Abstract
Alternative splicing shapes mammalian transcriptomes, with many RNA molecules undergoing multiple distant alternative splicing events. Comprehensive transcriptome analysis, including analysis of exon co-association in the same molecule, requires deep, long-read sequencing. Here we introduce an RNA sequencing method, synthetic long-read RNA sequencing (SLR-RNA-seq), in which small pools (≤1,000 molecules/pool, ≤1 molecule/gene for most genes) of full-length cDNAs are amplified, fragmented and short-read-sequenced. We demonstrate that these RNA sequences reconstructed from the short reads from each of the pools are mostly close to full length and contain few insertion and deletion errors. We report many previously undescribed isoforms (human brain: ∼13,800 affected genes, 14.5% of molecules; mouse brain ∼8,600 genes, 18% of molecules) and up to 165 human distant molecularly associated exon pairs (dMAPs) and distant molecularly and mutually exclusive pairs (dMEPs). Of 16 associated pairs detected in the mouse brain, 9 are conserved in human. Our results indicate conserved mechanisms that can produce distant but phased features on transcript and proteome isoforms.
Protein and RNA molecules can be very long, and multiple processes produce different kinds of isoforms. RNA molecules, specifically, may undergo removal of multiple introns, addition of the 5′ cap and the polyA-tail as well as RNA editing and modifications. Alternative splicing is crucial in shaping transcriptome variation1 and proteome diversity2 and is often associated with cancer3,4. Individual alternative splicing events can be studied in depth using microarrays5–7 and short-read RNA sequencing8–13. Estimates of the proportion of spliced genes that are alternatively spliced have increased over the years from 42%14 to 74%7, 86%15, 95%16 and 98–100%9. Each gene has an average of six transcript isoforms, a figure that is likely to increase10,17. Because long transcripts often undergo multiple alternative splicing events, it is usually unclear which exons are included in which transcript, and the true complexity of the transcriptome remains unknown. In particular, whether alternative (distant) exon pairs are included in a co-associated, mutually exclusive or independent manner into RNA molecules has not been covered in a comprehensive fashion and is difficult to address using microarrays and short-read sequencing because of fragment length limitations18 (Fig. 1a), although some distant alternative exons exhibit correlated inclusion when interrogated across multiple tissues6. Thus, accurate and quantitative full-length transcriptomes are lacking19 for nearly all eukaryotes that have multiple exons per gene. Short-read sequencing employs longer and longer reads20, and a number of studies have recently shown the feasibility of sequencing longer cDNA molecules of variable length18,21–24. Indeed, we previously monitored combinations of exons18,21,22 as well as combinations of heterozygous single-nucleotide variations (SNVs) and allele-specific expression and splicing directly from full-length cDNA-molecules18. However, these technologies (454 and Pacific Biosciences (PacBio))18,22,25 either lacked the ability to provide full-length sequences for each mRNA molecule (454), or it was difficult to generate large enough numbers of sequences (PacBio) so that comprehensive transcript diversity could not be deduced with statistical confidence, particularly for transcripts of low abundance.
Figure 1.
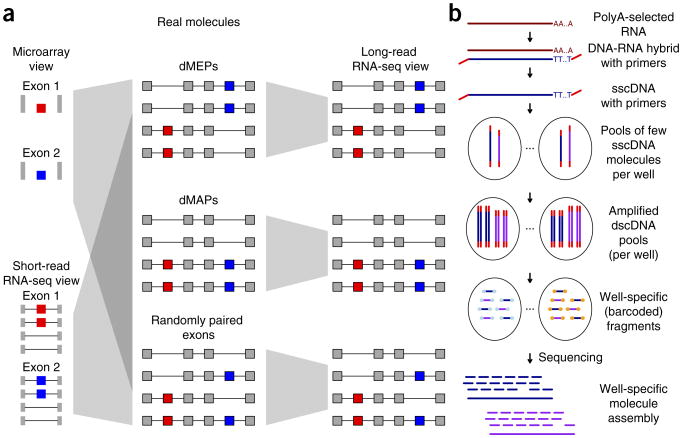
Illustration of purpose and strategy of this work. (a) Multiple and distant alternative exons (red and blue) can be combined in different ways to form RNA isoforms. On a molecular level they can be included in RNA molecules in an opposed (or mutually exclusive) manner (top middle, dMEPs), in a phased manner (center middle, dMAP) or in a randomly paired manner (bottom middle). Using traditional short-read sequencing (bottom left) or microarrays (top left), these three fundamentally different situations lead to the same observation, and thus cannot be distinguished. With long-read technologies (right) it is trivial to assign each group. (b) Outline of experimental procedure for SLR-RNA-seq.
Here we introduce a facile RNA-sequencing method, SLR-RNA-seq (Fig. 1b), which can be carried out by any laboratory with access to an Illumina sequencer and which is based on the Illumina MOLECULO technology. In this protocol 103–104 DNA molecules, of multiple kilobases each, are amplified and the resulting amplicons are fragmented and sequenced using shorter reads. This approach minimizes the occurrence of two DNA molecules from the same genomic region, which facilitates accurate reconstruction of the original molecules from short reads. For the human brain, we analyzed 4,224 pools of ∼1,000 molecules each in 11 lanes of HiSeq data (mouse: 3,072 pools in eight lanes). The MOLECULO technology has been used in genome sequencing to phase the GM12878-genome26 and to accurately place highly repetitive elements in de novo assembly of Drosophila melanogaster27. We adapt this technology to accurately deduce transcript structure and measure the molecular coordination between distant alternative splicing events in the mouse and human brain. Our results demonstrate that accurate and deep long-read transcriptomes can be obtained and show conservation of a significant fraction of distant splicing events as well as their coordinated regulation between human and mouse. These results indicate the existence of a phased proteome, in which distant peptides can be included into protein molecules in a coordinated manner.
