Conspectus
The catalytic diversity of living systems offers a broad range of opportunities for developing new methods to produce small molecule targets such as fuels, materials, and pharmaceuticals. In addition to providing cost-effective and renewable methods for large-scale commercial processes, the exploration of the unusual chemical phenotypes found in living organisms can also enable the expansion of chemical space for discovery of novel function by combining orthogonal attributes from both synthetic and biological chemistry. In this context, we have focused on the development of new fluorine chemistry using synthetic biology approaches. While fluorine has become an important feature in compounds of synthetic origin, the scope of biological fluorine chemistry in living systems is limited, with fewer than 20 organofluorine natural products identified to date. In order to expand the diversity of biosynthetically accessible organofluorines, we have begun to develop methods for the site-selective introduction of fluorine into complex natural products by engineering biosynthetic machinery to incorporate fluorinated building blocks. To gain insight into how both enzyme active sites and metabolic pathways can be evolved to manage and select for fluorinated compounds, we have studied one of the only characterized natural hosts for organofluorine biosynthesis, the soil microbe Streptomyces cattleya. This information provides a template for designing engineered organofluorine enzymes, pathways, and hosts and has allowed us to initiate construction of enzymatic and cellular pathways for the production of fluorinated polyketides.
Introduction
The remarkable facility of living systems for catalysis allows them to constantly evolve new ways to interface with their environment. Beyond the thousands of shared reactions of primary metabolism, cells can also tap into a large set of specialized transformations that enable a wide range of chemical phenotypes, such as the ability to fix CO2, break down recalcitrant plant biomass, or synthesize complex molecules. As such, cellular reaction chemistry offers unique opportunities to develop alternative approaches towards chemical synthesis by rewiring these metabolic networks in new ways for production of small molecule targets rather than cell biomass.1,2 However, the logic of cellular chemistry is quite different from synthetic chemistry as it depends on tight coupling between reactions to create metabolic pathways, which cannot always easily be disconnected from each other and recombined in the same way that independent synthetic reactions can. In this regard, our ability to construct new pathways and transplant chemistry from one host to another relies on understanding both the chemistry and biology of these systems. Towards this goal, we seek to study and elucidate the function of unusual chemical phenotypes that allow us to create synthetic connections between pathways in order to produce pharmaceuticals, fuels, and materials of interest.
A particularly interesting interface between biological and synthetic chemistry exists between natural products and organofluorines, which have each been important to the development of small-molecule therapeutics but rarely intersect in structural space. Fluorine’s small size and high electronegativity confer powerful effects on small molecule properties, and drug fluorination can improve bioavailability, reduce clearance, alter pKa and lipophilicity, block metabolism, or increase potency.3,4 As a result, fluorine is now found in 20-30% of therapeutics and agrochemicals and methods for generating diverse organofluorine structures continue to serve a growing number of applications.5 Despite these advantages, fluorine is rarely observed in naturally-occurring compounds. Using synthetic biology approaches to incorporate fluorine into natural product biosynthesis would combine some of the strengths of organic and biological chemistry and expand the structural space of natural products, which have provided the source for many human therapeutics (Figure 1). Towards this goal, we have studied the fluorine metabolism of Streptomyces cattleya,6 one of the few genetic hosts of fluorine metabolism, to elucidate the molecular basis for fluorine-selective enzymes and organisms in order to design and develop new fluorine biosynthetic pathways.
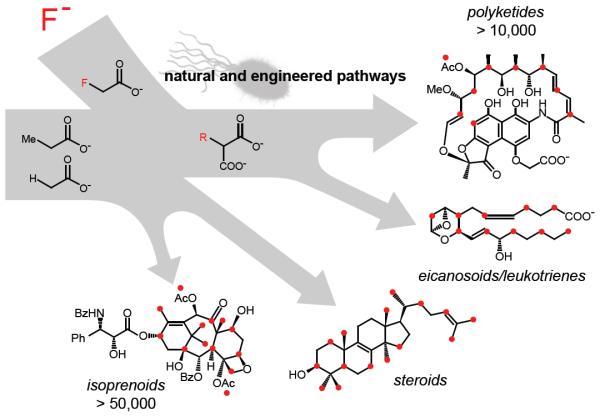
Figure 1.
Living systems offer the opportunity to mix natural and engineered pathways to produce new fluorinated compounds. Positions that could be fluorine-substituted are shown with red dots. R = fluorine or naturally occurring extender unit substituents.
Properties of organofluorines relevant to their biochemistry
Fluorine has been a useful design element not only for medicinal chemistry3,4,7 but also for enzymology8-10 and metabolism.11,12 Consequently, the physical organic and biochemical properties of synthetic organofluorines have been well studied and extensively reviewed.3,4,7-9,11,13-15 Here, we briefly summarize this work as it provides a foundation for understanding the behavior of fluorine with respect to our current efforts in the design of fluorine-specific active sites and pathways.
Fluorine is considered a sterically conservative substitution for hydrogen or oxygen; the C–F unit is similar in size to C=O.7,14 Since C–F falls between C–H and C–OH or C–CH3 in size, many enzymes that accept any of these substituents can accommodate fluorine as well. However, fluorine has very different properties with respect to electronegativitiy, polarizability, bond-strength, dipolar interactions, solvation, and conformation that can alter molecular recognition in protein binding sites.
