Abstract
Background:
The rising frequency of methicillin resistant Staphylococcus aureus (MRSA) has led to an increased use of antibiotics such as macrolide, lincosamide, streptogramin B (MLSB) for the treatment of S. aureus infections. Resistance to MLSB in S. aureus is commonly encoded by erm genes, which can be constitutive MLSB (cMLSB) or inducible MLSB (iMLSB). The purpose of this study was to determine the frequency of cMLSB, iMLSB, and MS phenotypes using D-test and polymerase chain reaction (PCR) methods.
Materials and Methods:
A total of 215 isolates of S. aureus were collected from January 2010 to May 2012 from Al-Zahra Hospital in Isfahan. PCR was performed for detection of mecA gene on all isolates using specific primers. The frequency of MLSB-resistant isolates was determined using D-test, and then a multiplex PCR was performed for detection of ermA, ermB, and ermC genes.
Results:
Among 215 S. aureus isolates examined, 82 (40.9%) were MRSA, and iMLSB, cMLSB, and MS resistance phenotypes had a frequency of 9 (4.18%), 58 (26.9%), and 11 (5.1%), respectively. Among nine isolates with iMLSB resistance phenotype, four isolates contained ermC gene, two isolates ermB gene, and one isolate ermA gene. Two isolates did not have any erm gene.
Conclusion:
In the current study, cMLSB was the most frequent phenotype and ermC was the most common gene in iMLSB resistant phenotypes.
Keywords: Clindamycin, D-test, erm genes, inducible resistance, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is one of the most frequent pathogens that cause both community and hospital-acquired infections worldwide. Development of drug resistance in S. aureus has led to the use of older antibiotics such as macrolide, lincosamide, and streptogramin B (MLSB) antibiotic.[1,2] However, extensive use of these antibiotics in serious staphylococcal infections has caused the emergence of S. aureus resistant to MLSB antibiotics.[3] There are three different mechanisms of resistance to MLSB antibiotics including: (1) Active efflux mechanism encoded by msr gene, (2) drug inactivation encoded by lun gene and (3) ribosomal binding site modification (by methylation or mutation in the 23s rRNA gene) encoded by erm genes (ermA, ermB, ermC, and ermF) among which, ermA and ermC are predominant genes responsible for resistance to MLSB antibiotics in staphylococci, which can be constitutive or inducible.[4,5,6,7,8] In vitro, S. aureus isolates with constitutive MLSB (cMLSB) resistance are resistant to erythromycin and clindamycin but isolates with inducible MLSB (iMLSB) resistance are resistant to erythromycin and susceptible to clindamycin. In this condition, treatment of patients with clindamycin can lead to the emergence of resistant mutants to cMLSB from iMLSB-resistant strains and treatment failure.[3,6] On the other hand, assigning all erythromycin-resistant S. aureus as clindamycin resistant strains may cause to avoid the use of clindamycin in the treatment of S. aureus infections. For this reason, careful screening of iMLSB-resistant strains is very important. While constitutive resistance is detectable by routine antimicrobial susceptibility tests, inducible resistance to clindamycin is not detectable by standard methods.[4,5] For detection of iMLS-resistant strains, Clinical and Laboratory Standards Institute (CLSI) developed a phenotypic method called the double disk diffusion test (D-test).[9,10,11,12] The aim of this study was to determine the frequency of inducible resistance to clindamycin using D-test and polymerase chain reaction (PCR) with specific primers to confirm the presence of the erm genes in these isolates.
MATERIALS AND METHODS
Bacterial strains and phenotypic testing
A total of 215 clinical isolates of S. aureus were collected from Al-Zahra Hospital in Isfahan from January 2010 to May 2012. Bacterial isolates were obtained from various clinical specimens including: Wound, blood, urine, sputum, etc., Early identification was performed based on Gram-staining and positive biochemical reactions such as catalase, coagulase, and DNase tests. D-test method was performed according to the CLSI guidelines using clindamycin (2 µg) and erythromycin (15 µg) disks (Himedia-India). For this purpose, suspensions of bacteria were prepared in the sterile saline (2 ml) equivalent to standard 0.5 McFarland and then two antibiotic disks placed on Muller-Hinton agar media in 15 mm distance (edge-to-edge). Plates were incubated at 35°C overnight. Strains with flat zone of growth inhibition of clindamycin near the erythromycin disk (D-shape) were classified as resistant phenotypes to iMLSB (D-test positive), while those with a circular zone were classified as MS resistant phenotypes (D-test negative) [Figure 1].