Results
Generation of SLR-RNA-seq libraries
We prepared single-stranded cDNA (sscDNA) that includes adapters containing PCR-primer sites at the beginning and the end of each cDNA molecule (Fig. 1b). Based on qPCR, ∼1,000 such sscDNA molecules were added into each well of a 384-well plate. In each well, sscDNA was amplified and the resulting double-stranded cDNA (dscDNA) molecules were fragmented and barcoded, so that each fragment could be assigned to its well. After sequencing using an Illumina HiSeq2000, 2 × 125-bp, paired-end reads were assembled into high-coverage SLRs in a well-specific fashion. For most genes, this minimized the possibility of a nonidentical molecule from the same locus interfering with the assembly (which we refer to as a ‘collision’ of two nonidentical transcripts of the same gene), and fixed tags at each end of each transcript further reduced the possibility of interference from nonidentical molecules27, as long as they do not have identical transcript starts and ends. Although this process involves amplification, we consider the resulting SLRs ‘quasi-single-molecule’, because a single RNA molecule cannot produce multiple redundant SLRs. By using multiple mini-libraries, 384-well plates and Illumina lanes, this technique allows us to sequence much deeper and at higher accuracy than in any previously published long-read RNA-sequencing experiments.
Analysis of external control sequence RNAs
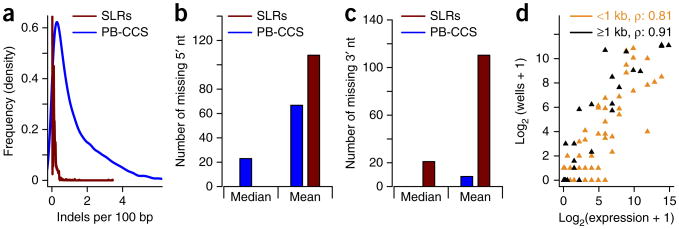
We first tested this procedure using the External RNA Controls Consortium (ERCC) control RNAs28, which is a mixture of variably sized control RNAs up to 2 kb in length. This mixture was spiked into a mouse brain sample from two 12-week-old mice. We produced libraries containing 3.7-million mouse brain and 19,000 ERCC synthetic long-reads (SLRs). The mixed SLRs were mapped to the mouse mm10-genome29 and the known ERCC-sequences simultaneously, using GMAP30, as described previously18,21,22. SLRs mapping to ERCC sequences were compared to the original ERCC sequences and to ERCC reads we previously sequenced using PacBio circular consensus sequences (PacBio-CCS)22—reads that lower PacBio's error rate from ∼15% to 1–2% by repeatedly sequencing the circular template. It is worth noting that there are methods for generating longer PacBio reads with relatively low error rates, but these methods sacrifice the single-molecule character of the resulting reads. 96% of SLRs are free of indels, which is only the case for 5.5% of PacBio-CCS (Fig. 2a). We then monitored the number of nucleotides missing at the 5′ end of the ERCC SLRs. PacBio-CCS missed a median of 23 5′ nucleotides; only 2.4% of all PacBio-CCS did not miss a single 5′ nucleotide. The majority of SLRs (54%), however, did not miss a single 5′ nucleotide, but those SLRs that did miss a 5′ nucleotide often missed many, presumably because incomplete assemblies can lead to SLRs representing only parts of the ERCC molecules (Fig. 2b); these are readily recognized as they lack an end-marker tag at either the 5′ or 3′ cDNA end. For the 3′ sequences we found similar characteristics (as compared to 5′) for SLRs, although the majority of SLRs miss ≥20 nucleotides, presumably due to removal of sequences around the polyA tail, which is recognized as a repeat in our assembly algorithms and is not included. As shown previously22, PacBio-CCS usually did not miss nucleotides on the 3′ end (Fig. 2c). The correlation between ERCC-annotated expression (the mixing frequency of these molecules) and observed SLRs for each ERCC sequence depended on the length of the ERCC sequence. For ERCC sequences shorter than 1 kb, the Spearman correlation was 0.81, but for ERCC sequences of 1 kb or longer it rose to 0.91 (Fig. 2d). This is very likely due to the bias against short molecules in the SLR-RNA-seq protocol (Supplementary Fig. 2 and Supplementary Results, section 2).
Figure 2.
Comparison of SLRs and PacBio-CCS on the ERCC sequences. (a) Distribution of indels (per 100 nt of mapping) in PB-CCS (blue) and SLRs (red) mapped to the ERCC-control RNAs. PacBio-CCS, PB-CCS. (b) Median and mean number of 5′ missing nucleotides for PB-CCS (blue) and SLRs (red) mapped to the ERCC-control RNAs. (c) Median and mean number of 3′ missing nucleotides for PB-CCS (blue) and SLRs (red) mapped to the ERCC-control RNAs. (d) Correlation of log-transformed given concentration for the ERCC sequences and the log-transformed number of wells, in which each ERCC sequence is observed.