A signature property of fluorine is its high electronegativity (4.0 on the Pauling scale). As a result, the C–F bond is extremely strong, largely because of its substantial ionic character (C∂+–F∂−).15 Fluorine substituents are therefore fairly inert, with substitution reactions uncommon and elimination requiring an E1CB mechanism with full carbanion formation.15 Fluorine substitution has also strong inductive effects on neighboring functional groups, activating electrophiles and increasing acidity. However, α-fluoro carbonyl compounds are less acidic than would be expected because of hybridization effects.7 While a fluoroenolate is stabilized by induction, it is concomitantly destabilized because fluorine prefers to be bonded to an sp3 rather than an sp2 carbon due to unfavorable overlap between the filled p orbitals on F and C-sp2 and reduction in σ-electron density that can be polarized toward F.7,15 A methyl group has the opposite hybridization preference, so these two substituents can confer similar acidity. By contrast, simple inductive effects make fluoroacetic acid over 2 pKa units more acidic than propionic or acetic acid.
Organofluorines are primarily stabilized by dipolar electrostatic interactions.14,15 The low polarizability of fluorine causes induced dipole effects to be weak and the C–F unit to be a poor hydrogen bond acceptor (half as strong as C–OH at most).7,15 However, the C–F dipole does interact favorably with centers of positive charge, as with the “fluorophilic” Arg side-chain, where the C–F bond adopts either a perpendicular or an out-of-plane parallel orientation to the plane of the guanidinium group, rather than the in-plane linear hydrogen bonding geometry.14 It is also favorable to orient the C–F dipole toward the carbonyl carbon of a backbone amide or Asn/Gln side-chain16 or toward the peptide Cα–H bond.
Because the C–F dipole can only form strong, favorable interactions within significant orientational constraints, fluorine substituents can be used to achieve target selectivity by formation of enthalpically favorable C–F dipole arrangements.14 Moreover, entropic effects are also important as solvation of the C–F unit is unfavorable, given its poor hydrogen-bonding ability and low polarizability.13 Since C–F is a poor replacement for H2O, only a few arrangements of water molecules will have their dipoles properly oriented to interact favorably with C–F, leading to solvent ordering. As a result, when the C–F unit is desolvated upon protein binding the release of ordered water provides an entropic driving force even in the absence of specific C–F dipolar interactions in the binding site. A very polar C–F unit can therefore bind favorably in a nonpolar pocket, leading to the description of organofluorines as “polar hydrophobic.”13
Studying fluorine selectivity in native enzymes: FlK as a model
A key part of redesigning enzyme active sites for fluorinated substrates includes understanding how organofluorines interact with their protein binding partners. While the design and optimization of synthetic organofluorine compounds for binding to macromolecular targets has yielded much information in this regard, we would like to explore the converse process of how an enzyme active site can be evolved to select for organofluorine substrates. However, there are fewer than 20 known organofluorine metabolites,17 with only a few enzymes identified that specialize on organofluorine substrates. The first organofluorine-selective enzymes to be characterized were the fluoroacetate dehalogenases, but they cleave the C–F bond without exhibiting highly selective C–F recognition.18 To find enzymes that recognize and also preserve the C–F unit, we turned our attention to S. cattleya, a soil microbe that biosynthesizes fluoroacetate and fluorothreonine (Figure 2).19,20
Figure 2.
The organofluorine biosynthetic pathway from S. cattleya and conversion of fluoroacetate into fluorocitrate.
The fluoroacetyl-CoA thioesterase from S. cattleya (FlK)
Fluoroacetate is a potent toxin that depends on the “lethal synthesis” of fluorocitrate, which inhibits the tricarboxylic acid (TCA) cycle via mechanism-based inhibition of aconitase (Figure 2).21 This process is initiated by activation of fluoroacetate to fluoroacetyl coenzyme A (CoA), the monofluorinated analog of the ubiquitous cellular building block, acetyl-CoA. A thioesterase (FlK) was identified within the gene cluster encoding fluorometabolite biosynthesis in S. cattleya and proposed to catalyze the specific hydrolysis of fluoroacetyl-CoA to reverse this activation and guard S. cattleya against self-poisoning.22-24 Interestingly, this hydrolysis is formally an acyl transfer reaction (to water), which is the same reaction class involved in the selection of acyl-CoA substrates by enzymes involved in polyketide and isoprenoid biosynthesis, which accept an acyl-CoA substrate by transacylation to form a covalent enzyme intermediate. Thus, we set out to characterize FlK in detail to investigate the molecular details of fluorine selectivity in acyl transfer.
