Abstract
We investigated the increased risk of Clostridium difficile infection (CDI) caused by the combined use of antibiotics and an immunosuppressive drug in a mouse model. Our data showed that an approximate return to pretreatment conditions of gut microbiota occurred within days after cessation of the antibiotic treatment, whereas the recovery of gut microbiota was delayed with the combined treatment of antibiotics and dexamethasone, leading to an increased severity of CDI. An alteration of gut microbiota is a key player in CDI. Therefore, our data implied that immunosuppressive drugs can increase the risk of CDI through the delayed recovery of altered gut microbiota.
Keywords: Microbiota, Clostridium difficile infection, antibiotics, immunosuppressive drug, gut
Clostridium difficile is a gram-positive, anaerobic, spore-forming bacillus, and is one of the leading causes of healthcare-associated diarrhea. The incidence of C. difficile infection (CDI) has increased dramatically over the past decade, and the majority of CDI cases are attributed to risk factors, such as antibiotics, immunosuppressive drugs, and advanced age, which result in alteration of gut bacterial communities and their functions [9, 10]. Therefore, the essential step toward a better understanding of CDI pathogenesis is to investigate the effects of risk factors in CDI.
It is well known that the intestinal microflora plays important roles in the health and well being of the host. The normal intestinal microflora has a protective mechanism against the incursion of pathogenic bacteria [1, 2]. As such, the gut microflora is a key player in CDI, and disruptions of human gut microflora are closely related with CDI. Therefore, another intriguing question regarding microbial functions in CDI is which microbial groups are active players in protecting the host against CDI, and which are detrimental. This question arose as different antibiotics target different subpopulations of gut microflora, and disruptions of bacterial subpopulations revealed varying magnitudes of CDI risk [16]. The immunosuppressive effects of glucocorticoids, including dexamethasone, are known to increase the severity of CDI [5, 23, 24]. However, it has not been well studied why the immunosuppressive effects of these agents lead to severe CDI. Despite recent progress in the understanding of the complexities involved in microbial community structures and functions, there is a lack of information concerning the modulation of microbial populations in C. difficile-induced host microenvironments affected by antibiotics or immunosuppressive drugs. It would be of importance to study such interactions in a model that would mimic the human reactions to antibiotic and immunosuppressive therapies in conjunction with CDI infection.
To the best of our knowledge, this is the first study to evaluate the effect of immunosuppressive drugs on C. difficile infection in relation to alterations of the gastrointestinal microbiota. We found that treatment of mice with the immunosuppressive drug dexamethasone delayed the recovery of gut bacterial populations altered by antibiotics, resulting in increased severity of disease.
The animal protocol was approved by the Tufts University Institutional Animal Care and Use Committee. Animals were cared according to criteria described by Renggaman et al. [18]. Fifteen 6-week-old C57BL/c mice purchased from Jackson Laboratory (MA, USA) were randomly separated into three groups with five mice in each group: a control group (G1: no antibiotics), an antibiotics-only group (G2: antibiotics + clindamycin), and antibiotics with dexamethasone mixture group (G3: antibiotics + clindamycin + dexamethasone). The epidemic strain C. difficile UK6 isolated in the United Kingdom was kindly provided by Dale Gerding and Abraham L. Sonenshein [11]. Sporulation of the C. difficile UK6 was induced as previously described [17, 21, 25]. Briefly, 20 ml of a C. difficile UK6 overnight culture in Columbia Broth was inoculated to 500 ml of Clospore medium, and incubated for 2 weeks at 37°C in an anaerobic incubator. After 2 weeks of incubation, the spore suspension was centrifuged at 1,000 ×g for 10 min. Then, the pellet was washed five times with sterile water and resuspended in 10 ml of ddH2O. In order to kill vegetative cells, the spore suspension was heated at 60°C for 20 min. The spore concentration was measured by 10× serial dilution on TCCFA plates [17, 21, 25]. The control group was treated with neither antibiotic solution nor dexamethasone before the spore inoculation with 1 × 106 spores of C. difficile UK6. For the antibiotics-only group, the antibiotic cocktail (kanamycin, gentamicin, colistin, metronidazole, and vancomycin) and clindamycin were used as previously described [4, 22]. Then the mice were challenged orally with 1 × 106 spores of C. difficile UK6. Mice in the antibiotics with dexamethasone mixture treatment group received dexamethasone in the drinking water (100 mg/l) along with the same treatment as in the antibiotics-only group. Then the mice were challenged with 1 × 106 spores of C. difficile UK6. The weight of mice and progression of the disease were monitored after the C. difficile challenge.