The ERCC sequences that we used here have been widely and successfully employed for RNA-seq method validation, but they represent molecules of up to 2 kb only. Given that 25% (∼1.3 M) of human brain SLRs are 2.5 kb or longer, there is, however, a clear need for longer control sequences. Overall, these results indicate that our SLRs generate nearly full-length reads and excel at sequencing very long molecules.
Comparison of long-read technologies on natural human RNA
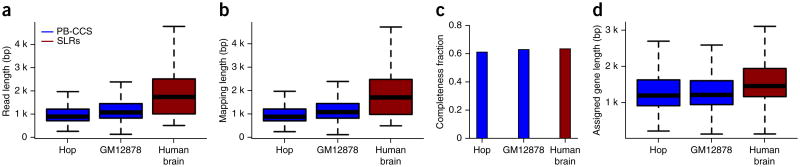
To assess how SLRs perform in the presence of longer and complex, alternatively spliced RNA molecules not represented in the ERCC set, we used 3.7 M SLRs we generated from mouse brain and 5.2 M from human brain (mapping statistics for the human brain RNA can be found in Supplementary Fig. 1). Comparing these samples to the PacBio-CCS we had previously sequenced18,22 in an RNA sample of multiple human organs and individuals as well as in the GM12878 cell line, we found our SLRs to be about 71% (human brain SLRs, average 1,907 bp) and 66% (mouse brain SLRs, average 1,849 bp) longer than the PacBio-CCS (weighted average 1,112 bp—for the pooled PacBio-CCS from ref. 18, which averaged 1,178 bp for GM12878 and from ref. 22, which averaged 999 bp). SLRs in all data sets were consistently longer than PacBio-CCS (Fig. 3a). Taken together with published PacBio-CCS in human brain31, which averaged 1,289 bp, a picture emerges of 1,000- to 1,300-bp-long CCS (depending on sample, laboratory, chemistry and polymerase version) and 1,850- to 1,900-bp-long SLRs (Supplementary Table 1). This difference in read length carried forward to mapping length, showing that SLRs do not frequently contain incorrectly assembled sequence, which cannot be aligned collinearly to the same chromosome and strand (Fig. 3b). For each spliced SLR and each spliced PacBio-CCS, we assessed its completeness with respect to the human genome annotation32. Overall, we found 61–64% (human organ panel PacBio-CCS: 61.2%; GM12878 PacBio-CCS: 63.0%; human brain SLR-RNA-seq: 63.5%) of the molecules represented full-length molecules—that is, extending from the first splice site of an annotated transcript to the last splice site of an annotated transcript (Fig. 3c). Although a slight advantage for SLRs was observed, the difference between the technologies was not as pronounced as for sequence and mapping length. This apparent discrepancy can be explained by differences in the sets of genes that the two technologies interrogate. Indeed, we found that SLRs were assigned more often to longer genes than PacBio-CCS (Fig. 3d), presumably because (i) very long cDNA-molecules (e.g., ≫ 4 kb) are less likely to be loaded into a PacBio well in the presence of numerous shorter molecules (e.g., 600 bp to 1 kb), and (ii) as cDNA length increases, it becomes less likely for a read to be at least twice as long as the cDNA insert, which is required for PacBio-CCS-sequencing but not for SLR-RNA-seq. For SLRs the only limitation is whether the entire cDNA molecule is covered sufficiently to allow its full-length assembly.
Figure 3.
Comparison of SLRs and PacBio-CCS on human and mouse transcriptomes. (a) Read length obtained for a human organ panel (Hop) and in the GM12878 cell-line using single-molecule PacBio-CCS, and for a human brain sample using SLR-RNA-seq. (b) Mapping length for the same data sets as in a. (c) Percentage of reads that could be classified as full-length in the same data sets as in a. (d) Distributions of mature gene length to which spliced reads were assigned for the same data sets as in a.
Human gene expression measurements in molecules per million (MPM; Supplementary Fig. 2a and Supplementary Data 1) correlated highly with fragments per kilobase per million fragments mapped (FPKM) measurements deduced from human brain RNA short-read sequencing31. Spliced genes found more highly expressed in short-read sequencing (N = 6,540) were enriched with pseudogenes and tended to be shorter than genes found with both technologies—presumably these two observations stem from some short reads mapping and contributing to the expression-estimation of multiple locations and from the bias against very short molecules during SLR-RNA-seq (Supplementary Fig. 2b and Supplementary Results). Mouse MPMs (Supplementary Fig. 2c and Supplementary Data 2–4) correlated also very highly between the two mice sequenced (Supplementary Fig. 2d). Calculating percent-spliced-in (Ψ) values for all observed splice sites as the fraction of all overlapping molecules (spliced at least once) that use the splice site and percent-isoform (Π) values for full-length isoforms, we found that Ψ-values and major-isoform Π-values correlate highly between the two sequenced mice (Supplementary Fig. 3 and Supplementary Data 5–12).