Figure 1.
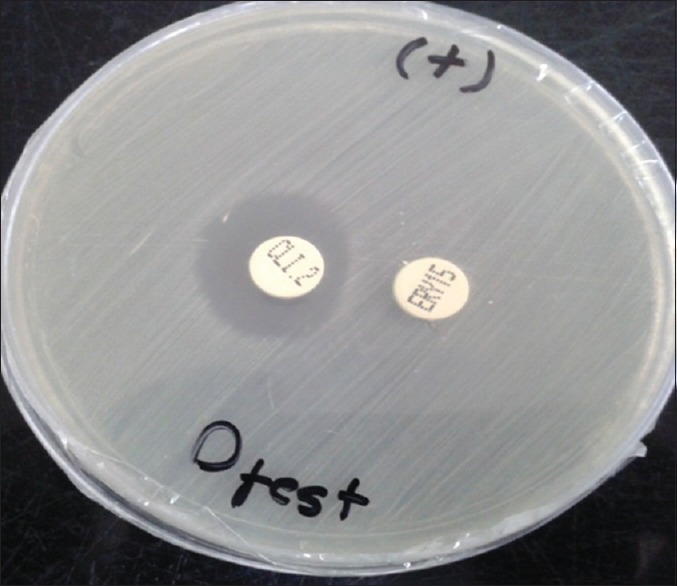
D-shape zone of growth inhibition around clindamycin disk (inducible macrolide, lincosamide, streptogramin B phenotype)
Molecular detection of mecA gene
DNA was extracted from 215 S. aureus isolates using Fermentas K0512 DNA kit (Fermentas-USA) in accordance with the manufacturer's protocol. PCR reaction was carried out for the amplification of the 310 bp fragment of mecA gene using primers as exhibited in Table 1. PCR amplification reaction mixture (25 μL) contained 4 µL of DNA template, 2.5 µL of PCR buffer (×10), 0.75 µL Mgcl2 (50 mM), 0.5 µL of dNTPs (10 mM), 1 µL of each primers (2 μL totally), 0.25 µL of Ex-Taq DNA polymerase (5u/µL) and 15 µL distill water. PCR conditions were as follows: Initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s, and final extension at 72°C for 7 min.
Table 1.
Primers used in this study

Multiplex polymerase chain reaction for erm gene
Multiplex PCR was performed for detection of erm gene in D-test positive isolates using specific primers for the ermA, B and C genes as exhibited in Table 1. Each PCR was performed in a final volume of 25 µL consisting of 5 µL of DNA template, 2.5 µL of PCR buffer (×10), 1 µL Mgcl2 (50 mM), 0.5 µL of dNTPs (10 mM), 0.75 µL of each primers (2 μL totally), 0.25 µL of Ex-Taq DNA polymerase (5 u/µL), 11.25 µL distill water. DNA was amplified on a thermocycler (Ependorf-Germany), and PCR conditions were as follows: Initial denaturation at 94°C for 10 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 60 s, followed by a final extension at 72°C for 10 min.
RESULTS
In this study, 215 isolates of S. aureus were collected from various clinical specimens, wound 53 (24.6%), blood 49 (22.79%), urinary tract infection 30 (13.9%), sputum 35 (16.27%), abscess 21 (9.76) and others 27 (12.55%), from Al-Zahra Hospital in Isfahan. The patient's average age was 47 years (ranged 1–88 years). The mecA gene screening in all isolates showed that 82 (40.9%) of the 215 tested isolates were methicillin resistant S. aureus (MRSA) and mecA positive [Figure 2]. Furthermore, double disk diffusion test results revealed that 134 (62.3%) of the isolates were susceptible to both clindamycin and erythromycin and 81 (37.7%) were shown to have four different resistance phenotypes in which 58 (26.9%) isolates were resistant phenotype to cMLSB(resistant to both erythromycin and clindamycin), 9 (4.18%) isolates were resistant phenotype to iMLSB (resistant to erythromycin and susceptible to clindamycin), 11 (5.1%) isolates were MS resistance phenotype (susceptible to clindamycin and resistant to erythromycin) and finally, 3 (1.39%) isolates were susceptible to erythromycin and resistant to clindamycin [Figure 3]. Among nine isolates with iMLSB resistance phenotype, 5 (55.5%) were MRSA. Nine staphylococcal isolates with iMLSB resistance phenotype were tested for the presence of the erm genes, the ermA gene in 1 (11.1%) isolate, the ermB gene in 2 (22.2%) isolates, the ermC gene in 4 (44.4%) isolates was detected and two isolates did not have any erm genes [Figure 3].
