/fl mice. Scale bar = 100 μm. (E) wap‐cre‐ganp fl/fl mice developed mammary gland tumors with lung metastasis after aging (a,b). Scale bar = 100 μm (a) and 200 μm (b). Incidence of mammary gland tumors in primiparous female mice. Mice with tumors are represented by white columns; those with lung metastasis are represented by black columns (c).
Ganp‐homodeficient mice established by a conventional homologous recombination strategy were embryonic lethal by day 12 and could not be used for further analysis.17 Thus, we carried out long‐term monitoring of the ganp +/d mice. Female ganp +/d mice developed mammary gland tumors with atypical cell growth upon reaching a multiparous state (multiple pregnancies; Fig. 5A). GANP expression levels in the ganp +/d mice were lower compared with those in the wild‐type control mice, but they were not completely absent. This finding may suggest that GANP expression from both alleles of the chromosome is necessary for the suppression of mammary tumors. The tumors showed various expression levels of ERα, PgR, Her2, and Ki67 (Fig. 5B, Table S6). This expression profile suggested that the mouse mammary tumors developed with diverse subtypes, similar to those observed in human cases. The tumor cells showed the multiplication of genes on chromosome 14 (red) and chromosome 16 (yellow), which were detected by dual‐color FISH analysis (Fig. 5C; arrowheads). Short‐term culturing of the mammary gland tumor cells revealed aneuploidy as determined by chromosome analysis (Fig. 5D), which was further confirmed by the presence of a high‐rate of chromosomal instability with chromosome truncation and translocation in the long‐term cultured cell lines (Fig. 5E; arrowheads). Female multiparous ganp +/d mice generated mammary gland tumors with a high incidence (30%, 19/63) after aging (>60 weeks as indicated by the red triangles; Fig. 5F). The life spans of the ganp +/d mice were shorter than those of the ganp +/+ mice. The female multiparous ganp +/d mice had a significantly lower survival time (n = 63) compared with the female wild‐type C57BL/6 mice (n = 55). The survival of the tumor‐free mice did not differ between the wild‐type ganp +/+ (male, n = 36; female, n = 42) and ganp +/d (male, n = 33; female, n = 40) mice (Fig. 5G), indicating that the lower survival time was due to tumor development. GANP is essential for preventing mammary gland tumor development in multiparous female mice and to preserve a normal life span.
Figure 5.
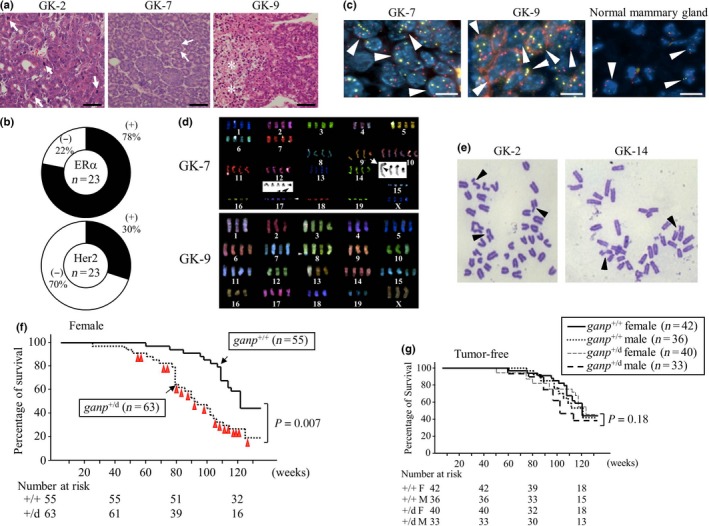
Female multiparous ganp +/d mice developed mammary gland tumors after aging. (A) Representative images of H&E staining of breast cancer tumors isolated from ganp +/d mice. Mitotic cells and necrotic lesions are indicated by arrows and asterisks, respectively. Scale bar = 100 μm. GK‐2, GK‐7, and GK‐9 are tumor specimens from different mice. (B) Expression levels of estrogen receptor α (ERα) and human epidermal growth factor receptor 2 (Her2) in the tumors. ERα and Her2 were examined by immunohistochemistry, and the frequencies of positive tumors are displayed in pie charts (n = 23). (C) Polyploidy in tumor sections. Dual centromere signals of mouse chromosome 14 (red) and 16 (yellow) were detected in the control mammary gland specimen (arrowheads). Two independent tumors (GK‐7 and GK‐9) showed more than two signals per cell (arrowheads). Scale bar = 20 μm. (D) Multicolor FISH analysis of cell lines. Abnormalities included increases in the numbers of chromosomes and translocations, as shown for chromosomes 9 and 17 (arrowheads in GK‐7). (E) Chromosome analysis of long‐term cultured tumor cell lines. Chromosomal breakages are shown (arrowheads). (F) Survival rates of female ganp +/+ (littermates) and ganp +/d mice. The life spans of the ganp +/d mice were shorter than those of the ganp +/+ mice (log–rank [Cox–Mantel] test). (G) Survival of individual tumor‐free mice was not significantly altered. Data were analyzed as in (F).
