Abstract
Cadmium is an established human lung carcinogen with weak mutagenicity. However, the mechanisms underlying cadmium-induced carcinogenesis remain obscure. It has been suggested that epigenetic mechanisms may play a role in cadmium-induced carcinogenesis. In this study, we investigated the effects of cadmium on histone methylation and histone demethylases, and the role of histone methylation in transformation of immortalized normal human bronchial epithelial (BEAS-2B) cells. Exposure to 0.625, 1.25, 2.5, and 5.0 μM of cadmium for 6, 24, and 48 h increased global trimethylated histone H3 on lysine 4 (H3K4me3) and dimethylated histone H3 on lysine 9 (H3K9me2) in BEAS-2B cells compared with untreated cells, and most of these changes remained after the removal of cadmium (P < .05 or P < .01 for most modifications). Meanwhile, cadmium inhibited the activities of histone H3 on lysine 4 (H3K4) and histone H3 on lysine 9 (H3K9) demethylases which were detected by histone demethylation assay. However, there was no significant change in the protein levels of the H3K4 demethylase lysine-specific demethylase 5A (KDM5A) and the H3K9 demethylase lysine-specific demethylase 3A (KDM3A). Interestingly, during transformation of BEAS-2B cells by 20 weeks of exposure to 2.0 μM cadmium as assessed by anchorage-independent growth in soft agar, global H3K4me3, and H3K9me2 were significantly increased at 4 weeks (P < .05 or P < .01), whereas no significant change was observed at 8, 12, 16, and 20 weeks compared with control. Our study suggests that cadmium increases global H3K4me3 and H3K9me2 by inhibiting the activities of histone demethylases, and aberrant histone methylation that occurs early (48 h) and at 4 weeks is associated with cadmium-induced transformation of BEAS-2B cells at the early stage.
Keywords: cadmium, histone methylation, histone demethylase, epigenetics, carcinogenesis
Cadmium is highly toxic to almost all forms of life. It is released into the environment through natural and anthropogenic activities. Natural sources originate from volcanic activity, forest fires, fossil fuel burning, and soil particles contaminated with cadmium (Joseph, 2009). Cadmium is extensively used in industries such as nickel-cadmium battery manufacturing, electroplating, welding, smelting and refining, pigments, and plastic stabilizers (Joseph, 2009). Inhalation through occupational exposure, cigarette smoking, and indoor inhalable particles contaminated with cadmium is one of the main routes of exposure to cadmium in humans, with more than 90% of a dose being absorbed through the lung after inhalation of cadmium (Nawrot et al., 2010; Waalkes, 2003). Both epidemiological and experimental studies have identified cadmium as a lung carcinogen in humans (IARC, 2012). Due to persistence in the environment and a long biological half-life in humans, cadmium has always been a serious public health concern. Even though much work has been carried out to elucidate the molecular mechanisms of cadmium-induced carcinogenesis, the exact mechanisms still remain unclear.
The ability of cadmium to induce gene mutations in bacteria was limited (Beyersmann and Hartwig, 1994), and no clear association between cadmium exposure and cytogenetic endpoint in humans has been found (Verougstraete et al., 2002), both of which implied that cadmium-induced carcinogenesis may be mediated through nongenotoxic or indirect genotoxic mechanisms (Beyersmann and Hechtenberg, 1997; Bolognesi et al., 1999). It has been proposed that epigenetic mechanisms may play a role in cadmium-induced carcinogenesis (Waalkes, 2003). Over the last decade or two, accumulating evidences have shown that cadmium is able to alter DNA methylation both at the global and gene-specific levels, which may play a role in carcinogenesis. Takiguchi et al. (2003) reported that after 10 weeks of exposure to cadmium, TRL1215 rat liver cells showed indications of transformation and significant increases in genomic DNA methylation and DNA methyltransferase (DNMT) activity. In cadmium-transformed human prostate epithelial RWPE-1 cells, the tumor suppressor genes RASSF1A and p16 were inactivated due to DNA hypermethylation at their promoter regions (Benbrahim-Tallaa et al., 2007). Another study showed that chronic exposure to cadmium in human embryo lung fibroblast cells resulted in increases in global DNA methylation and DNMTs activities (Jiang et al., 2008). A recent study reported that in cadmium-transformed human bronchial epithelial (16HBE) cells, global DNA methylation, and DNA methylation at the promoter regions of DNA repair genes (hMSH2, ERCC1, XRCC1, and hOGG1) were increased (Zhou et al., 2012). These findings suggested that cadmium disrupts DNA methylation, which may be involved in cadmium-induced carcinogenesis.