Figure 2.
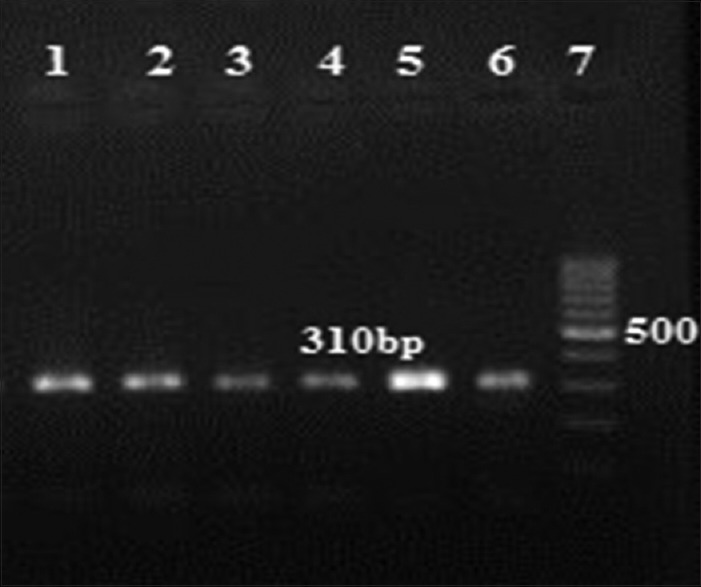
Gel electrophoresis of mecA gene. Lanes 1–5: 310 bp fragment, Lane 6: positive control of methicillin resistant Staphylococcus aureus strains ATCC 33591, Lane 7: DNA Ladder 100 bp
Figure 3.
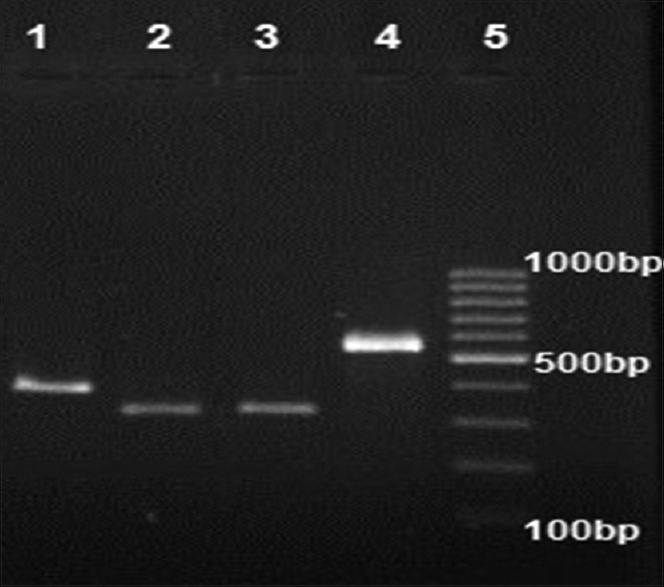
Gel electrophoresis of erm genes. Lane 1: ermA positive (421 bp), Lane 2 and 3: ermB positive (359 bp), Lane 4: ermC positive (572 bp), Lane 5: DNA Ladder 100 bp
DISCUSSION
D-test results in our study demonstrated that 134 (62.3%) isolates were sensitive to both erythromycin and clindamycin; the frequency of cMLSB, iMLSB, and MS phenotypes were found to be 58 (26.9%), 9 (4.18%), and 11 (5.1%), respectively. In addition, the frequency of ermC, ermB, and ermA genes among isolates with iMLSB phenotype was determined to be 44.4%, 22.2%, and 11.1% respectively. Clindamycin due to its advantages including low-cost, low side effects, and good tissue penetration is used for the treatment of S. aureus infections. Although it is a good alternative in allergic patients instead of β-lactam antibiotics;[1,9,14,15] however, excessive use of this antibiotic has an important role in bacterial resistance to clindamycin. Since the treatment of infected patients with resistant strains to iMLSB can lead to the expansion of constitutive resistance (cMLSB) and therapy failure with clindamycin, detection of resistant strains to iMLSB is important from other resistance phenotypes. Since the frequency of cMLSB, iMLSB, and MS phenotypes varies in different geographical areas, even among different hospitals, awareness of regional frequency of MLSB resistant isolates is important for laboratories to decide for performing the D-test routinely or reporting all erythromycin-resistant S. aureus as clindamycin resistant.[7,10,12,16]
In the current study, 82 (40.9%) isolates were found to be MRSA that is, comparable with a study conducted by Seifi et al.[5] Also, 6.09% of MRSA isolates had resistant phenotype to iMLSB, which is lower than those reported by Shoja et al.[17] In the current study, 134 (62.3%) isolates were sensitive to both erythromycin and clindamycin and the frequency of cMLSB, iMLSB and MS phenotypes were found to be 58 (26.9%), 9 (4.18%), and 11 (5.1%) respectively. Similar results were reported by Aslanimehr et al.[18] In the present study, the frequency of cMLSB phenotype was higher than iMLSB phenotype. Similar results were obtained by Memarian et al.[19] and Mahesh et al.[20] In contrast, Reddy and Suresh found the frequency of iMLSB phenotype to be higher than cMLSB phenotype.[3] In our study, the frequency of MS resistance phenotype was shown to be higher than iMLSB phenotype, which was concordant to some previous studies.[3,5,7] Incidentally, we detected 3 (1.39%) isolates resistant to clindamycin and susceptible to erythromycin, similar results were also obtained by Coutinho et al.[10] In addition, Seifi et al. reported 6 (2.84%) S. aureus isolates with such a phenotype.[5] This phenotype can be created by lincosamide nucleotide transferase enzyme that only inactivates lincosamide (clindamycin). Therefore, we investigated erm gene distribution among isolates with iMLSB phenotype. Our results revealed the frequency of ermC, ermB, and ermA genes among isolates with iMLSB phenotype to be 44.4%, 22.2%, and 11.1%, respectively. Two isolates with iMLSB phenotype were negative in genotypic test.