Discussion
Children with Down syndrome (DS) are at a high risk of developing hematological malignancies, and for affected individuals of all ages; however, there is a decreased risk of developing solid tumors, with an over 50% reduction in their incidence.1 For DS patients, the incidence of breast cancer is significantly decreased, and the mortality rate of this disease is markedly reduced (by 10–25‐fold) compared with age‐matched control groups.20 These findings suggest that human chromosome 21 encodes molecule(s) with a tumor‐suppressive or regulatory function(s). The majority of genes on chromosome 21 are located on the long arm (21q), which contains 261–364 protein‐coding genes, as revealed by the combined analysis of data obtained using computational methods, EST sequencing, laboratory verification, and comparative genome analysis.21
The gene network for breast cancer showed four significant interactions between chromosome 21 and chromosomes 2, 7, 13, and 20, respectively.22 The tumor‐suppressive effect of trisomy 21 has been studied in mouse models of Ts65Dn and Tc1. Ts65Dn harbors three copies of the mouse counterpart on chromosome 16. Tc1 is a trans‐chromosomal mouse model that contains ~90% of the genes on human chromosome 21. These mouse models show reduced tumor progression with decreases in angiogenesis.23
To identify the genes that are biologically relevant to DS, an ideal approach would be to establish a transgenic mouse model that expresses one extra copy of the orthologous mouse gene under physiological regulation by its own transcriptional element in the genome. An alternative approach is to examine the expression of genes on chromosome 21q in human solid tumors of various organs and then to investigate the spontaneous development of tumors in mice that are heterodeficient in candidate genes. One tumor suppressor gene on human chromosome 21q that has been identified is the single‐minded 2 (SIM2), which is known to downregulate MMP‐3 and abrogate breast cancer cell invasion.24
This study assessed the function of ganp/MCM3AP as a tumor suppressor in both mice and humans. GANP is involved in the affinity maturation of antigen‐specific B cells12, 25 with the facilitated recruitment of AID to the rearranged immunoglobulin variable region loci.26, 27 Structurally, GANP carries a homologous region (580–1240 a.a.) to Sac3p that is involved in mRNA export in Saccharomyces cerevisiae and is an essential core protein of the transcription‐exportation 2 (TREX2) complex. It also plays a role in the regulation of DNA recombination.28, 29 In yeast cells, mutations in sub2, yra, sac3, or thp1 cause defects in transcription and a high frequency of transcription‐dependent DNA recombination.30 GANP carries an RNA‐recognition motif and an MCM3‐binding/histone acetyltransferase region that associates with the minichromosome maintenance complex, which possesses DNA helicase activity. GANP has been shown to inhibit DNA recombination in NIH3T3 cells using a β‐galactosidase tandem‐repeat construct for both extrachromosomal and genome‐integrated substrate DNA.17 GANP interacts with DNA‐dependent protein kinase, catalytic subunit (DNA‐PKcs) and may regulate the DNA repair pathway during homologous recombination,31 suggesting that this protein plays roles in DNA repair and genomic stability.