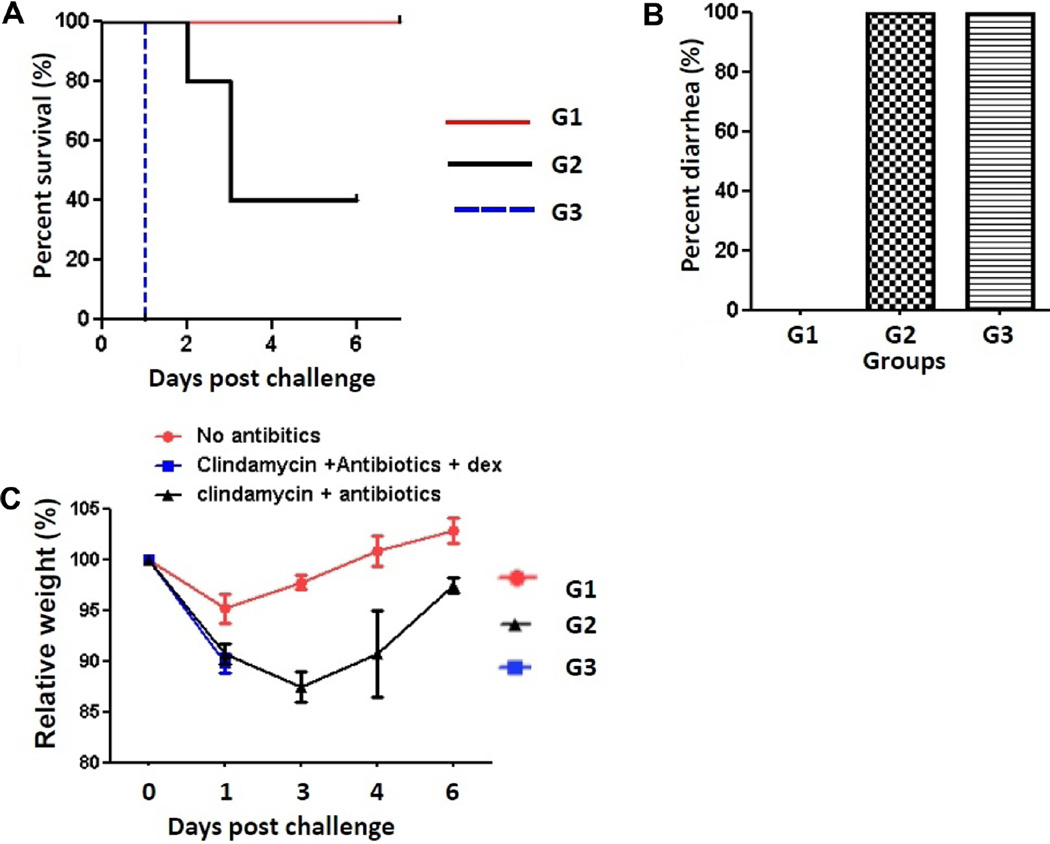
Whereas all mice treated with either the antibiotics only (G2) or the antibiotics with dexamethasone mixture (G3) developed diarrhea (Fig. 1B), the mortality rate of the antibiotics-only group was lower than that of the antibiotics with dexamethasone mixture group (Fig. 1A). All mice in the antibiotics with dexamethasone mixture treatment group (G3) followed by a challenge with 1 × 106 spores of C. difficile UK6 became moribund after 1 day post inoculation (DPI). On the other hand, 40% of the mice in the antibiotics-only group (G2) survived until the end of the experiment, and all mice in the control group (G1) also survived (Fig. 1A). The mean relative weight of the mice declined sharply within 2–3 DPI. However, survived mice began to gain weight by 3–4 DPI, and returned to their normal weight by 6 DPI (Fig. 1C). Overall, our data indicate that the immunosuppressive drug dexamethasone increased the severity of CDI (Fig. 1).
Fig. 1. Clinical presentation of mice in each group.
Data on survival (A), diarrhea (B), and weight loss (C) of each group of mice are shown. Mice in group 1 (G1) were not treated with anything. Mice in groups 2 and 3 (G2 and G3) were treated with antibiotics or antibiotics with dexamethasone mixture for 5 days, respectively. Then the mice were challenged with 1 × 106 spores of C. difficile UK6. The weight of mice and progression of the disease were monitored after the C. difficile challenge. G1: No antibiotics. G2: Clindamycin+antibiotics. G3: Clindamycin+antibiotics+dexamethasone.