Many novel isoforms for current human annotations
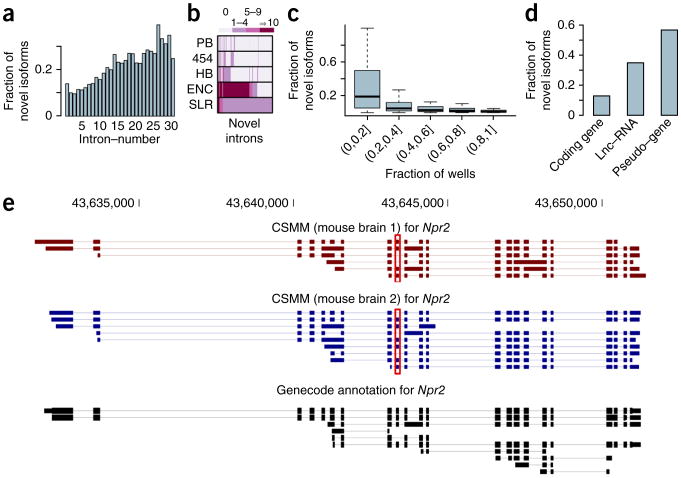
On the basis of 454 and PacBio RNA-seq data from a diverse set of cell lines and tissues18,21,22, we have recently shown that a large fraction of spliced RNA molecules cannot be interpreted as corresponding to entirely or partially annotated transcripts from GENCODE32. In general, the more exons an RNA molecule contains, the more likely it is to show a novel splicing pattern. Given that SLRs excel at representing long RNA molecules, we tested SLRs for novelty (see ref. 21 for an exact definition of “novel isoforms”). Overall, 14.5% of all human brain spliced reads exhibit splice-site combinations that are novel with respect to the GENCODE version-15 annotation. Consistent with our previous research18,21,22, molecules with large numbers of introns were more likely to represent novel isoforms. Specifically, SLRs with 20 to 30 introns, which were only rarely identified with PacBio-CCS, had a very large fraction of novel isoforms (Fig. 4a). Comparing spliced reads from mouse brain to the mouse GENCODE M3 annotation, we found very similar results, although spliced mouse reads had a higher fraction of novel transcripts—likely because the human GENCODE annotation is more advanced than the mouse GENCODE annotation (Supplementary Fig. 4a,b). For novel isoforms with novel introns (one or both splice sites novel, or two known splice sites forming an unannotated intron, such as in the case of novel exon skipping), 73.8% can also be found in published data sets (Fig. 4b) such as RNA-seq data from the ENCODE10 project or the ABRF (Association of Biomolecular Resource Facilities) study31. For a single data set of matched shifted introns, we found a background validation of 1.7% in the same short-read data sets. Different approaches of generating random introns using only real splice sites yielded 9.9–24.1%, numbers that are far below the 73.8% found above (Supplementary Results, section 3).
Figure 4.
Analysis of novel isoforms revealed by SLR-RNA-seq. (a) Fraction of mapped reads that show a novel splice pattern with respect to the GENCODE annotation, broken up by the number of introns in the mapping. (b) Heatmap of novel introns (with respect to GENCODE) determined by SLR-RNA-seq showing the number of times each intron was observed in the combined set of short-read ENCODE-RNA-seq data sets (ENC)10, in a human brain sample (HB)31, and in our previous data using the Roche-454 platform (454)21 and the PacBio-platform (PB)18,22. (c) Fraction of mapped reads that show a novel splice pattern with respect to the GENCODE annotation, broken up by gene expression of the gene to which the read was mapped. Gene expression is here given as the fraction of wells, in which the gene was detected. (d) Fraction of mapped reads that show a novel splice pattern with respect to the GENCODE annotation, for mappings assigned to coding genes, lncRNA genes or to pseudogenes. (e) Illustration of novel isoforms revealed by SLR-RNA-seq for Npr2. Note, that we show isoforms from only four lanes of sequencing (our first round of sequencing). Some isoforms are novel, because of intron retention events, which can be easily observed. Others are novel, because they skip an exon in long transcripts (see red box)—a skipping event that occurs only in short transcripts according to the annotation. CSMM, consensus split mapped molecule, a read mapping for which all splits respect both the donor consensus and the acceptor consensus.
A concern with SLR-RNA-seq is that, for highly expressed genes, different RNA isoforms might be present in the same microtiter well and eventually be assembled into a false-positive novel isoform. If so, one would expect a higher fraction of novel SLRs for highly expressed genes. We therefore calculated for each human GENCODE gene separately the fraction of novel isoforms and binned genes according to their expression (measured by the fraction of wells among the total 4,224 wells, in which the gene was found). In general, we did not observe a tendency for more novel SLRs in highly expressed genes; instead, we observed the opposite (Fig. 4c). Overall, 98.9% all human brain SLRs (96.2% for mouse) could be attributed to a spliced annotated gene (the one with which it shared most splice sites21) of the human GENCODE annotation15,32. Consistent with the lower expression of long, noncoding RNAs (lncRNAs), especially in the cytosol33, we found relatively few human spliced reads (∼28,000, or 1%) and mouse spliced reads (∼2,400, 0.6%) corresponding to known lncRNA genes. Notably, whereas most spliced reads belonged to protein coding genes (human: 97.8%, mouse 95.9%), other gene types such as “processed transcript” and “pseudogene,” also received a substantial number of spliced reads. The percentage of human novel-isoform reads was higher for pseudogenes and lncRNAs than for protein coding genes (Fig. 4d), presumably because these categories of RNA-producing genes (in contrast to protein coding genes) have only recently attracted much interest and thus are not as well annotated as protein coding genes. This