In our extensive search of GANP‐binding proteins using yeast two‐hybrid and proteomics analyses, no conventional molecules of cancer‐associated pathways were identified. GANP deficiency induces transcription‐coupled DNA damage, as detected by the Comet assay in cancer cell lines,32 which is followed by a DNA damage response with immediate p53 upregulation in normal fibroblasts.14 As the mammalian TREX2 complex is essential for mRNA export, GANP is presumably associated with the other members of PCID2 and DSS1. Recently, insufficiency of the TREX2 complex has been raised as a novel cause of genomic instability, inducing tumor development. For example, Bhatia et al. reported that DSS1‐associated BRCA2 prevented R‐loop accumulation.33 Their data suggested the functional interplay between the BRCA2 and TREX2 complex. Further analysis will be needed to clarify how GANP is linked to well‐known DNA repair pathways, especially to those involved in homologous recombination.
The association of GANP insufficiency with the development of breast cancer was experimentally proven using two different animal models. Conditional targeting of the ganp gene in the mammary gland led to the development of tumors at similar incidences (35%) in primiparous mice and caused lung metastasis in aged mice (Fig. 4). We used wap‐cre mice for mammary‐specific deletion; however, this system may not be perfect because the fact that pregnancy is required for cre deletion may influence mammary tumorigenesis. However, ganp heterodeficiency, without wap‐cre‐mediated gene targeting, led to the development of mammary gland tumors at a high incidence in female mice after multiparous episodes (Fig. 5). The frequency of tumor generation in single‐gene (ganp) heterodeficiency is approximately 30% in mice of the C57BL/6 background. This frequency of tumor generation was extremely high in mice with the C57BL/6 background, which are resistant to tumor development in the presence of heterodeficiencies in many types of tumor suppressor genes. The two models (ganp‐heterodeficient mice and wap‐cre‐mediated conditional targeted mice) showed similar rates of tumor generation after pregnancy. Therefore, the sporadic generation of mammary gland tumors is likely related to low GANP expression.
We propose a model of the sporadic generation of mammary gland tumors through two‐step processes. Pregnant mice show rapid differentiation and the development of mammary glands with changes in the hormonal environment that inevitably cause estrogen‐induced DNA damage. The increased GANP expression in mammary gland cells might be essential for reducing DNA damage (Figs 1A,3B), particularly at transcription‐competent gene loci, in accordance with the accelerated cell proliferation. The abnormality of mammary gland development presumably underlies the background of tumor development in conditionally ganp‐deficient mice, which is in agreement with the similar phenotypes observed in mammary‐specific Brca1‐deficient mice that have genomic instability in mammary epithelia, resulting in abnormal mammary gland development. In a study by other investigators,16 the mammary‐specific Brca1‐deficiency alone did not cause tumors, but the combined deficiency of Trp53 and Brca1 resulted in a high incidence of tumor development, as observed in our study of the wap‐cre‐ganp fl/fl mice. The wap‐cre‐ganp fl/fl mice spontaneously developed mammary tumors without p53 deficiency. The wap‐cre‐ganp fl/fl murine system is a useful model for studying the spontaneous development of breast tumors that have a single gene deficiency and may be necessary to elucidate how GANP and BRCA1 regulate the development of mammary gland tumors.
It remains to be investigated how GANP overexpression counteracts the oncogenic process of tumor initiation, promotion, or development. The level of GANP expression does not directly affect tumor migratory and invasive properties (Fig. S3). More importantly, we do not have direct evidence that GANP is upregulated in cells with trisomy 21. DNA injuries occurring during cell proliferation are usually repaired promptly, but cells with excessive DNA damage may remain unrepaired and eventually undergo apoptosis in the presence of wild‐type p53. Oncogenesis is initiated with increased genomic instability, causing mutations of various oncogenes and tumor suppressor genes, and chromosomal translocations.