The function of FlK in clearing of low levels of toxic fluoroacetyl-CoA (R = F) while maintaining integrity of the substantial (mM) acetyl-CoA pool (R = H) requires both specificity for fluorine and discrimination against hydrogen at the α-position. In vivo experiments in Escherichia coli indicated that expression of FlK can protect against fluoroacetate poisoning without causing the growth defect that would be expected if the acetyl-CoA pool were also hydrolyzed.23 In vitro characterization of FlK revealed a surprisingly high 106-fold selectivity based on the single fluorine substitution,23 in contrast to other members of the hot dog fold thioesterase superfamily which tend to exhibit substrate promiscuity.25,26
Structural studies of FlK revealed that it shares the catalytic cluster of other hot-dog thioesterases in the active site (Thr42, His76, and Glu50) as well as their overall fold.23,27 However, it also contains a unique hydrophobic lid structure, including Phe33, Phe36, Val23, Leu26 and Val39, which may desolvate the C–F unit, freeing ordered water to make binding more entropically favorable (Figure 3A). Of these, Phe36 appears to be especially important and may serve as a gate that excludes water from the active site.23 Within this hydrophobic environment, the C–F dipole interacts electrostatically with Arg120 (with the C–F bond oriented perpendicularly to the guanidinium group) and the Gly69 backbone amide.27
Figure 3.
Enzymatic fluorine selectivity in the fluoracetyl-CoA thioesterase (FlK). (A) The FlK active site includes fluorine-specific interactions and catalysis elements that result in 106-selectivity for a fluorinated substrate over the hydrogen analog. (B) Divergent mechanisms for the fluorinated and non-fluorinated substrate occur within the same active site.
The origin of the 106-fold discrimination between F and H mainly resides in the increased rate of hydrolysis (kcat) of the fluorinated substrate (104-fold) rather than differences in KM (102-fold).23 This rate enhancement is higher than the intrinsic activation conferred by the fluorine substituent, which is only 10-fold for uncatalyzed thioester hydrolysis at neutral pH, suggesting that additional mechanisms of rate acceleration within the FlK active site contribute an additional factor of 103. The rate of acetyl-CoA hydrolysis by FlK is especially slow compared to related thioesterases, implying a catalytic deficiency with unactivated substrates. Notably, FlK provides only a single hydrogen bond to activate the thioester carbonyl,25,27 which may result in weak oxyanion hole catalysis compared to related hydrolases (Figure 3A).
A combination of mutagenesis, Hammett analysis, pre-steady state kinetic analysis, kinetic isotope studies, and trapping experiments revealed that fluorine selectivity in FlK relies on the existence of two different mechanisms of thioester hydrolysis within the same active site (Figure 3B).22 For both substrates, a common covalent acyl-enzyme intermediate is generated on Glu50, with a rate constant that depends on the inductive polarization of the substrate carbonyl.24 However, the next step diverges depending on the substrate. The acetyl acyl-enzyme intermediate undergoes slow hydrolysis by attack of water on the carbonyl moiety through a canonical thioesterase mechanism. In contrast, the fluoroacyl-enzyme intermediate can access an accelerated Cα-deprotonation pathway because of its lower pKa, using His76 as a base in a mechanism analogous to the MaoC dehydratases.28,29 Based on chemical model systems,30,31 the fluoroenolate is thought to break down via a transient ketene intermediate, which could rapidly hydrate to form fluoroacetate.
FlK provides a design template for engineering fluorine selectivity in an acyltransferase active site. In this case, most of the discrimination between F and H is provided not by molecular recognition of fluorine but encoded instead by the reactivity of the substrate itself where the presence of a single fluorine atom opens up a new pathway for catalysis. The active site is tuned to enhance reactivity with the more activated substrate while being deactivated for a canonical reaction pathway with the unactivated substrate. The stereospecific deprotonation of the fluoroacyl intermediate suggests that FlK may interact directly with fluorine.24 However, preliminary studies indicate that the difference in KD between fluorinated and nonfluorinated substrates is only approximately 10-fold. This approach to fluorine selectivity allows FlK to distinguish low levels of the fluorinated substrate over high concentrations of the endogenous hydrogen analog, a situation that might be encountered in an engineered system.
Other enzymes involved in organofluorine metabolism
Additional enzymes participating in organofluorine management in native producers may also be candidates for studying interaction between evolved enzyme-organofluorine pairs. For example, the fluorinase (FlA),32,33 5′-fluorodeoxyadenosine phosphorylase (FlB), and threonine transaldolase appear to be dedicated to fluorine metabolism but do not necessarily distinguish F from H (Figure 2).34 In addition, the fluoroacetaldehyde dehydrogenase does prefer fluoroacetaldehyde over acetaldehyde, although not over other substrates.35 Furthermore, the growing availability of new microbial genome sequences may provide additional orthologs of known genes or even enzymes from novel organofluorine biosynthetic pathways.36
Elucidating fluorine physiology in native organofluorine-producing hosts
Studying native organofluorine producers allows us to explore how organofluorines and their potential crosstalk with the rest of their host metabolism can be handled in vivo. Information from these studies will facilitate understanding of molecular mechanisms of organofluorine resistance and provide a potential template for design of engineered hosts that can maintain orthogonal pools of fluorinated and non-fluorinated building blocks for downstream biosynthetic pathways.