To date, much of the work has been focused on DNA methylation. Whether cadmium induces aberrant histone methylation has yet to be investigated. In general, DNA methylation may act as a template for some histone modifications following DNA replication but more likely histone methylation can aid in directing DNA methylation patterns (Cedar and Bergman, 2009). Therefore, it’s likely that cadmium is able to induce aberrant histone methylation. Histone H3 on lysine 4 (H3K4) and H3K9 are the regular sites of lysine methylations, both of which can be mono-, di-, or trimethylated. In general, trimethylated H3K4 (H3K4me3) is always found at the promoter regions of transcriptionally activated genes (Santos-Rosa et al., 2002), whereas dimethylated H3K9 (H3K9me2) is located in the regulatory regions of transcriptionally silent genes (Rice et al., 2003). Aberrant modifications of H3K4me3 and H3K9me2 have been found to be closely associated with carcinogenesis. H3K4me3 has been found to be increased at the promoter region of MT-3 in cadmium-transformed human urothelial cells compared with that in parental human urothelial cells (Somji et al., 2011). Activation of H3K4me3 has also been found to be associated with overexpression of LAMB3 and LAMC2 genes in gastric cancer cell line, which may play an important role in gastric carcinogenesis (Kwon et al., 2011). Gain of H3K9me2 has been observed in silencing RASSF1A in prostate cancer (Kawamoto et al., 2007). However, to the best of our knowledge, no one has investigated the effects of cadmium on global H3K4me3 and H3K9me2 and their potential roles in cadmium-induced carcinogenesis.
In this present study, we investigated the effects of cadmium on global H3K4me3 and H3K9me2 in immortalized normal human bronchial epithelial (BEAS-2B) cells and whether histone demethylases played a role in cadmium-induced histone modifications. BEAS-2B cells are similar to normal human lung cells in characteristics and cellular responses to carcinogens (Jing et al., 2012; Son et al., 2012; Wang et al., 2011), thus they are often used to establish the model of cell transformation. Since cell transformation assay is considered as a predictive test for carcinogenicity (Barrett et al., 1984), we further studied whether cadmium modulated global H3K4me3 and H3K9me2 during cadmium-induced transformation of BEAS-2B cells. Our work would further contribute to the understanding of the mechanisms of cadmium-induced carcinogenesis.
MATERIALS AND METHODS
Materials
CdCl2 was purchased from Sigma (St Louis, MO).
Cell culture
BEAS-2B cells, which were a generous gift from Dr. Chuanshu Huang (New York University), were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, Invitrogen corporation, Beijing, China) supplemented with 10% heat-inactivated fetal bovine serum (Sijiqing, Hangzhou, China) and 1% penicillin-streptomycin (Solarbio, Beijing, China). The cells were grown in an incubator at 37°C with a humidified atmosphere containing 5% CO2.
Colony survival assay
After treatment with cadmium, BEAS-2B cells were plated in 100-mm diameter culture dishes at 1000 cells/dish. The cells were then cultured in a cadmium-free environment for 2 weeks. Following staining with Giemsa (Amresco, OH), the number of colonies was counted. The experiment was performed in triplicate.
Histone extraction
Histones were extracted from BEAS-2B cells according to Chen et al. (2006). In brief, cells were lysed in ice-cold radioimmunoprecipitation assay buffer [50 mM Tris-HCl, pH 7.4, 1% octyl phenoxypolyethoxylethanol (NP-40), 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM ethylene diamine tetraacetic acid (EDTA)] containing protease inhibitor cocktail (Roche Applied Sciences, Mannheim, Germany) for 10 min on ice. Following centrifugation, the pellet was washed with a buffer containing 10 mM Tris-HCl (pH 7.4) and 37 mM EDTA (pH 8.0) and resuspended in 0.4 N H2SO4 on ice for 1.5 h. The supernatant was collected after centrifugation and incubated with ice-cold acetone overnight at −20°C. Following centrifugation and one wash with ice-cold acetone, the histones were collected and resuspended in 4 M urea.
Preparation of whole cell lysate
Cells were lysed in radioimmunoprecipitation assay buffer containing protease inhibitor cocktail for 20 min on ice. The supernatant was collected after centrifugation and kept at −20°C.
Western blot
The concentrations of the proteins were determined using Bio-Rad detergent-compatible protein assay (Bio-Rad, Hercules, CA). The whole cell lysates containing 50 μg of proteins were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and 5 μg of histones were separated by 15% SDS-PAGE gel. Following gel electrophoresis, the gels were transferred to polyvinylidene difluoride membranes (Bio-Rad) and were then stained with Coomassie blue (Guoyao, Shanghai, China) to assess the loading of histones. After blocking, the membranes were incubated with primary antibodies including lysine-specific demethylase 5 A (KDM5A, 1:5000; Abcam, Cambridge, MA), lysine-specific demethylase 3 A (KDM3A, 1:1000; Abcam), β-actin (1:5000; Abmart, Shanghai, China), tri-methyl H3K4 (1:5000; Abcam), di-methyl H3K9 (1:10 000; Abcam), and then incubated with horse-radish peroxidase-conjugated anti-rabbit (1:5000; Jackson ImmunoResearch, West Grove, PA) or anti-mouse secondary antibodies (1:5000; Jackson ImmunoResearch). The proteins were detected by an enhanced chemiluminescence kit (Multisciences Biotech, Hangzhou, China) and then scanned with a GeneGnome chemiluminescent imaging system (Syngene, Frederick, MD). The relative intensities of the bands were analyzed using Image J software and were normalized to those of control.