Translocations of chromosomes occur during the repair of double‐strand DNA breaks, particularly in cancer cells, which may drive tumorigenesis in human colorectal carcinomas, breast cancer, and acute lymphoblastic leukemia. Tumor cells, however, contain a large number of complex translocations with interchromosomal and intrachromosomal rearrangements, which preclude drawing definite conclusions about the mechanism or extent by which any one of these individual translocations contributes to the malignancy of the cancer cell.34 The tumor cells generated in the ganp‐heterodeficient mammary glands harbored obvious chromosome translocations (Fig. 5D). Moreover, GANP interacts with DNA‐PKcs and is involved in the choice of DNA repair pathway toward higher fidelity homologous recombination by the suppression of error‐prone non‐homologous end‐joining of the camptothecin‐induced double‐strand DNA breaks.31 These results clearly support that GANP is an essential molecule to counteract at the very early stage of tumor initiation by preventing extensive changes in the genome associated with oncogenic mutations and chromosomal translocations during the growth and development of mammary gland cells.
The GANP protein encoded by the ganp gene on human chromosome 21 is a candidate that may help to elucidate the mechanisms underlying tumor resistance in individuals with DS.
Disclosure statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Representative images of GANP expression in normal mammary gland and breast cancer tissues. The staining positivity in all cases is almost homogeneous. Scale bar = 200 μm.
Fig. S2. A weak trend of GANP expression among TNM classification. A comparison of GANP expression is shown by immunohistochemistry scores using box plots with whiskers defining the range of scores. n.d., not determined.
Fig. S3. No obvious change was noted in migratory and invasive properties in E2‐stimulated ganp‐knockdown and ganp‐overexpressing MCF7 cells. The migration and invasion of the cells with or without E2 treatment were detected by Transwell‐based migration and invasion assays (upper panel). Migratory and invasive cells were stained and quantified at OD 560 nm after extraction (lower panel). The data are representative of three independent experiments.
Table S1. Distribution of clinical parameters in 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort) and 92 female patients (1995–2000) treated at Nagoya City University Hospital (NCU cohort).
Table S2. Background information of clinical parameters for normal samples in 92 female patients (1995–2000) treated at Nagoya City University Hospital (NCU cohort).
Table S3. Association between GANP expression and clinicopathological parameters in invasive breast cancer of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S4. Univariate and multivariate analysis for breast cancer‐specific survival of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S5. Univariate and multivariate competing risk analyses for breast cancer‐specific survival of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S6. Subtype of mammary gland tumors from ganp +/d mice.
Doc. S1. Supplementary materials and methods.
Acknowledgments
We are grateful to Y. Azakami‐Ogasawara for excellent technical assistance. This work was supported by JSPS Kakenhi (Grant Nos. 24590388 and 15K10083) and by grants from the Kanzawa Medical Research Foundation, Aichi Cancer Research Foundation, the 24th General Assembly of the Japanese Association of Medical Sciences, and Uehara Memorial Foundation.
Cancer Sci 107 (2016) 469–477
Funding Information
Japan Society for the Promotion of Science; Kanzawa Medical Research Foundation; Aichi Cancer Research Foundation; 24th General Assembly of the Japanese Association of Medical Sciences; Uehara Memorial Foundation.