is intriguing because pseudogenes may gain lncRNA-like functions, as appears to have occurred with XIST34. When determining the fraction of human genes that had at least one novel spliced read for each of the three gene types, we found similar fractions of novel isoforms between protein coding genes, lncRNAs and pseudogenes (Supplementary Fig. 4c,d). Thus comparably few new-isoform molecules affect a large number of protein coding genes, whereas comparably many new-isoform molecules affect a large number of lncRNA genes and pseudogenes. This suggests that novel isoforms for protein coding genes are more often minor isoforms than they are for the other gene classes. Notably, the majority of the detected spliced genes in all three gene types had at least one novel isoform (protein coding genes: 86%; lncRNA genes: 86%; pseudogenes: 91%)—although these data alone cannot prove the functional significance of each of these molecules. Additionally ∼36,000 human spliced reads (mouse ∼30,000) did not share a splice site with any known gene, and presumably often represent novel lncRNA genes or pseudogenes. Note that mapping criteria were generally very stringent, so that any mapping with unclear origin (pseudogene or parent gene) was discarded (Supplementary Fig. 5). Consistent with previous research35, most introns exhibited little intron retention but the majority of spliced genes (61% in mouse, 55% in human) that had ten or more spliced reads had at least one intron with a percent intron retention (PIR)35 of 0.1 or higher. As an example, mouse Npr2 (for which mouse GENCODE M3 annotated isoforms as well as our SLRs had up to 22 exons) had a large number of novel isoforms (Fig. 4e). The majority of aligned Npr2 isoforms differed from the annotation both by the (easily observable) retention of introns and also by two exon-skipping events, one of which (see red boxes in Fig. 4e) affected every third molecule for this gene. Note that the skipping of this exon per se is annotated in a short GENCODE transcript; however, our results indicate that this skipping can also occur in long transcripts that are not annotated. This long-distance information is observable only using long-read technologies.
We tested whether novel isoforms could stem from two nonidentical isoforms of the same gene in one well, which potentially (but not necessarily) could produce chimeric SLRs, which may in turn represent false-positive novel isoforms. We calculated an upper bound on the collision probability for each gene separately (Supplementary Results, section 6, and Supplementary Fig. 6), revealing 86% of all genes had at least twice as many novel spliced reads as predicted by the upper bound on the collision probability (and the assumption that no collisions were detected and removed; Supplementary Fig. 7).
Molecular co-association of distant human alternative exons
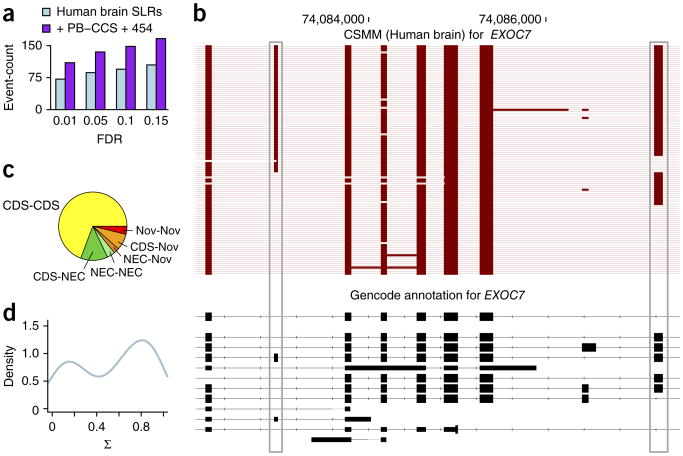
The length of SLRs and their large potential sequencing depth makes SLR-RNA-seq ideal to investigate the question of molecular co-association of distant alternative exons. We first determined alternative exons, with inclusion levels of at least 5% and at most 95%. For genes with multiple such alternative exons, we tested ∼36,000 pairs of distant exons for molecular co-association using a Fisher's exact test. More specifically, we counted the number of SLRs that included both alternative exons, the number that included only the first, the number that included only the second, as well as the number that included none. At an FDR, in the sense of Benjamini-Yekutieli36, of 0.05, we found a total of 86 different, significantly dependent exon pairs (FDR = 0.01: 71 exon pairs; FDR = 0.10: 94; FDR = 0.15: 104; Supplementary Data 13). Adding previously published PacBio-CCS18,22 and 454-reads21 increased these numbers to up to 165 (FDR = 0.15; Fig. 5a and Supplementary Data 14). These significant events included both distant dMAPs and dMEPs. The latter (dMEPs) share some similarity to mutually exclusive exons, but always harbor at least one exon in between the two alternative exons. Six dMAPs (occurring in BIN1, CAPN7, ABCD4, EXOC7, MAP4 and NRCAM) were independently validated, by sequencing PCR products from their genes using the PacBio platform (Supplementary Table 2 and Supplementary Results, section 7).
Figure 5.
Analysis of distant molecularly associated exon pairs in the human brain transcriptome. (a) Number of distinct distant alternative exon pairs (that is separated by at least one constitutive exon) at different FDR values. (b) All spliced reads (from the first four lanes) overlapping the two alternative exons (gray boxes) in EXOC7. CSMM, consensus split mapped molecule. (c) Pie chart of distant alternative exon pairs at FDR = 0.05, broken up by exon kind (CDS: the RNA-deduced exon is annotated as an entirely coding exon; NEC (not entirely coding): the RNA-deduced exon is annotated as an exon, but not as an entirely coding exon; Nov: the RNA-deduced exon has at least one novel splice site). (d) Density of score of intragenic molecular association (Σ) for distant alternative exon pairs at FDR = 0.05.