In vitro histone demethylation assay
In vitro histone demethylation assay was performed with minor modifications as described previously (Zhou et al., 2010). Nuclear extracts were prepared according to the instructions of CelLytic NuCLEAR extraction kit (Sigma). In brief, 50 μg of nuclear extracts from BEAS-2B cells were incubated with 5 μg of histones in histone demethylation buffer (0.1 mM dl-dithiothreitol, 2 μg/ml bovine serum albumin, 50 mM N-2-hydroxyethylpiperazine-N-ethane-sulphonicacid, pH 8.0, 100 μM FeSO4, 2 mM ascorbate, 1 mM 2-ketoglutarate, 1 mM phenylmethanesulfonyl fluoride, and protease inhibitor cocktail). The reaction mixture was in a total volume of 50 μl. After overnight incubation on ice or at 37°C, EDTA was added into the reaction mixture to a final concentration of 1 mM to terminate the reaction. Then the reaction mixture was subjected to Western blot using trimethyl H3K4 or dimethyl H3K9 antibodies. Three independent experiments were performed.
Soft agar assay
5 × 103 cells were suspended in 3 ml of 0.35% agar and poured onto a 2.5 ml of 0.5% agar bed in 6-well plates. Colonies were stained with 0.04% crystal violet (Guoyao) and photographed after 3 weeks. The number of colonies was counted using Image J software (size: 50−infinity; circularity: 0.6−1.0). The plates were prepared in triplicate.
Statistical analysis
Statistical analysis was performed with two-tailed Student's t-test. Difference was considered statistically significant at P < .05 and statistically highly significant at P < .01. Data were presented as the means ± SD.
RESULTS
Effect of Cadmium on Colony Survival of BEAS-2B Cells
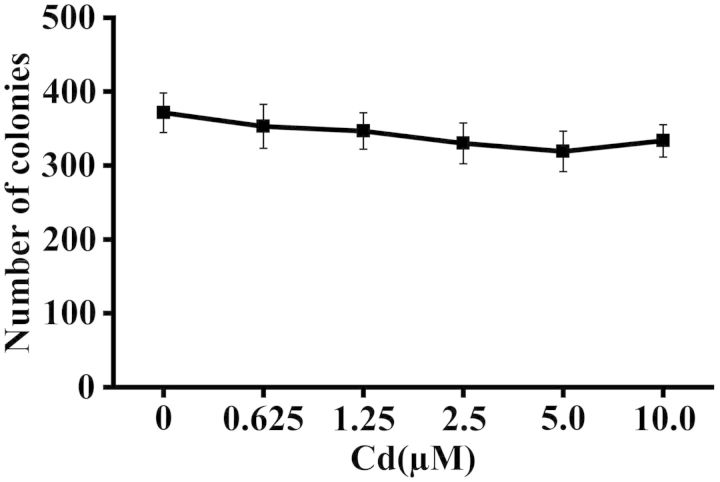
Colony survival assay was first conducted to determine the doses of cadmium to treat BEAS-2B cells. The cells were exposed to cadmium at 0, 0.625, 1.25, 2.5, 5.0, and 10.0 μM for 24 h and then allowed to form colonies for 2 weeks. As shown in Figure 1, there was no significant change in the number of colonies formed by the cells treated with up to 10.0 μM of cadmium compared with that of untreated cells.
FIG. 1.
Cadmium had no effect on the colony survival of BEAS-2B cells. BEAS-2B cells were treated with cadmium at 0, 0.625, 1.25, 2.5, 5.0, and 10.0 μM for 24 h, and subjected to colony survival assay. After 2 weeks, the colonies were stained with Giemsa, and the number of colonies was counted. The experiment was performed in triplicate. Data were expressed as the means ± SD.