References
- 1. Yang Q, Rasmussen SA, Friedman JM. Mortality associated with Down's syndrome in the USA from 1983 to 1997: a population‐based study. Lancet 2002; 359: 1019–25. [DOI] [PubMed] [Google Scholar]
- 2. Gardiner K, Davisson M. The sequence of human chromosome 21 and implications for research into Down syndrome. Genome Biol 2000; 1: REVIEWS0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haydar TF, Blue ME, Molliver ME, Krueger BK, Yarowsky PJ. Consequences of trisomy 16 for mouse brain development: corticogenesis in a model of Down syndrome. J Neurosci 1996; 16: 6175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reeves RH, Irving NG, Moran TH et al A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet 1995; 11: 177–84. [DOI] [PubMed] [Google Scholar]
- 5. Sago H, Carlson EJ, Smith DJ et al Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA 1998; 95: 6256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sago H, Carlson EJ, Smith DJ et al Genetic dissection of region associated with behavioral abnormalities in mouse models for Down syndrome. Pediatr Res 2000; 48: 606–13. [DOI] [PubMed] [Google Scholar]
- 7. Gahtan E, Auerbach JM, Groner Y, Segal M. Reversible impairment of long‐term potentiation in transgenic Cu/Zn‐SOD mice. Eur J Neurosci 1998; 10: 538–44. [DOI] [PubMed] [Google Scholar]
- 8. Peled‐Kamar M, Degani H, Bendel P, Margalit R, Groner Y. Altered brain glucose metabolism in transgenic‐PFKL mice with elevated L‐phosphofructokinase: in vivo NMR studies. Brain Res 1998; 810: 138–45. [DOI] [PubMed] [Google Scholar]
- 9. Whitaker‐Azmitia PM, Wingate M, Borella A, Gerlai R, Roder J, Azmitia EC. Transgenic mice overexpressing the neurotrophic factor S‐100 beta show neuronal cytoskeletal and behavioral signs of altered aging processes: implications for Alzheimer's disease and Down's syndrome. Brain Res 1997; 776: 51–60. [DOI] [PubMed] [Google Scholar]
- 10. Lana‐Elola E, Watson‐Scales SD, Fisher EM, Tybulewicz VL. Down syndrome: searching for the genetic culprits. Dis Model Mech 2011; 4: 586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chrast R, Scott HS, Madani R et al Mice trisomic for a bacterial artificial chromosome with the single‐minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum Mol Genet 2000; 9: 1853–64. [DOI] [PubMed] [Google Scholar]
- 12. Kuwahara K, Yoshida M, Kondo E et al A novel nuclear phosphoprotein, GANP, is up‐regulated in centrocytes of the germinal center and associated with MCM3, a protein essential for DNA replication. Blood 2000; 95: 2321–8. [PubMed] [Google Scholar]
- 13. Fujimura S, Xing Y, Takeya M et al Increased expression of germinal center‐associated nuclear protein RNA‐primase is associated with lymphomagenesis. Cancer Res 2005; 65: 5925–34. [DOI] [PubMed] [Google Scholar]
- 14. Ohta K, Kuwahara K, Zhang Z et al Decreased expression of germinal center‐associated nuclear protein is involved in chromosomal instability in malignant gliomas. Cancer Sci 2009; 100: 2069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuwahara K, Fujimura S, Takahashi Y et al Germinal center‐associated nuclear protein contributes to affinity maturation of B cell antigen receptor in T cell‐dependent responses. Proc Natl Acad Sci USA 2004; 101: 1010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu X, Wagner KU, Larson D et al Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumor formation. Nat Genet 1999; 22: 37–43. [DOI] [PubMed] [Google Scholar]
- 17. Yoshida M, Kuwahara K, Shimasaki T, Nakagata N, Matsuoka M, Sakaguchi N. GANP suppresses DNA recombination, measured by direct‐repeat beta‐galactosidase gene construct, but does not suppress the type of recombination applying to immunoglobulin genes in mammalian cells. Genes Cells 2007; 12: 1205–13. [DOI] [PubMed] [Google Scholar]
- 18. Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Amer Stat Assoc 1999; 94: 496–509. [Google Scholar]
- 19. Chan‐on W, Kuwahara K, Kobayashi N et al GANP, involved in immunoglobulin V‐region diversification, is co‐expressed with AID in cholangiocarcinomas associated with long‐term inflammation caused by liver fluke infestation. Int J Oncol 2009; 35: 287–95. [PubMed] [Google Scholar]
- 20. Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet 2000; 355: 165–9. [DOI] [PubMed] [Google Scholar]