S. cattleya
Despite the observation of fluoroacetate production in plants, S. cattleya was the only genetic host known to specifically utilize fluorine as a key element in its life cycle and we sought to explore how it manages organofluorine toxicity. While the FlK thioesterase provides a clear fluoroacetate resistance mechanism, knockout studies showed that the gene encoding FlK is not strictly required for survival of S. cattleya during fluoroacetate biosynthesis and that other modes of resistance likely exist.37 One possible alternative is that the enzymes involved in the lethal synthesis of fluorocitrate or the aconitase itself exclude fluorinated substrates to a higher level than those from fluoroacetate-sensitive hosts (Figure 2). If true, these enzymes could be used to engineer fluoroacetate resistance in a synthetic host while maintaining a pool of fluoroacetyl-CoA. However, we found that the enzymes from S. cattleya do not exhibit any significant difference in behavior with fluorinated substrates in terms of kcat or KM compared to orthologs from a related fluoroacetate-sensitive species, Streptomcyes coelicolor, or the fluoroacetate-sensitive model bacterium, E. coli.37
In contrast, we found that organofluorine toxicity was mediated by transcriptional coordination where biosynthesis is activated upon the onset of stationary phase when the TCA cycle is downregulated. Interestingly, we observed changes in the global transcriptional landscape in response to fluoride, indicating that S. cattleya senses either fluoride or an organofluorine to regulate transcription and that organofluorine biosynthesis plays an important role in its physiology. We also observed that S. cattleya does not appear to support use of the glyoxylate shunt pathway, which may protect against fluoroacetate activation and minimize its incorporation into other cellular metabolites. Overall, these observations suggest that organofluorines are difficult to exclude from acetate metabolism and that simultaneous use of fluoroacetate and the TCA cycle should be avoided.
Discovery of new organofluorine hosts
The rapidly escalating effort to sequence and mine microbial diversity has provided new information about distribution of organofluorine biosynthetic pathways. Microbes other than S. cattleya have since been found to contain genes encoding fluorinase (FlA) orthologs,38 and some also produce new and unidentified organofluorines.39 Interestingly, the different organization of fl gene clusters compared to S. cattleya, where only a few biosynthetic genes are found adjacent to the flA gene, may help to further understand fluoroacetate and fluorothreonine biosynthesis.38 In addition, other organisms may reveal completely distinct organofluorine biosynthesis pathways. For example, the production of the 4′-fluoronucleoside antibiotic nucleocidin by Streptomyces calvus may indicate that C–F bond formation is different in this system.36 These few examples indicate that the scope of organofluorine diversity is not limited to fluoroacetate and fluorothreonine pathways and may serve as a rich area for discovery.
Engineering organofluorine biochemistry
Synthetic biology approaches provide a framework for bringing new diversity to molecular structure by mixing chemistry from living systems with that from synthetic methodology. In this regard, the intersection of natural products and organofluorine compounds is an especially interesting one to explore as they are each very important in medicinal chemistry but their overlap has been underexplored for practical reasons. A mechanism for expanding this scope is to engineer the incorporation of fluorinated building blocks using natural product biosynthetic machinery to generate complex fluorinated compounds, such as polyketides, isoprenoids, alkaloids, and ribosomal or non-ribosomal peptides (Figure 1). Indeed, previous work has shown that fluorinated building blocks can be accommodated by some of these pathways,17 while semisynthesis of fluorinated natural product analogs has shown that a fluorine substituent can improve pharmaceutical function.40 An advantage of this approach is that it would enable the production of value-added fluorinated products by microbial fermentation, either by endogenously produced or exogenously provided fluorinated building blocks in the form of fluoride or relatively inexpensive organofluorines, respectively. Towards this goal, we have focused on studying the uptake and activation of fluorinated building blocks by cells and enzymes as well as the engineering of pathways for site-selective incorporation of fluorinated building blocks into natural products in vitro and in vivo.
Fluorine and polyketides
We chose to begin by investigating the addition of the fluorine substituent to polyketide biosynthesis using fluoroacetate as a monomer. Polyketides represent an expansive family of over 10,000 diverse structures with a broad range of applications in human health, including antibiotic, anti-cancer, anti-inflammatory, and immune suppression.41 Given this diversity, it is surprising that polyketide backbones are largely derived from just a few monomers, most commonly acetate and propionate. The starter unit may be more chemically diverse, and a few polyketides incorporate a specialized alternative extender unit at a particular position, making it possible to replace these starters and specialized extenders with non-native analogs, including fluorinated species.17 Indeed, fluoroacetate has been successfully used as a starter unit for the minimal actinorhodin polyketide synthase (PKS), supporting formation of a full-length polyketide.42 However, modular polyketide synthases maintain much stricter substrate selectivity at the acetate- and propionate-derived extender unit positions that form the bulk of the polyketide backbone, limiting the diversity of accessible analogs.
We therefore set out to develop methods for the site-selective introduction of fluoroacetate in place of acetate or propionate, providing a new monomer that could greatly expand the structural diversity of this family of natural products. In this regard, fluoroacetate is a particularly attractive building block because it can be produced using the S. cattleya pathway, enabling total biosynthesis from fluoride. We have focused our initial efforts on the type I modular PKS family, which coordinates the processive biosynthesis of a covalently-tethered polyketide using a modular assembly-line organization (Figure 4).41 In these systems, each module typically encodes the information for each chain elongation step and thus provides an internal design for regioselective fluorine introduction.
Figure 4.