Acute Exposure to Cadmium Increased H3K4me3 and H3K9me2 at the Global Level
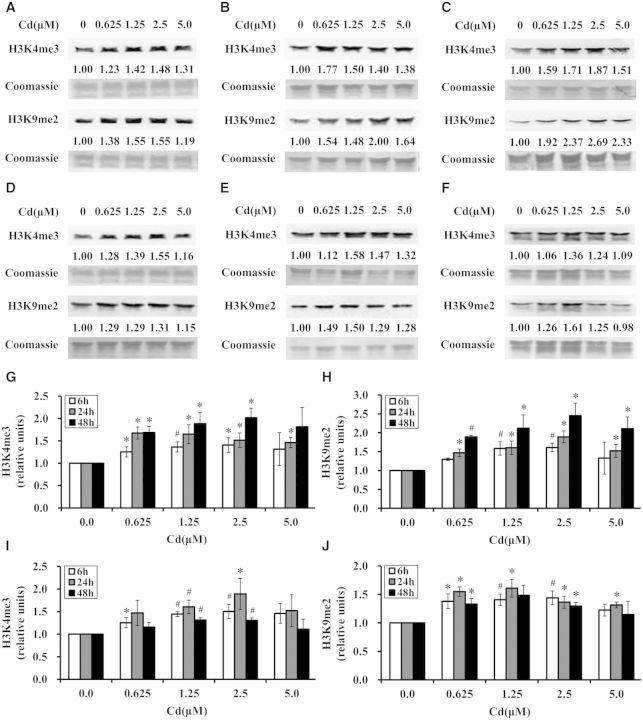
To investigate the effects of cadmium on global H3K4me3 and H3K9me2, BEAS-2B cells were treated with cadmium at 0, 0.625, 1.25, 2.5, and 5.0 μM for 6, 24, and 48 h. After cadmium exposure, histones were extracted, and global levels of H3K4me3 and H3K9me2 were measured by Western blot with antibodies against trimethyl H3K4 or dimethyl H3K9. The results showed that except for the cells treated with 5.0 μM cadmium for 6 and 48 h, global H3K4me3 were significantly increased after exposure to 0.625, 1.25, 2.5, and 5.0 μM cadmium for 6, 24, and 48 h (Figs. 2A–C, respectively, and G) compared with those of untreated cells (P < .05 or P < .01). Meanwhile, except for the cells treated with 0.625 and 5.0 μM cadmium for 6 h, exposure of BEAS-2B cells to 0.625, 1.25, 2.5, and 5.0 μM cadmium for 6, 24, and 48 h (Figs. 2A–C, respectively, and H) also significantly increased global H3K9me2 compared with untreated cells (P < .05 or P < .01). In general, global H3K4me3 and H3K9me2 were elevated as the time of the treatment increased. It was noted that the levels of the increases of global H3K4me3 and H3K9me2 were lower at 5.0 μM than those at 2.5 μM. These results demonstrated that acute exposure of BEAS-2B cells to cadmium elevated global H3K4me3 and H3K9me2.
FIG. 2.
Cadmium increased global H3K4me3 and H3K9me2 in BEAS-2B cells. BEAS-2B cells were exposed to cadmium at 0, 0.625, 1.25, 2.5, and 5.0 μM for 6 (A), 24 (B), and 48 h (C) and then allowed to recover for 12 (D), 48 (E), and 96 h (F), respectively, following the removal of cadmium. Histones were extracted and subjected to Western blot with antibodies against trimethyl H3K4 or dimethyl H3K9. To assess the loading of histones in each lane, the gels were stained with Coomassie blue. Three independent experiments were performed and representative results are shown. The relative intensities of the bands were quantified using Image J software (shown below the bands) and were expressed as the means ± SD in histograms. (G, H) Quantification of global H3K4me3 and H3K9me2 in BEAS-2B cells after exposure to cadmium. (I, J) Quantification of global H3K4me3 and H3K9me2 in BEAS-2B cells after removal of cadmium. *, P < .05 and #, P < .01 compared with control.
We next examined if cadmium-induced global H3K4me3 and H3K9me2 were stable after the removal of cadmium. Following exposure to cadmium at 0, 0.625, 1.25, 2.5, and 5.0 μM for 6, 24, and 48 h, BEAS-2B cells were allowed to proliferate in the absence of cadmium for another 12, 48, and 96 h, respectively (Figs. 2D–F, respectively, I and J). Strikingly, the results showed that compared with those of untreated cells, global H3K4me3 remained significantly elevated after the removal of cadmium (P < .05 or P < .01) except for the cells treated with 0.625 μM cadmium for 24 and 48 h and 5.0 μM cadmium for 6, 24, and 48 h (Figs. 2D–F and I). Meanwhile, except for the cells treated with 1.25 μM cadmium for 48 h and 5.0 μM cadmium for 6 and 48 h (Figs. 2D–F and J), global H3K9me2 remained significantly increased after the removal of cadmium (P < .05 or P < .01). Moreover, the increases of global H3K4me3 and H3K9me2 induced by cadmium were less at 5.0 μM than those at 2.5 μM after the removal of cadmium as well. These results indicated that cadmium-induced increases of H3K4me3 and H3K9me2 remained after the removal of cadmium.