An example of a dMAP can be observed in the EXOC7 (Fig. 5b). Two distant alternative exons are separated by five almost constitutive exons. Their co-association is not perfect, as all four combinations of exon inclusion of the two alternative exons exist. However, 78% of the SLRs that spanned both alternative exons either included both exons or skipped both exons (P < 2 × 10−19 after Benjamini-Yekutieli correction). Notably, the fourth isoform (inclusion of alternative exon 1 and exclusion of alternative exon 2) is missing in the GENCODE annotation (Fig. 5b).
We then investigated the effects of dMAPs and dMEPs on their encoded proteins. For about two-thirds (54 of 78 pairs at FDR = 0.05, with uniquely defined outer splice sites), both exons were annotated as entirely protein coding (“CDS-CDS”; Fig. 5c), and in 59% of these cases, the summed length of the alternative exons was divisible by three. This suggests that many dMAPs produce molecularly phased proteins, for example, a protein isoform that had two additional peptide sequences that were distant in the amino acid chain, and another protein isoform that was missing both. In fewer cases, one or both of the alternative exons was annotated as an exon but not entirely coding (NEC; Fig. 5c) or even novel exons (“Nov”; Fig. 5c).
To quantify the molecular phasing for each exon pair, we defined the score of intragenic molecular association (Σ) as the ratio of the number of SLRs that included both exons or skipped both exons to the total number of SLRs for the exon pair. A Σ of 0 indicates that exactly one of two exons is included in each RNA molecule and a Σ of 1 indicates that only the “both-exons-included” and the “both-exons-skipped “isoforms can be observed. Of note, although related, this measure does not correspond directly to the Fisher's exact test P-value. For significantly associated exon pairs (at FDR = 0.05), Σ-values between 0.7 and 1.0 were most common, with at least one additional isoform usually observed at low level (Fig. 5d). A combination of the STAR-mapper37, Samtools38 and Cufflinks39 with the unassembled short-reads of SLR-RNA-seq performed reasonably well in predicting Σ-values, thus allowing a separation of dMAPs from dMEPs (Supplementary Fig. 8).
Conservation of molecular co-association in mouse brain
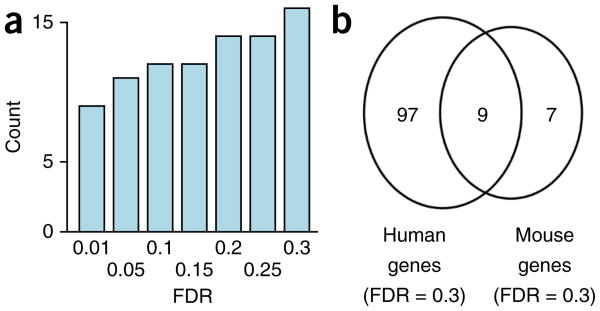
To investigate whether co-association of exons is evolutionarily conserved between mouse and humans, we also analyzed co-associated exons in mouse brain. We found 11 dMAPs and dMEPs at an FDR of 0.05 and 16 at an FDR of 0.3 (Fig. 6a and Supplementary Data 15). Nine of these genes (Abi2, Ank2, Bin1, Dnm1l, Dock9, Map4, Nfasc, Nrcam and Ppfia1) also appeared in the list of human dMAPs and dMEPs (at FDR = 0.3, two-sided Fisher's exact test P-value for overlap: 5.5 × 10−4; Fig. 6b); using liftOver40, we found exact splice-site positions to be conserved. For the remaining events, the sequencing depth employed did not allow us to judge conservation.
Figure 6.
Conservation of distant molecularly associated exon pairs between human and mouse. (a) Number of distant alternative exon pairs (that is, separated by at least one constitutive exon), which show nonrandom co-inclusion patterns, at different FDR values for the mouse brain. (b) Overlap between affected genes between human (at FDR = 0.3) and mouse (at FDR = 0.3).
Discussion
Alternative splicing, heterozygous SNVs, insertion-deletions and RNA editing may introduce multiple kinds of variation into RNA molecules. An ideal sequence-determining technology therefore would be able to (i) determine all residues of each molecule, including each varying residue; and (ii) do so for a sufficient number of molecules so that all potential dependencies between the different kinds of variations can be statistically tested. Short-read RNA-seq, which currently produces >200 million reads per sample, allows statistical comparison of many sources of variation between samples. However, its current read length (≤150 bp) cannot determine all alternative splicing events and other sources of variation along single RNA molecules. Recent advances using the 454 and the PacBio platforms enabled the recording of alternative splicing18,21,22 and SNVs18, but the achieved sequencing depth only rarely suffices to deduce quantitative results. SLR-RNA-seq achieves a strong improvement in read length (with an average read length of 1,907 for the human brain sample) in comparison to PacBio-CCS as well as a dramatic increase in sequencing depth. These improvements allowed us to determine a number of statistically linked pairs of alternative splicing events.
Alternative splicing shapes the flow of genetic information in the cell in multiple ways, including (but not limited to) the production of two different protein isoforms41, the downregulation of gene expression through alternative splicing-coupled nonsense-mediated decay42 and the retention of specific isoforms in the nucleus43. Thus, single alternative splicing events can have profound consequences for the fate of an RNA-molecule and the effect of n distant alternative splicing events in a gene (potentially encoding 2n possible isoforms) is even more difficult to forecast. Although pairs of alternative exons have been shown to correlate across tissues6, it has remained unclear whether (or which of) these instances stem from (i) two upregulated isoforms, both of which include one alternative exon but not the other or (ii) from one isoform that contains both exons and from another that skips both. The molecularly co-associated exon pairs that we determined here demonstrate that the second kind of co-regulation (“both-exons-included” and “both-exons-excluded”) exists. In our study, they are mostly pairs of protein coding exons, probably because for protein coding genes, we more often have the necessary sequencing depth to determine significance, due to the higher expression of protein coding genes in comparison to lncRNAs33.