Mechanistic steps in a single chain extension cycle catalyzed by a modular PKS (light grey, upstream module; light blue, active module; red, downstream module).
Reactivity of fluorinated building blocks
The presence of a fluorine substituent at the α-position creates unique challenges by altering the chemistry and reactivity of monomer intermediates and the growing polyketide chain itself (Figure 5). Indeed, it is interesting to note that fluoroacetate is only found in nature to be incorporated into fatty acids at the starter position via fluoroacetyl-CoA6 rather than at any chain extension positions via fluoromalonyl-CoA (Figure 2). Fluorine activates pathway intermediates as electrophiles, which makes CoA thioesters and thioester-tethered biosynthetic intermediates more labile to hydrolysis. For example, fluoroacetyl phosphate hydrolyzes 63-fold faster than the already highly activated acetyl phosphate at pH 7.43 Fluoroacetyl-CoA is hydrolyzed only 10-fold faster than acetyl-CoA at pH 7, but is up to 110-fold more susceptible when hydroxide ion is readily available at pH 11,22 a scenario that may mimic enzyme-catalyzed hydrolysis. Thus, the stability of the more labile intermediates in polyketide biosynthesis, particularly α-fluoro β-keto thioesters, will need to be addressed.
Figure 5.
Possible derailment pathways of fluoroacetyl-CoA during polyketide synthesis.
In addition to hydrolytic stability, fluorine substitution will affect the kinetics and thermodynamics of extender unit decarboxylation, enolate generation and C–C bond formation during chain extension. Defluorination reactions have been documented in enzyme active sites (Figure 6A-E);44 however, a source of electrons adjacent to fluorine, directly or via conjugation, are typically required for fluoride elimination, which is a poor leaving group in water. The α-fluorinated intermediates and reactions in polyketide synthesis should avoid such configurations. The active site base that removes the α-proton to generate an enolate could also serve as a nucleophile for fluoride displacement and alkylation, leading to covalent enzyme inactivation. In this regard, cysteine but not serine proteases react with fluoromethyl ketones (Figure 6FG), suggesting that there are specific enzyme active site requirements for reactivity.45 Finally, the stability and activity of the fluoroenolate towards carbon-carbon bond formation may also cause issues within a PKS assembly line.
Figure 6.
Reactions of enzymes with organofluorine substrates resulting in loss of fluoride or covalent enzyme inhibition. Only the key step(s) are shown. (A) Cytochrome P450 enzymes. (B) PLP-dependent enzymes. (C) Thymidylate synthase. (D) Ribonucleotide reductase. (E) Aconitase. (F) Serine proteases. (G) Cysteine proteases.
Uptake and activation of organofluorine building blocks
To activate fluoroacetate as an extender unit for PKSs, it must be converted to the CoA thioester and carboxylated to generate fluoromalonyl-CoA. We investigated the naturally occurring pathways for acetate activation (Figure 4), which can be carried out by either acetyl-CoA synthetase (ACS) or acetate kinase/phosphotransacetylase (AckA/Pta). Our previous studies had shown that neither the E. coli nor the S. cattleya ACS is competent to form substantial amounts of fluoroacetyl-CoA,37 so we used the AckA/Pta pathway, which accepts fluoroacetate with only a 20-fold rate reduction across both steps.46 To carboxylate fluoroacetyl-CoA, we used the E. coli acetyl-CoA carboxylase, and found that it accepts fluoroacetyl-CoA with a modest decrease in efficiency.46 Another approach is to directly use the advanced intermediate, fluoromalonate, which can be activated in a single step using MatB.47 Because malonyl-CoA is itself committed to either fatty acid or polyketide biosynthesis, we reasoned that fluoromalonyl-CoA might bypass the toxicity associated with fluoroacetyl-CoA. Although we found that S. coelicolor MatB has a 300-fold reduction in kcat/KM for fluoromalonate, it is efficient enough to build a fluoromalonyl-CoA pool in vitro or in vivo for polyketide biosynthesis.46 In the course of these studies, we found no evidence that any of these enzymes catalyze defluorination or are covalently inhibited by fluorinated substrates. While the stereochemistry of the fluoromalonyl-CoA produced by MatB and the ACCase has not been established, it can be racemized both non-enzymatically and by methylmalonyl-CoA epimerase.
Incorporation of fluoromalonyl extenders by polyketide synthases
We first set out to examine C–C bond formation using fluoromalonyl-CoA as an extender unit by testing standalone proteins, NphT7 and PhaB, which are respectively homologous to the ketosynthase (KS) and ketoreductase (KR) polyketide synthase domains (Figure 7A). We found that NphT7 catalyzed condensation of acetyl-CoA with fluoromalonyl-CoA with no change in KM and only a 6-fold reduction in kcat, and that PhaB was competent to reduce the resulting α-fluoro β-keto CoA, indicating that there is no intrinsic chemical barrier to these steps.46 With this information in hand, we then turned our attention to exploring the behavior of fluoromalonyl-CoA with a modular PKS (Figure 4). In these systems, extender units are transferred from CoA to the acyl carrier protein (ACP) by the acyl transferase domain (AT) before chain extension takes place. We tested two different modules from the well-studied erythromycin PKS (6-deoxyerythronolide B synthase, DEBS), each of which carries out one chain extension on a synthetic diketide substrate, normally using a methylmalonyl extender, to produce a triketide lactone (Figure 7B). We were encouraged to find that fluorotriketide could be formed by either module in a one-pot reaction starting from fluoroacetate or fluoromalonate.46
Figure 7.