Cadmium Increased Global H3K4me3 and H3K9me2 by Inhibiting the Activities of H3K4 and H3K9 Demethylases, Respectively
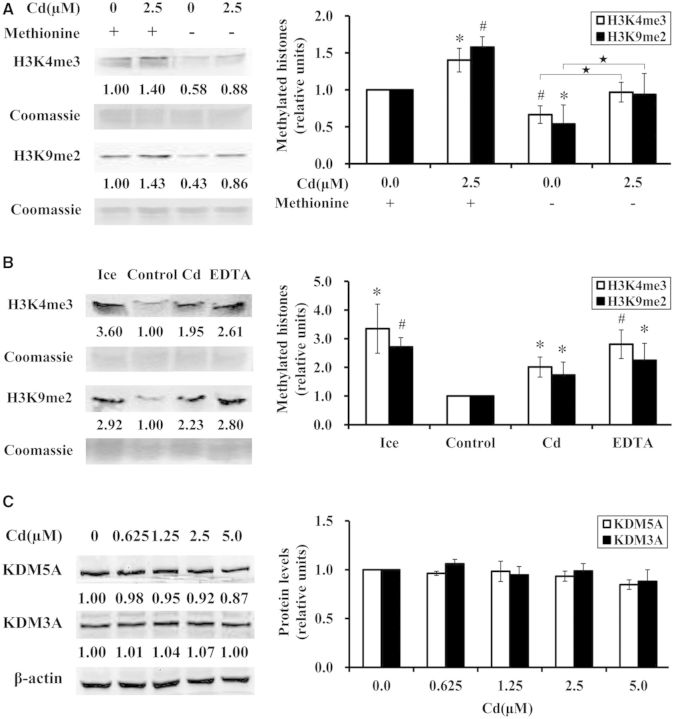
The homeostasis of histone methylation was maintained by both histone methyltransferases and demethylases (Shi et al., 2004). Histone methyltransferases catalyze the transfer of the methyl groups to histone residues whereas histone demethylases remove the methyl groups. Therefore, inhibition of histone demethylases would result in an increase in histone methylation. It has been reported that nickel increased global H3K9me2 by inhibiting histone demethylase in human lung carcinoma A549 cells (Chen et al., 2006). We next investigated whether cadmium increased global H3K4me3 and H3K9me2 by inhibiting the activities of histone demethylases. Since S-adenosyl-methionine (SAM), a methyl donor, has an extremely short half-life in the cells and its synthesis requires methionine, the absence of methionine in DMEM would result in low content of SAM in the cells, which would inhibit methyltransfer reactions. To investigate the role of histone demethylases in cadmium-induced increases of global H3K4me3 and H3K9me2, BEAS-2B cells were pretreated with DMEM or methionine-deficient DMEM for 4 h prior to cadmium exposure. As a result of the removal of methionine in DMEM, the methylating process was inhibited leading to significant decreases of global H3K4me3 and H3K9me2 in BEAS-2B cells (P < .05 or P < .01) (Fig. 3A). However, treatment with 2.5 μM cadmium in methionine-deficient DMEM still significantly increased global H3K4me3 and H3K9me2 compared with untreated cells (P < .05) (Fig. 3A). These results suggested that cadmium increased global H3K4me3 and H3K9me2 at least by inhibiting histone demethylation.
FIG. 3.
Cadmium increased global H3K4me3 and H3K9me2 by inhibiting the demethylating process in BEAS-2B cells. (A) BEAS-2B cells were preincubated with complete or methionine-deficient DMEM for 4 h and treated with 2.5 μM cadmium for 24 h. Histones were extracted and subjected to Western blot with antibodies against trimethyl H3K4 or dimethyl H3K9. (B) 50 μg of nuclear extracts from BEAS-2B cells were incubated with 5 μg of histones overnight at 37°C in the reaction mixture containing iron, ascorbate, and 2-ketoglutarate, in the absence or presence of 2.5 μΜ cadmium, or in the presence of 1 mM EDTA, or at ice bath temperature. To assess the loading of histones in each lane, the gels were stained with Coomassie blue. (C) BEAS-2B cells were treated with cadmium at 0, 0.625, 1.25, 2.5, and 5.0 μM for 24 h. Whole cell lysates were prepared and subjected to Western blot with antibodies against KDM5A or KDM3A. The same membrane was reblotted with β-actin as a loading control. Three independent experiments were performed and representative results are shown. The relative intensities of the bands were quantified using Image J software (shown below the bands) and were expressed as the means ± SD in histograms. *, P < .05 and #, P < .01 compared with control in complete DMEM, and  , P < .05 compared with control in methionine-deficient DMEM (A). *, P < .05 and #, P < .01 compared with control (B, C).
, P < .05 compared with control in methionine-deficient DMEM (A). *, P < .05 and #, P < .01 compared with control (B, C).