It must be noted that the frequency of erm genes is variable in different studies. According to our findings, the ermC gene was the most prevalent gene, similar study was performed by Aktas et al. in Turkey,[7] while in a study conducted by Saderi et al. ermA gene was prevalent (60%) among erythromycin-resistant S. aureus.[2] An interesting point to notice in our study was the high frequency of ermB gene, Similar results were shown in some studies.[21,22]
CONCLUSION
This report has investigated the frequency of inducible resistance to clindamycin using D-test and PCR methods. This was the first study to investigate the frequency of MLSB phenotypes in Isfahan which demonstrated cMLSB resistance to be the most prevalent resistance phenotype, ermC gene as the most common gene among iMLS-resistant S. aureus and iMLSB phenotype having a low frequency. Therefore, we do not recommend the routine performance of D-test but since the frequency of different resistance phenotype may change through time with the emergence of strains with different antibiotic susceptibility patterns, it is recommended that local periodic survey be performed.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Vivek JS, Rajesh GN, Mukesh S, Manpreet K, Manpreet K, Misra RN, et al. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates in a tertiary care hospital in India. Biomed Res. 2011;22:465–9. [Google Scholar]
- 2.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med Sci Monit. 2011;17:BR48–53. doi: 10.12659/MSM.881386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy SP, Suresh R. Phenotypic detection of inducible clindamycin resistance among the clinical isolates of staphylococcus aureus by using the lower limit of inter disk space. J Microbiol Biotechnol Res. 2012;2:258–64. [Google Scholar]
- 4.Zelazny AM, Ferraro MJ, Glennen A, Hindler JF, Mann LM, Munro S, et al. Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in staphylococci: A CLSI collaborative study. J Clin Microbiol. 2005;43:2613–5. doi: 10.1128/JCM.43.6.2613-2615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seifi N, Kahani N, Askari E, Mahdipour S, Naderi NM. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iran J Microbiol. 2012;4:82–6. [PMC free article] [PubMed] [Google Scholar]
- 6.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43:1716–21. doi: 10.1128/JCM.43.4.1716-1721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococci isolated in Istanbul, Turkey. J Microbiol. 2007;45:286–90. [PubMed] [Google Scholar]
- 8.Schmitz FJ, Sadurski R, Kray A, Boos M, Geisel R, Köhrer K, et al. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45:891–4. doi: 10.1093/jac/45.6.891. [DOI] [PubMed] [Google Scholar]