Polyketide synthesis from fluoromalonyl-CoA. (A) Standalone KS and KR enzymes catalyze a single chain extension. (B) In a single chain extension cycle by a complete PKS module, cis-AT activity can be complemented by a trans-AT (DszsAT) to incorporate a fluorinated monomer. (C) A bimodular system carries out two chain extensions to form tetraketides. Fluorine can be regioselectively incorporated by one module or the other using cis-AT mutations and complementation by a trans-AT (DszsAT).
These results showed that the fluoromalonyl extender could be accommodated by a methylmalonyl-specific PKS module. However, the efficiency of chain extension with fluoromalonyl-CoA remained very low compared to the native substrate, even though we found no evidence of covalent enzyme inhibition. Furthermore, we were interested in pursuing a strategy for fluorine introduction that would enable it to be incorporated site-selectively. In this regard, we observed that the methylmalonyl-CoA-selective AT domain catalyzes substantial hydrolysis of fluoromalonyl-CoA, presumably in preference to transacylation, giving rise to the hypothesis that malonyl-CoA-selective ATs may demonstrate improved behavior with the fluorinated extender. Previous studies have shown that the native AT of a DEBS module can be inactivated via an active site mutation and complemented with a trans-acting, malonyl-specific AT.48 Thus, the use of a trans-AT could allow targeting of fluorine to desired modules simply by inactivating their ATs, bypassing the challenge of engineering fluorine recognition in each individual AT domain of interest. Indeed, we found that using an AT-inactivated module in combination with a malonyl-CoA specific trans-AT (DszsAT), fluoromalonyl-CoA incorporation could be amplified to up to 30% of wild-type extender yields, depending on the module (Figure 7B). We were also able to use this approach to target the fluoromalonyl-CoA extender to a specific module in a bimodular system that catalyzes two consecutive chain extension steps, resulting in regioselective fluorine incorporation (Figure 7C).46
We would like to now extend this initial work to produce full-length polyketides, achieve robust yields of fluorinated products, and also examine β-carbon processing. In addition to work in vitro, which will be augmented by the recent reconstitution of the complete DEBS assembly line,49 we are also interested in developing systems for in vivo production. Our initial efforts in E. coli show that fluoromalonate can be fed without significant toxicity and MatB can produce sufficient levels of fluoromalonyl-CoA to carry out PKS-catalyzed chain extension in vivo. However, since PKS expression in E. coli can be difficult, we also plan to explore alternative hosts.
Conclusions
The chemistry of living systems can provide both the inspiration and the tools to transform the production of molecular targets of interest to society. Along these lines, the divergence of structure and function between compounds of biological or synthetic origin also creates new opportunities to expand molecular diversity by merging these two areas using synthetic biology approaches. Towards this goal, the study of naturally-occurring enzymes and organisms with unusual chemical phenotypes allow us not only to elucidate the design principles that may enable these goals but also to increase our fundamental understanding of how living systems function.
ACKNOWLEDGMENT
We are highly indebted to our talented colleagues and collaborators who have contributed to the work described here. We would particularly like to thank Dr. Mark Walker and Dr. Amy Weeks for their work establishing this research area in our laboratory. This research was funded by a National Institutes of Health New Innovator Award (1 DP2 OD008696). B.W.T. would like to acknowledge a National Institutes of Health NRSA Training Grant (1 T32 GMO66698), Gerald K. Branch Predoctoral Fellowship, UC Cancer Research Council Committee Predoctoral Fellowship, and Amgen Fellowship in Organic Chemistry for support.
Biography
Benjamin Thuronyi graduated from Swarthmore College in 2007 with a B.A. in Chemistry. After two years in the RNAi Medicinal Chemistry group at Merck, he entered the Ph.D. program in the Department of Chemistry at the University of California, Berkeley. His graduate research with Michelle Chang focuses on engineering new organofluorine biosynthesis.
Michelle Chang graduated from the University of California, San Diego in 1997 with a B.S. in Biochemistry and B.A. in French Literature. She then obtained her Ph.D. in Chemistry in 2004 at the Massachusetts Institute of Technology as a joint student with Daniel Nocera and JoAnne Stubbe. After postdoctoral studies with Jay Keasling at the University of California, Berkeley, she started her independent career in 2007 with a joint appointment in the Departments of Chemistry and Molecular and Cell Biology at UC Berkeley.