The number of exon pairs for which we found molecularly co-associated splicing is so far relatively small and thousands of exon pairs give nonsignificant p-values. For many exon-pairs, too few read counts are available and for many other exon pairs, nonrandom pairing may simply not apply. For the moment it is difficult to draw any general conclusions as to how coordinated inclusion is achieved, but the following nonexclusive models seem possible. First, some cells (or cell types) may express the set of splicing regulators that exerts the combinatorial control for both exons44,45, whereas other cells (or cell types) may lack these splicing factors. Second, the two exons of a coordinated alternative exon pair may simply have similar enough sequence surroundings, which may force the splicing process to occur similarly at both exons. Third, the removal of an intron neighboring one of the exons may create a splicing enhancer or silencer that influences splicing at the other exon. Fourth, the RNA-binding of splicing factors close to one exon might favor the RNA-binding of splicing factors close to the second exon. Finally, while introns are thought to be removed mostly co-transcriptionally46–48, splicing at alternative exons appears to occur later46, especially in the case of exon-skipping49 – which would considerably reduce the distance (in nucleotides) between the two alternative exons at the time of splicing.
Distant alternative splicing events are usually connected either through complicated mathematical models39, whose accuracy for this purpose is unknown due to the lack of a gold standard, or by relying on single-molecule, long-read sequencing, which so far produces fewer reads than needed for thorough statistical analysis. We believe that the technology presented here will be key to the development of true isoform biology and help us further understand the complexity of the transcriptome, the means by which it is generated and its functional implications.
Online Methods
Sample preparation, RNA isolation and mRNA purification
Mouse brain tissue was obtained from 3-month-old C57BL/6 male mice, immediately frozen in liquid nitrogen and stored at −80 °C until use. Total RNA was extracted using TRIzol LS reagent (Life Technology, Cat No: 10296-028) according to the manufacturer's instructions. Extracted RNA was subjected to DNase I digestion to minimize genomic DNA contamination. DNase-treated human brain total RNA (Ambion First Choice Human Brain Reference RNA, Cat No: AM6050) was collected from multiple donors and several brain regions.
The FastTrack MAG mRNA Isolation kit (Life Technologies, Cat No: K1580-01) was used for polyA+ RNA selection from total RNA, following the manufacturer's protocol. The RNA integrity of all samples was assessed with the Agilent RNA 6000 Nano Assay kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Cat No: 50674626). ERCC ExFold RNA Spike-In Mix 1 (Life Technology, Cat No: 4456739) was used as an external RNA control.
First-strand cDNA synthesis
cDNA was generated from mRNA and Spike-In Mix 1 in independent reactions using a modified version of the Clontech SMART methodology developed internally at Illumina. Briefly, 500 ng of mRNA was used as the standard input amount for both mouse and human mRNA. ERCC Spike-In Mix 1 was diluted tenfold before use. Each input RNA was incubated with poly-dT oligos containing additional sequence specific to the TruSeq Synthetic Long-Read Library Prep method (Illumina, Cat No: 15047264) at 72 °C for 3 min and transferred immediately to ice. The Clontech SMARTScribe Reverse Transcriptase and 5X First-Strand Buffer combined with DTT, dNTPs and an oligo containing additional Illumina-specific sequences was then added to the input +RNA mixture, mixed by gentle pipetting, and incubated for 90 min at 42 °C and 10 min at 70 °C.
cDNA quantification and library preparation
Following cDNA synthesis, products were subjected to a SPRI bead clean-up and quantified by qPCR, as described in the Illumina TruSeq Synthetic Long-Read DNA Library Prep Guide (Illumina, Cat No: 15047264). Samples were quantified by comparison to a standard curve generated from a four-log serial dilution of a standard template (Illumina, Cat No: 15048791).
After quantification, an estimated 3 fg cDNA was seeded in each well of a 384-well plate. Long-range amplification was performed using Long-Amp Master Mix (Illumina, Cat No: 15046513) and Long-Amp Primer Mix (Illumina, Cat No: 15046508). PCR conditions were 60 s at 94 °C; 23 cycles of 30 s at 94 °C, 30 s at 65 °C, and 10 min at 68 °C; and 10 min at 68 °C. The products of four wells were removed and pooled for visual quantification against a twofold serial dilution of a 10 kb DNA fragment on a 1% eGel (Life Technologies, Cat No: G402001) to confirm adequate amplification of the cDNA product.
Each of the remaining 380 wells of amplified cDNA product was then fragmented and barcoded before pooling, size-selection and library validation as described in the Illumina TruSeq Synthetic Long-Read DNA Library Prep Guide (Illumina, Cat No: 15047264). Finally, each validated library product was diluted to 12 pM and clustered on one lane of a v3 HiSeq flowcell for sequencing. HiSeq read data was assembled into contigs upon completion of sequencing using the Illumina TruSeq Synthetic Long Reads analysis pipeline, as previously described27 in detail.