In an effort to determine whether cadmium inhibited the demethylating process by inhibiting the activities of H3K4 and H3K9 demethylases, we performed an in vitro histone demethylation assay. The methyl groups in H3K4me3 and H3K9me2 can be removed by members of Jumonji-domain-containing histone demethylases. It is essential for these demethylases, which belong to the dioxygenase family, to have adequate concentrations of iron, ascorbate and 2-ketoglutarate as cofactors (Tsukada et al., 2006). In this study, 50 μg of nuclear extracts from BEAS-2B cells were incubated with 5 μg of histones overnight at 37°C in the reaction mixture containing iron, ascorbate and 2-ketoglutarate in the absence or presence of 2.5 μΜ cadmium. Then Western blot was performed with antibodies against trimethyl H3K4 or dimethyl H3K9. EDTA is an iron chelator, which can inhibit the activities of histone demethylases, we therefore used EDTA to terminate all of the reactions. We also incubated the mixture with EDTA as a positive control. Since the optimum temperature for the enzymatic activities of histone demethylases is 37°C, we used the mixture incubated on ice to abolish the activities of the demethylases and examined the histone methylation levels. As shown in Figure 3B, global H3K4me3 and H3K9me2 were much lower in control that were incubated without cadmium at 37°C compared with those in the reaction mixture incubated on ice (P < .05 or P < .01), suggesting that the histone demethylases were active in control to reduce the methylation levels. Nevertheless, global H3K4me3 and H3K9me2 were significantly increased by cadmium compared with control, but they were still lower than those in the reaction mixture incubated on ice (P < .05 or P < .01). Altogether, these results indicated that cadmium partly inhibited the activities of H3K4 and H3K9 demethylases, which resulted in increases of H3K4me3 and H3K9me2, respectively.
A family of 4 lysine-specific demethylases (KDM5) can catalyze the removal of the methyl groups from di- and trimethylated H3K4 (Rasmussen and Staller, 2014). It has been previously reported that the transcriptional level of KDM5A was the highest of the KDM5 family demethylases in BEAS-2B cells and it might be the major demethylase of H3K4me3 in this cell type (Zhou et al., 2010). KDM3A is a histone demethylase specific to mono- and dimethylated H3K9 (Yamane et al., 2006). To investigate if cadmium increased global H3K4me3 and H3K9me2 by modulating the protein levels of KDM5A and KDM3A, BEAS-2B cells were treated with cadmium at 0, 0.625, 1.25, 2.5, and 5.0 μM for 24 h. Whole cell lysates were extracted and then subjected to Western blot using antibodies against KDM5A or KDM3A. As shown in Figure 3C, there was no significant change in the protein levels of KDM5A and KDM3A after 24 h of exposure to cadmium compared with those of untreated cells.
Cadmium Transformed BEAS-2B Cells
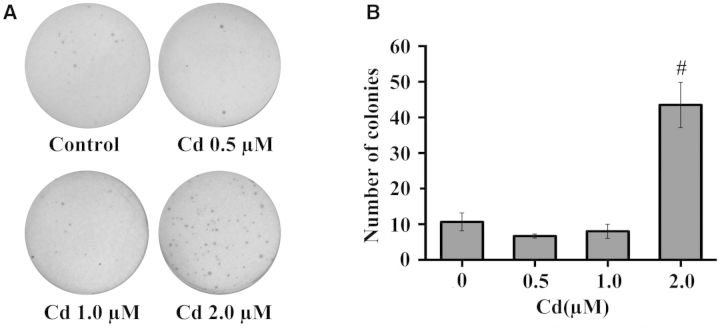
Anchorage-independent growth as measured by acquired ability of normal cells to grow in soft agar is often used as an indicator of cell transformation (Shin et al., 1975). Hence, we performed soft agar colony formation assay to examine whether cadmium can transform BEAS-2B cells following chronic exposure. The doses of chronic cadmium exposure were chosen based on the work of Son et al. (2012). Accordingly, BEAS-2B cells were continuously treated with cadmium at 0, 0.5, 1.0, and 2.0 μM for 20 weeks. BEAS-2B cells passaged at the same time with cadmium-exposed cells were used as control. We observed no significant change in the number of colonies after exposure to cadmium for 8, 12, and 16 weeks compared with that of control (data not shown). After 20 weeks of exposure, 0.5 and 1.0 μΜ cadmium did not significantly induced colonies in BEAS-2B cells compared with control (Figs. 4A and B). However, there was an approximate of 4.5-fold increase in the number of colonies formed by the cells exposed to 2.0 μΜ cadmium for 20 weeks compared with control (P < .01). The results indicated that 20 weeks of exposure to cadmium at 2.0 μΜ was capable of transforming BEAS-2B cells to acquire the ability of anchorage independent growth.
FIG. 4.