REFERENCES
- (1).Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- (2).Weeks AM, Chang MCY. Constructing de novo biosynthetic pathways for chemical synthesis inside living cells. Biochemistry. 2011;50:5404–5418. doi: 10.1021/bi200416g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hagmann W. The many roles for fluorine in medicinal chemistry. J. Med. Chem. 2008;51:4359–4369. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- (4).Purser S, Moore P, Swallow S, Gouverneur V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 2008;37:320–330. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- (5).Furuya T, Kamlet AS, Ritter T. Catalysis for fluorination and trifluoromethylation. Nature. 2011;473:470–477. doi: 10.1038/nature10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Harper D, O’Hagan D, Murphy C. Fluorinated natural products: Occurrence and biosynthesis. The Handbook of Environmental Chemistry: Natural Production of Organohalogen Compounds. 2003;3/3P:141–169. [Google Scholar]
- (7).Smart BE. Fluorine substituent effects (on bioactivity) J. Fluorine Chem. 2001;109:3–11. [Google Scholar]
- (8).Goldstein J, Cheung Y, Marletta M, Walsh CT. Fluorinated substrate analogs as stereochemical probes of enzymatic reaction mechanisms. Biochemistry. 1978;17:5567–5575. doi: 10.1021/bi00618a037. [DOI] [PubMed] [Google Scholar]
- (9).O’Hagan D, Rzepa HS. Some influences of fluorine in bioorganic chemistry. Chem. Commun. 1997:645–652. [Google Scholar]
- (10).Poulter CD. Mechanistic studies of the prenyl transfer reaction with fluorinated substrate analogs. ACS Symp. Ser. 1996;639:158–168. [Google Scholar]
- (11).Vulpetti A, Dalvit C. Fluorine local environment: From screening to drug design. Drug Discovery Today. 2012;17:739–746. doi: 10.1016/j.drudis.2012.03.014. [DOI] [PubMed] [Google Scholar]
- (12).Vulpetti A, Dalvit C. Design and generation of highly diverse fluorinated fragment libraries and their efficient screening with improved 19F NMR methodology. ChemMedChem. 2013;8:2057–2069. doi: 10.1002/cmdc.201300351. [DOI] [PubMed] [Google Scholar]
- (13).Biffinger JC, Kim HW, Dimagno SG. The polar hydrophobicity of fluorinated compounds. ChemBioChem. 2004;5:622–627. doi: 10.1002/cbic.200300910. [DOI] [PubMed] [Google Scholar]
- (14).Muller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- (15).O'Hagan D. Understanding organofluorine chemistry. An introduction to the C–F bond. Chem. Soc. Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- (16).Fischer FR, Schweizer WB, Diederich F. Molecular torsion balances: Evidence for favorable orthogonal dipolar interactions between organic fluorine and amide groups. Angew. Chem. Int. Ed. 2007;46:8270–8273. doi: 10.1002/anie.200702497. [DOI] [PubMed] [Google Scholar]
- (17).Walker MC, Chang MCY. Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev. 2014;43:6527–6536. doi: 10.1039/c4cs00027g. [DOI] [PubMed] [Google Scholar]
- (18).Goldman P. The enzymatic cleavage of the carbon-fluorine bond in fluoroacetate. J. Biol. Chem. 1965;240:3434–3438. [PubMed] [Google Scholar]
- (19).Deng H, O'Hagan D, Schaffrath C. Fluorometabolite biosynthesis and the fluorinase from Streptomyces cattleya. Nat. Prod. Rep. 2004;21:773–784. doi: 10.1039/b415087m. [DOI] [PubMed] [Google Scholar]
- (20).Sanada M, Miyano T, Iwadare S, Williamson JM, Arison BH, Smith JL, Douglas AW, Liesch JM, Inamine E. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya. The Journal of Antibiotics. 1986;39:259–265. doi: 10.7164/antibiotics.39.259. [DOI] [PubMed] [Google Scholar]
- (21).Lauble H, Kennedy M, Emptage M, Beinert H, Stout C. The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex. Proc. Natl. Acad. Sci. USA. 1996;93:13699–13703. doi: 10.1073/pnas.93.24.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Weeks AM, Chang MCY. Catalytic control of enzymatic fluorine specificity. Proc. Natl. Acad. Sci. USA. 2012;109:19667–19672. doi: 10.1073/pnas.1212591109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Weeks AM, Coyle SM, Jinek M, Doudna JA, Chang MCY. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry. 2010;49:9269–9279. doi: 10.1021/bi101102u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Weeks AM, Keddie NS, Wadoux RDP, O'Hagan D, Chang MCY. Molecular recognition of fluorine impacts substrate selectivity in the fluoroacetyl-CoA thioesterase FlK. Biochemistry. 2014;53:2053–2063. doi: 10.1021/bi4015049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry. 2009;48:1293–304. doi: 10.1021/bi801879z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Divergence of function in the hot dog fold enzyme superfamily: The bacterial thioesterase YciA. Biochemistry. 2008;47:2789–96. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- (27).Dias MV, Huang F, Chirgadze DY, Tosin M, Spiteller D, Dry EF, Leadlay PF, Spencer JB, Blundell TL. Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK. J. Biol. Chem. 2010;285:22495–22504. doi: 10.1074/jbc.M110.107177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hisano T, Tsuge T, Fukui T, Iwata T, Miki K, Doi Y. Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate biosynthesis. J. Biol. Chem. 2003;278:617–24. doi: 10.1074/jbc.M205484200. [DOI] [PubMed] [Google Scholar]