Mapping against ERCC RNAs and human and mouse genomes
GMAP-indices were built including all the regular chromosomes of the genome in question and 92 extra chromosomes corresponding to the ERCC-control RNA sequences (from which we removed the polyA tails).
Search for distant molecularly coordinated exon pairs
We first counted for each internal exon (from all mapped SLRs) the number of reads that included the exons and the number of reads that excluded them. The ratio for the former number to the sum of both numbers was used as the percent-spliced-in value (PSI). We then discarded exons:
That corresponded to retained introns with respect to the GENCODE v15 annotation
Whose PSI-value was greater than 0.95 or lower than 0.05
Which were the only one in their gene surviving the previous filters
We then considered all possible exon pairs within each gene and discarded exon pairs:
For which more than 2% of the reads extending into each exon, had no exon in between the alternative exon pair
Which had less than two reads extending into each of the two alternative exons
For the remaining exons we counted the number of reads that:
Used the donor of the first alternative exon and the acceptor of the second alternative exon along with an exon in between the two alternative exons
Used the donor of the first alternative exon and not the acceptor of the second alternative exon but an exon in between the two alternative exons
Did not use the donor of the first alternative exon but did use the acceptor of the second alternative exon along with an exon in between the two alternative exons
Did not use the donor of the first alternative exon and did not use the acceptor of the second alternative exon but an exon in between the two alternative exons
A 2 × 2 table was constructed based on the above four numbers and the table was submitted to a two-sided Fisher's exact test.
Molecule per million (MPM) calculation
For all annotated spliced genes we determined the number of spliced reads that could be attributed to them (by identity of at least one splice site21) and normalized by the number of overall uniquely mapped reads in millions.
Percent-isoform (Π) calculation
For all spliced genes, we determined all spliced reads that were classified18 (at least once) as full length. For each such isoform we divided its count by the sum of all such full-length isoforms for the gene in question.
Percent-spliced-in (Ψ) value calculation
For observed splice sites, we counted the ratio of all spliced reads that used this splice site to all reads (spliced at least once in the read) that overlapped the splice site.
Percent-intron-retention (PIR) calculation
For all annotated and observed introns we counted the number of reads (which had to be spliced at least once) that contained an exonic block, covering the entire intron as well as the number of spliced reads that used the exact intron. The ratio of the first to the sum of both was defined as the PIR. Note that for very long introns or those with very repetitive sequences, the PIR may be underestimated. Similarly when there is alternative splicing around the splice sites, readers may consider redefining the PIR according to their needs.
Read-mapping
Reads were mapped using GMAP30 as described previously21. The exact command line was gmap -D directoryWith-GMAPdatabase -d a_gmap__database_withGenome_and_ERCC_ afterPolyARemoval –min-intronlength = 25 -t 8 -f 3. This exports the result into gff3. For convenience for the reader we will also provide sam files (see “Accession codes”). Uniquely mapping reads were determined as described previously21.
Probability calculations
Please see Supplementary Figures 1–8, Figure 6 and Supplementary Results, section 6.
Supplementary Material
Acknowledgments
We thank N. Spies and F.A. Bava for a thorough reading of this manuscript and valuable comments and S. Shringarpure, V. Kuleshov, C.S. Foo and H. Tang for valuable comments on statistics. We thank A. Brunet for providing mice and S. Munro for valuable comments on this manuscript. We also thank the Genetics Bioinformatics Service Center at Stanford for providing a well-working computing cluster. M.R. is paid by grant 12-131829 from the Danish Council for Independent Research. This work was supported by grant 5U01HL10739304 (to M.S. as co-PI), 1P50HG007735-01 (to M.S. as co-PI) and 5P01GM09913004 (to M.S.).
Footnotes
Author Contributions: H.T., T.B., F.C. and M.P.S. devised the project. F.J., T.B., E.J., A.M. and M.R. carried out experiments. I.H. euthanized mice and extracted brains. H.T. carried out computational analysis. C.D.B. and M.P.S. supervised the project and provided financial support. H.T. wrote the first version of the manuscript. H.T., F.J., M.R. and M.P.S. wrote the final version of the manuscript with contributions from the other authors.
Accession codes. SRA: SRP049776 (human brain transcriptome: Ambion AM6050) and SRP050183 (mouse brain). Additional data will be made available at http://stanford.edu/∼htilgner/2014_humanMouseBrain_SLR_RNA_Seq/index_SLRseq.html.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Competing Financial Interests: The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Kornblihtt AR, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2014;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 4.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Dov C, Hartmann B, Lundgren J, Valcárcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- 6.Fagnani M, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 8.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 12.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 14.Modrek B, Resch a, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(suppl. 1):S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci USA. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho H, et al. High-resolution transcriptome analysis with long-read RNA sequencing. PLoS ONE. 2014;9:e108095. doi: 10.1371/journal.pone.0108095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilgner H, et al. Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G. 2013;33:387–397. doi: 10.1534/g3.112.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Au KF, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci USA. 2013;110:E4821–E4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 26.Kuleshov V, et al. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy RC, et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS ONE. 2014;9:e106689. doi: 10.1371/journal.pone.0106689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The External RNA Controls Consortium. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 29.Chinwalla AT, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 30.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 31.Li S, et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol. 2014;32:915–925. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 35.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 37.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng J, et al. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 42.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 43.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 45.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 46.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dujardin G, et al. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Vargas DY, et al. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.