For the fecal microbiome analysis, the fecal samples of mice in different groups were collected before the antibiotic treatment, after the antibiotic treatment, and before the UK6 challenge.
DNA extraction and barcoded library preparation from the mice in each group were conducted as previously described with one modification [14, 26]. The modification was to use the universal primer set 27F and 338R to amplify the V1–V2 hypervariable regions of the 16S rRNA genes. The pooled barcoded libraries were sequenced in a HiSeq2000 Illumina sequencer at the Tufts Genomics Core Facility (Tufts University, Boston, USA). Quality control analysis was conducted as previously described to obtain only high-quality sequences for the analysis [12, 13]. In order to identify chimeras, UCHIME was used. Phylogenetic assessments were performed using the RDP classifier implemented in QIIME with a bootstrap cutoff of 80%, and diversity indices were calculated with an operational taxonomic unit definition at an identify cutoff of 97% [3, 8].
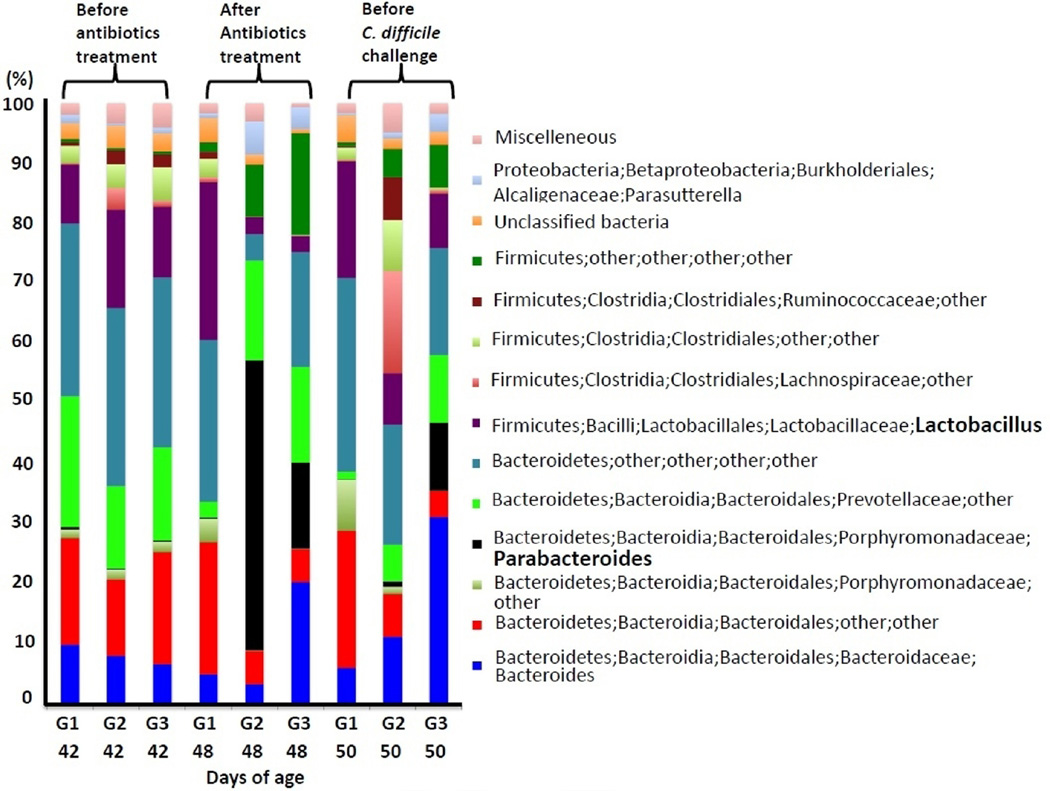
The average numbers of sequence reads generated per group were 144,649 after the quality control implemented in this study. The composition of the fecal microbiome altered by antibiotics (G2) or antibiotics with dexamethasone treatment (G3) was shown at the genus level (Fig. 2). Perturbation of gut microbial bacteria belonging to the genus Lactobacillus and Parabacteroides was prominent. Gram-positive lactobaccilli are members of the lactic acid bacteria, and are considered to be beneficial for the hosts [19]. Therefore, it will be of importance to evaluate how reduction of the Lactobacillus population in the gut affects the pathogenesis of CDI. Interestingly, antibiotics (G2) or combined treatments of antibiotics and dexamethasone (G3) markedly decreased the concentrations of Lactobacillus (Fig. 2). Meanwhile, proportions of bacterial members belonging to Parabacteroides increased concurrently with the reduction of Lactobacillus after antibiotics (G2) or combined treatments of antibiotics and dexamethasone (G3) (Fig. 2). Parabacteroides has been isolated from hospitalized patients with Crohn’s disease and those with organ transplantation [6]. However, information about the pathogenesis of Parabacteroides in patients is still lacking. An approximate return to pretreatment conditions also occurred in this study within days after cessation of treatments, as described by others using different antibiotics [7, 15]. The proportion of Lactobacillus increased while those of Parabacteroides decreased days after cessation of the treatments (Fig. 2).