- (29).Song F, Thoden JB, Zhuang Z, Latham J, Trujillo M, Holden HM, Dunaway-Mariano D. The catalytic mechanism of the hotdog-fold enzyme superfamily 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. Biochemistry. 2012;51:7000–7016. doi: 10.1021/bi301059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Holmquist B, Bruice TC. The carbanion (E1cB) mechanism of ester hydrolysis. I. Hydrolysis of malonate esters. J. Am. Chem. Soc. 1969;91:2993–3002. [Google Scholar]
- (31).Holmquist B, Bruice TC. The carbanion mechanism of ester hydrolysis. II. o-Nitrophenyl α-cyano- and α-dimethylsulfonioacetate esters. J. Am. Chem. Soc. 1969;91:3003–3009. [Google Scholar]
- (32).Dong C, Huang F, Deng H, Schaffrath C, Spencer JB, O'Hagan D, Naismith J. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- (33).Zhu X, Robinson D, McEwan A, O'Hagan D, Naismith J. Mechanism of enzymatic fluorination in Streptomyces cattleya. J. Am. Chem. Soc. 2007;129:14597–14604. doi: 10.1021/ja0731569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhao C, Li P, Deng Z, Ou H-Y, McGlinchey RP, O'Hagan D. Insights into fluorometabolite biosynthesis in Streptomyces cattleya DSM46488 through genome sequence and knockout mutants. Bioorg. Chem. 2012;44:1–7. doi: 10.1016/j.bioorg.2012.06.002. [DOI] [PubMed] [Google Scholar]
- (35).Murphy C, Moss S, O'Hagan D. Isolation of an aldehyde dehydrogenase involved in the oxidation of fluoroacetaldehyde to fluoroacetate in Streptomyces cattleya. Appl. Environ. Microbiol. 2001;67:4919. doi: 10.1128/AEM.67.10.4919-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kalan L, Gessner A, Thaker MN, Waglechner N, Zhu X, Szawiola A, Bechthold A, Wright GD, Zechel DL. A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem. Biol. 2013;20:1214–1224. doi: 10.1016/j.chembiol.2013.09.006. [DOI] [PubMed] [Google Scholar]
- (37).Walker MC, Wen M, Weeks AM, Chang MCY. Temporal and fluoride control of secondary metabolism regulates cellular organofluorine biosynthesis. ACS Chem. Biol. 2012;7:1576–1585. doi: 10.1021/cb3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Deng H, Ma L, Bandaranayaka N, Qin Z, Mann G, Kyeremeh K, Yu Y, Shepherd T, Naismith JH, O'Hagan D. Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining. ChemBioChem. 2014;15:364–368. doi: 10.1002/cbic.201300732. [DOI] [PubMed] [Google Scholar]
- (39).Huang S, Ma L, Tong MH, Yu Y, O'Hagan D, Deng H. Fluoroacetate biosynthesis from the marine-derived bacterium Streptomyces xinghaiensis NRRL B-24674. Org. Biomol. Chem. 2014;12:4828–4831. doi: 10.1039/c4ob00970c. [DOI] [PubMed] [Google Scholar]
- (40).Llano-Sotelo B, Dunkle J, Klepacki D, Zhang W, Fernandes P, Cate JHD, Mankin AS. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob. Agents Chemother. 2010;54:4961–4970. doi: 10.1128/AAC.00860-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Staunton J, Weissman KJ. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- (42).Hong H, Spiteller D, Spencer JB. Incorporation of fluoroacetate into an aromatic polyketide and its influence on the mode of cyclization. Angewandte Chemie International Edition. 2008;47:6028–6032. doi: 10.1002/anie.200801100. [DOI] [PubMed] [Google Scholar]
- (43).Marcus A, Elliott W. Fluoroacetyl phosphate; preparation and properties. J. Am. Chem. Soc. 1958;80:4287–4291. [Google Scholar]
- (44).Walsh CT. Suicide substrates: Mechanism-based enzyme inactivators. Tetrahedron. 1982;38:871–909. doi: 10.1146/annurev.bi.53.070184.002425. [DOI] [PubMed] [Google Scholar]
- (45).Powers J, Asgian J, Ekici Ö, James K. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. (Washington, DC, U. S.) 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- (46).Walker MC, Thuronyi BW, Charkoudian LK, Lowry B, Khosla C, Chang MCY. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science. 2013;341:1089–1094. doi: 10.1126/science.1242345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Hughes Amanda J., Keatinge-Clay A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: Structural and functional showcasing of Streptomyces coelicolor MatB. Chem. Biol. 2011;18:165–176. doi: 10.1016/j.chembiol.2010.12.014. [DOI] [PubMed] [Google Scholar]
- (48).Kumar P, Koppisch AT, Cane DE, Khosla C. Enhancing the modularity of the modular polyketide synthases: Transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J. Am. Chem. Soc. 2003;125:14307–14312. doi: 10.1021/ja037429l. [DOI] [PubMed] [Google Scholar]
- (49).Lowry B, Robbins T, Weng C-H, O’Brien RV, Cane DE, Khosla C. In vitro reconstitution and analysis of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc. 2013;135:16809–16812. doi: 10.1021/ja409048k. [DOI] [PMC free article] [PubMed] [Google Scholar]