- 21. Antonarakis SE, Epstein CJ. The challenge of Down syndrome. Trend Mol Med 2006; 12: 473–9. [DOI] [PubMed] [Google Scholar]
- 22. Emmert‐Streib F, de Matos Simoes R, Mullan P, Haibe‐Kains B, Dehmer M. The gene regulatory network for breast cancer: integrated regulatory landscape of cancer hallmarks. Front Genet 2014; 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reynolds LE, Watson AR, Baker M et al Tumour angiogenesis is reduced in the Tc1 mouse model of Down's syndrome. Nature 2010; 465: 813–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single‐minded 2s. Carcinogenesis 2007; 28: 259–66. [DOI] [PubMed] [Google Scholar]
- 25. Sakaguchi N, Kimura T, Matsushita S et al Generation of high‐affinity antibody against T cell‐dependent antigen in the Ganp gene‐transgenic mouse. J Immunol 2005; 174: 4485–94. [DOI] [PubMed] [Google Scholar]
- 26. Maeda K, Singh SK, Eda K et al GANP‐mediated recruitment of activation‐induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA. J Biol Chem 2010; 285: 23945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh SK, Maeda K, Eid MM et al GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy. Nat Commun 2013; 4: 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fischer T, Strässer K, Rácz A et al The mRNA export machinery requires the novel Sac3p‐Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 2002; 21: 5843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gallardo M, Luna R, Erdjument‐Bromage H, Tempst P, Aguilera A. Nab2p and the Thp1p‐Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J Biol Chem 2003; 278: 24225–32. [DOI] [PubMed] [Google Scholar]
- 30. Aguilera A, García‐Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012; 46: 115–24. [DOI] [PubMed] [Google Scholar]
- 31. Eid MM, Maeda K, Almofty SA, Singh SK, Shimoda M, Sakaguchi N. GANP regulates the choice of DNA repair pathway by DNA‐PKcs interaction in AID‐dependent IgV region diversification. J Immunol 2014; 192: 5529–39. [DOI] [PubMed] [Google Scholar]
- 32. Phimsen S, Kuwahara K, Nakaya T et al Selective cell death of p53‐insufficient cancer cells is induced by knockdown of the mRNA export molecule GANP. Apoptosis 2012; 17: 679–90. [DOI] [PubMed] [Google Scholar]
- 33. Bhatia V, Barroso SI, García‐Rubio ML, Tumini E, Herrera‐Moyano E, Aguilera A. BRCA2 prevents R‐loop accumulation and associates with TREX‐2 mRNA export factor PCID2. Nature 2014; 511: 362–5. [DOI] [PubMed] [Google Scholar]
- 34. Bunting SF, Nussenzweig A. End‐joining, translocations and cancer. Nat Rev Cancer 2013; 13: 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative images of GANP expression in normal mammary gland and breast cancer tissues. The staining positivity in all cases is almost homogeneous. Scale bar = 200 μm.
Fig. S2. A weak trend of GANP expression among TNM classification. A comparison of GANP expression is shown by immunohistochemistry scores using box plots with whiskers defining the range of scores. n.d., not determined.
Fig. S3. No obvious change was noted in migratory and invasive properties in E2‐stimulated ganp‐knockdown and ganp‐overexpressing MCF7 cells. The migration and invasion of the cells with or without E2 treatment were detected by Transwell‐based migration and invasion assays (upper panel). Migratory and invasive cells were stained and quantified at OD 560 nm after extraction (lower panel). The data are representative of three independent experiments.
Table S1. Distribution of clinical parameters in 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort) and 92 female patients (1995–2000) treated at Nagoya City University Hospital (NCU cohort).
Table S2. Background information of clinical parameters for normal samples in 92 female patients (1995–2000) treated at Nagoya City University Hospital (NCU cohort).
Table S3. Association between GANP expression and clinicopathological parameters in invasive breast cancer of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S4. Univariate and multivariate analysis for breast cancer‐specific survival of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S5. Univariate and multivariate competing risk analyses for breast cancer‐specific survival of 376 female patients (2001–2008) treated at Kumamoto University Hospital (KUH cohort).
Table S6. Subtype of mammary gland tumors from ganp +/d mice.
Doc. S1. Supplementary materials and methods.