- 9.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740–4. doi: 10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho Vde L, Paiva RM, Reiter KC, de-Paris F, Barth AL, Machado AB. Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates. Braz J Infect Dis. 2010;14:564–8. doi: 10.1016/s1413-8670(10)70113-6. [DOI] [PubMed] [Google Scholar]
- 11.Sedighi I, Yousefi Mashouf R, Pak N, Seif Rabiee MA. D-test method for detection of inducible clindamycin resistance in Staphylococcus aureus. Iran J Pediatr. 2009;19:293–7. [Google Scholar]
- 12.O'Sullivan MV, Cai Y, Kong F, Zeng X, Gilbert GL. Influence of disk separation distance on accuracy of the disk approximation test for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2006;44:4072–6. doi: 10.1128/JCM.01632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–6. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim HS, Lee H, Roh KH, Yum JH, Yong D, Lee K, et al. Prevalence of inducible clindamycin resistance in staphylococcal isolates at a Korean tertiary care hospital. Yonsei Med J. 2006;47:480–4. doi: 10.3349/ymj.2006.47.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42:2777–9. doi: 10.1128/JCM.42.6.2777-2779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis JS, 2nd, Jorgensen JH. Inducible clindamycin resistance in Staphylococci: Should clinicians and microbiologists be concerned? Antimicrob Resist. 2005;40:280–5. doi: 10.1086/426894. [DOI] [PubMed] [Google Scholar]
- 17.Shoja S, Nahaei M, Nahaei M. Detection of inducible clindamycin resistance in Staphylococcus aureus and Staphylococcus epidermidis by using D-test. Pharma Sci. 2009;15:1–8. [Google Scholar]
- 18.Aslanimehr M, Yaghobfar R, Peymani A. Detection of MLSB phenotypes and inducible clindamycin resistance in Staphylococcus aureus isolates in-patients of Qazvin and Tehran teaching hospitals. J Qazvin Univ Med Sci. 2014;18:30–6. [Google Scholar]
- 19.Memariani M, Pourmand MR, Shirazi MH, Soltan Dallal MM, Abdolsamadi Z, Mardani N. The importance of inducible clindamycin resistance in enterotoxin positive S. aureus isolated from clinical samples. Tehran Univ Med J. 2009;67:250–6. Persian. [Google Scholar]
- 20.Mahesh CB, Ramakant BK, Jagadeesh VS. The prevalence of inducible and constitutive clindamycin resistance among the nasal isolates of staphylococci. J Clin Diagn Res. 2013;7:1620–2. doi: 10.7860/JCDR/2013/6378.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emaneini M, Eslampour MA, Sedaghat H, Aligholi M, Jabalameli F, Shahsavan S, et al. Characterization of phenotypic and genotypic inducible macrolide resistance in staphylococci in Tehran, Iran. J Chemother. 2009;21:595–7. doi: 10.1179/joc.2009.21.5.595. [DOI] [PubMed] [Google Scholar]
- 22.Zmantar T, Chaieb K, Ben Abdallah F, Ben Kahla-Nakbi A, Ben Hassen A, Mahdouani K, et al. Multiplex PCR detection of the antibiotic resistance genes in Staphylococcus aureus strains isolated from auricular infections. Folia Microbiol (Praha) 2008;53:357–62. doi: 10.1007/s12223-008-0055-5. [DOI] [PubMed] [Google Scholar]