Fig. 2. Taxonomic classification of the sequences at the genus level.
Phylogenetic assessments were performed using the RDP classifier implemented in QIIME with a bootstrap cutoff of 80%. Phylum, class, order, family, and genus are presented in each legend. “Other” represents the unclassified taxon at each level of classification. The antibiotics treatments were continued for 5 days between 44 and 48 days of age. G1: No antibiotics. G2: Clindamycin+antibiotics. G3: Clindamycin + antibiotics + dexamethasone.
Diversity indices were used to calculate the diversity of the microbial communities [20]. These values decreased concurrently with the antibiotics or combined treatments of antibiotics and dexamethasone (Table 1). Patterns of gut bacterial alterations between antibiotics-only (G2) and antibiotics with dexamethasone treatment (G3) groups were similar to each other; however, combined treatments of antibiotics and dexamethasone (G3) delayed the recovery of gut bacterial populations compared with antibiotics-only treatment (G2). The diversity indices increased to approximate pretreatment conditions within days after cessation of the treatment; however, it was faster in the antibiotics-only group (G2) than the combined treatment group with antibiotics and dexamethasone (G3) (Table 1). Therefore, we speculated that the immunosuppressive drug dexamethasone delayed the recovery of gut bacterial populations altered by antibiotics, resulting in increased severity of disease. Effects of immunosuppressive drugs, including dexamethasone, should be investigated further to elucidate their potential roles in the pathogenesis of CDI.
Table 1.
Diversity indices of the 16S rRNA gene sequence reads of each group.
| Group | Days of age | Chao1 | ACE | No. of OTUs | Shannon | Simpson |
|---|---|---|---|---|---|---|
| G1 | 42 | 280 | 280 | 280 | 4.83 | 0.94 |
| G2 | 42 | 400 | 400 | 400 | 5.15 | 0.94 |
| G3 | 42 | 381 | 381 | 381 | 5.06 | 0.94 |
| G1 | 48 | 388 | 388 | 388 | 4.61 | 0.91 |
| G2 | 48 | 84 | 84 | 84 | 3.17 | 0.76 |
| G3 | 48 | 99 | 99 | 99 | 4.11 | 0.92 |
| G1 | 50 | 265 | 265 | 265 | 4.43 | 0.92 |
| G2 | 50 | 297 | 297 | 297 | 5.59 | 0.96 |
| G3 | 50 | 140 | 140 | 140 | 4.38 | 0.93 |
Diversity indices were calculated with an operational taxonomic unit definition at an identify cutoff of 97%. The antibiotics treatments were continued for 5 days between 44 and 48 days of age.
G1: No antibiotics.
G2: Clindamycin+antibiotics.
G3: Clindamycin+antibiotics+dexamethasone.
42 days of age: Before antibiotics treatment.
48 days of age: After antibiotics treatment.
50 days of age: Before C. difficile challenge.
This study was designed to evaluate if treatment with the immunosuppressive drug dexamethasone alters gut bacterial populations resulting in symptomatic CDI or increased severity of disease. In summary, an analysis of the mouse gut microbiome showed that it had shifted after antibiotics or combined treatment with antibiotics and dexamethasone. Moreover, our data suggested that the dexamethasone treatment delayed the recovery of gut bacterial populations altered by antibiotics, resulting in increased severity of disease.
The sequences used in this paper are publicly available at Galaxy, https://usegalaxy.org/u/kim-et-al/h/unnamed-history-1.