Cadmium transformed BEAS-2B cells following treatment at 2.0 μM for 20 weeks. (A) BEAS-2B cells were continuously treated with cadmium at 0, 0.5, 1.0, and 2.0 μM for 20 weeks, and then subjected to soft agar assay. Colonies were stained with crystal violet and photographed after 3 weeks. The experiment was carried out in triplicate and representative results are shown. (B) The number of colonies was analyzed with Image J software, which was expressed as the means ± SD in the graph. #, P < .01 compared with control.
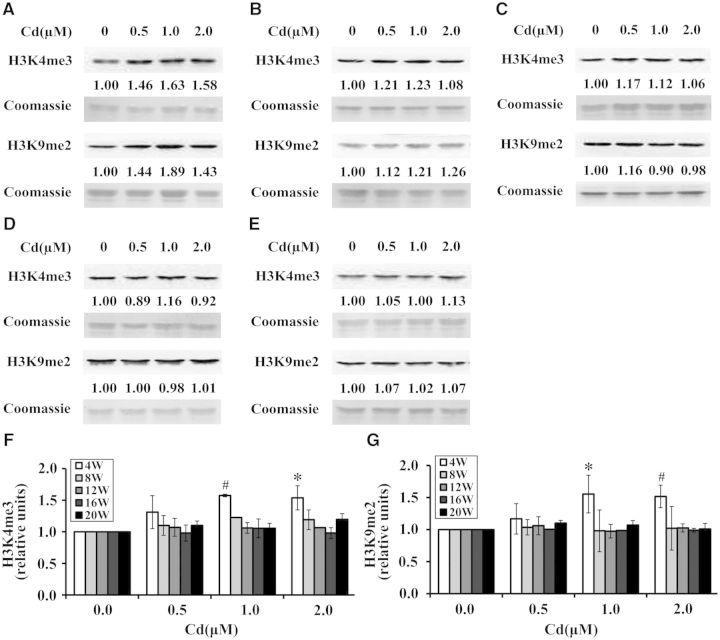
Alterations of Global H3K4me3 and H3K9me2 During Cadmium-Induced Transformation of BEAS-2B Cells
We next examined if global H3K4me3 and H3K9me2 were altered in the process of cadmium-induced transformation of BEAS-2B cells. During transformation, histones were extracted at 4, 8, 12, 16, and 20 weeks and Western blot was performed with antibodies against trimethyl H3K4 or dimethyl H3K9. Interestingly, compared with control, exposure to cadmium at 1.0 and 2.0 μM for 4 weeks significantly increased global H3K4me3 and H3K9me2 (P < .05 or P < .01) (Figs. 5A, F, and G). However, there was no significant change in global H3K4me3 and H3K9me2 following exposure to cadmium at 0.5 μM for 4 weeks and at 0.5, 1.0, and 2.0 μM for 8, 12, 16, and 20 weeks (Figs. 5A–E, respectively, F, and G).
FIG. 5.
Cadmium elevated global H3K4me3 and H3K9me2 in BEAS-2B cells after 4 weeks of exposure. BEAS-2B cells were continuously treated with cadmium at 0, 0.5, 1.0, and 2.0 μM for 20 weeks. Histones were extracted at 4 (A), 8 (B), 12 (C), 16 (D), and 20 weeks (E) and subjected to Western blot with antibodies against trimethyl H3K4 or dimethyl H3K9. Coomassie blue was used to assess the loading of histones. Two independent experiments were performed and representative results are shown. The relative intensities of the bands were quantified using Image J software (shown below the bands) and were expressed as the means ± SD in histograms (F, G). *, P < .05 and #, P < .01 compared with control.
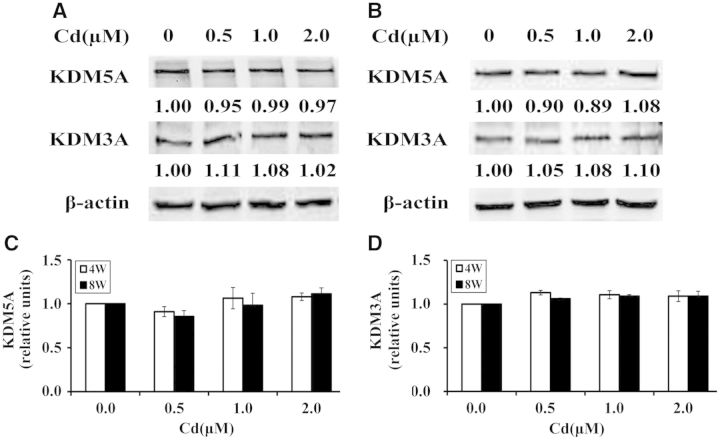
In order to determine the roles of KDM5A and KDM3A in cadmium-induced increases of H3K4me3 and H3K9me2 during the early stage of cadmium-induced cell transformation, the protein levels of KDM5A and KDM3A were examined in BEAS-2B cells after exposure to cadmium at 0, 0.5, 1.0, and 2.0 μM for 4 and 8 weeks (Figs. 6A, B, respectively, C, and D). The results showed that there was no significant change in the protein levels of KDM5A and KDM3A in cadmium-treated cells compared with control.