Acknowledgments
The present research was supported by NIH R21 AI113470 (X. Sun), Startup fund from the University of South Florida (X. Sun), Tufts Collaborates in 2013 V330421 (X. Sun), NRF-2015R1D1A1A01061268 from the Ministry of Education, Republic of Korea (H.B. Kim), and the research fund of Dankook University (BK21 Plus) in 2014 (H.B. Kim).
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 3.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Das R, Feuerstadt P, Brandt LJ. Glucocorticoids are associated with increased risk of short-term mortality in hospitalized patients with Clostridium difficile-associated disease. Am. J. Gastroenterol. 2010;105:2040–2049. doi: 10.1038/ajg.2010.142. [DOI] [PubMed] [Google Scholar]
- 6.De Cruz P, Kang S, Wagner J, Buckley M, Sim WH, Prideaux L, et al. Association between specific mucosa-associated microbiota in Crohn's disease at the time of resection and subsequent disease recurrence: a pilot study. J. Gastroenterol. Hepatol. 2015;30:268–278. doi: 10.1111/jgh.12694. [DOI] [PubMed] [Google Scholar]
- 7.De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J. Clin. Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachrimanidou M, Malisiovas N. Clostridium difficile infection: a comprehensive review. Crit. Rev. Microbiol. 2011;37:178–187. doi: 10.3109/1040841X.2011.556598. [DOI] [PubMed] [Google Scholar]
- 10.Kelly CP. Current strategies for management of initial Clostridium difficile infection. J. Hosp. Med. 2012;7:S5–S10. doi: 10.1002/jhm.1909. [DOI] [PubMed] [Google Scholar]
- 11.Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 2008;46:431–437. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, et al. Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. USA. 2012;109:15485–15490. doi: 10.1073/pnas.1205147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HB, Isaacson RE. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015;177:242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Kim HB, Zhang Q, Sun X, Beamer G, Wang Y, Tzipori S. Beneficial effect of oral tigecycline treatment on Clostridium difficile infection in gnotobiotic piglets. Antimicrob. Agents Chemother. 2014;58:7560–7564. doi: 10.1128/AAC.03447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lode H, Von der Hoh N, Ziege S, Borner K, Nord CE. Ecological effects of linezolid versus amoxicillin/clavulanic acid on the normal intestinal microflora. Scand. J. Infect. Dis. 2001;33:899–903. doi: 10.1080/00365540110076714. [DOI] [PubMed] [Google Scholar]
- 16.Owens RC, Jr, Donskey CJ, Gaynes RP, Loo VG, Muto CA. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin. Infect. Dis. 2008;46:S19–S31. doi: 10.1086/521859. [DOI] [PubMed] [Google Scholar]
- 17.Perez J, Springthorpe VS, Sattar SA. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J. AOAC Int. 2011;94:618–626. [PubMed] [Google Scholar]
- 18.Renggaman A, Choi HL, Sudiarto SI, Alasaarela L, Nam OS. Development of pig welfare assessment protocol integrating animal-, environment-, and management-based measures. J. Anim. Sci. Technol. 2015;57:1. doi: 10.1186/s40781-014-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riboulet-Bisson E, Sturme MH, Jeffery IB, O’Donnell MM, Neville BA, Forde BM, et al. Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS One. 2012;7:e31113. doi: 10.1371/journal.pone.0031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele J, Feng H, Parry N, Tzipori S. Piglet models of acute or chronic Clostridium difficile illness. J. Infect. Dis. 2010;201:428–434. doi: 10.1086/649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuck AE, Minder CE, Frey FJ. Risk of infectious complications in patients taking glucocorticosteroids. Rev. Infect. Dis. 1989;11:954–963. doi: 10.1093/clinids/11.6.954. [DOI] [PubMed] [Google Scholar]
- 24.Sun X, Wang H, Zhang Y, Chen K, Davis B, Feng H. Mouse relapse model of Clostridium difficile infection. Infect. Immun. 2011;79:2856–2864. doi: 10.1128/IAI.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YK, Yan YX, Kim HB, Ju X, Zhao S, Zhang K, et al. A chimeric protein comprising the glucosyltransferase and cysteine proteinase domains of toxin B and the receptor binding domain of toxin A induces protective immunity against Clostridium difficile infection in mice and hamsters. Hum. Vaccin. Immunother. 2015;11:2215–2222. doi: 10.1080/21645515.2015.1052352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Widmer G, Tzipori S. A pig model of the human gastrointestinal tract. Gut Microbes. 2013;4:193–200. doi: 10.4161/gmic.23867. [DOI] [PMC free article] [PubMed] [Google Scholar]