FIG. 6.
Cadmium had no effect on the protein levels of KDM5A and KDM3A in BEAS-2B cells. BEAS-2B cells were treated with cadmium at 0, 0.5, 1.0, and 2.0 μM for 4 weeks (A) and 8 weeks (B). Whole cell lysates were extracted and then subjected to Western blot with antibodies against KDM5A or KDM3A. β-actin was used as a protein loading control. Three independent experiments were performed and representative results are shown. The relative intensities of the bands were quantified using Image J software (shown below the bands) and were expressed as the means ± SD in histograms (C, D).
DISCUSSION
Over decades, mechanisms involved in cadmium-induced carcinogenesis such as induction of oxidative stress, repression of DNA repair, disruption of gene regulation, and suppression of apoptosis have been investigated (Joseph, 2009). However, the exact mechanism underlying cadmium-induced carcinogenesis remains unidentified. Since cadmium is a carcinogen with weak mutagenicity, it has been proposed that epigenetic modifications could be one of the major mechanisms underlying cadmium-induced carcinogenesis (Arita and Costa, 2009; Joseph, 2009; Waalkes, 2003). This notion has been supported by the findings demonstrating that cadmium disrupted DNA methylation, which was associated with cadmium-induced carcinogenesis (Benbrahim-Tallaa et al., 2007; Jiang et al., 2008; Takiguchi et al., 2003; Zhou et al., 2012). Therefore, we investigated whether histone modifications were involved in cadmium-induced carcinogenesis in this study.
We used 0−5.0 μM cadmium to acutely treat BEAS-2B cells because the results from colony survival assay demonstrated that there was no significant change in the number of colonies formed by the cells treated with up to 10.0 μM of cadmium compared with that of untreated cells. The result was consistent with a recent study showing that there was no significant cytotoxicity in BEAS-2B cells after treatment with 10.0 μM cadmium for 36 h but 20.0 μM cadmium significantly induced cytotoxicity after 36 h of exposure (Chen et al., 2014). Forti et al. (2010) also showed that the viability of human bronchial epithelial Calu-3 cells was significantly reduced by cadmium at the lowest dose of 30.0 μM after 24 h of exposure. Moreover, cadmium has been found to be accumulated in the lungs of nonsmokers at the concentrations of 0.9−6.0 μM (Jin et al., 2003). Thus, the doses of cadmium we used in this study were nontoxic and were comparable to the levels accumulated in the lungs of humans. Furthermore, the treatment doses were also consistent with the studies as reported by Jing et al. (2012) and Person et al. (2013).
Our results showed that acute exposure to cadmium increased global H3K4me3 and H3K9me2 and most of the increases remained after the removal of cadmium. However, some persistence was lost after recovery at later time points, such as the methylation levels of H3K4me3 in the cells treated with 0.625 cadmium for 24 and 48 h and 5 μM cadmium for 24 h, and the methylation levels of H3K9me2 in the cells treated with 1.25 and 5.0 μM cadmium for 48 h. These results suggested a transient effect of cadmium on histone methylations. It is worth noting that cadmium-induced elevations of global H3K4me3 and H3K9me2 were less at 5.0 μM than those at 2.5 μM, suggesting that cadmium may have induced a nonlinear dose-response. Waalkes et al. (1988) also observed a nonlinear dose-response in cadmium-induced prostatic tumor formation in rats. In this study, we found increases both in the global levels of the transcriptional activating mark H3K4me3 and the repressing mark H3K9me2. This phenomenon has been previously reported in A549 cells exposed to arsenite, chromate, and nickel (Sun et al., 2009; Zhou et al., 2008, 2009). As indicated by immunofluorescent staining in these studies, H3K4me3 and H3K9me2 were located at different regions in the nucleus. H3K4me3 mainly resides in euchromatin whereas H3K9me2 primarily exists at heterochromatin. Therefore, it is not paradoxical that exposure of BEAS-2B cells to cadmium could simultaneously increase global H3K4me3 and H3K9me2.
Since our study showed that actue exposure of BEAS-2B cells to cadmium inhibited the activities of H3K4 and H3K9 demethylases, cadmium appeared to increase global H3K4me3 and H3K9me2 by inhibiting demethylation. However, it is still not clear how cadmium inhibited the activities of H3K4 and H3K9 demethylases. The possible mechanisms are as followed. First, cadmium and zinc belong to IIB family of transition elements, they have many similarities in physical and chemical characteristics and have been considered to antagonize each other (Das et al., 1997). Kothinti et al. (2010) have reported that cadmium inhibited the DNA binding affinity of the transcription factor Sp1 by replacing zinc in its zinc finger DNA binding domain. Some of the histone demethylases, such as KDM5A and KDM3A, contain zinc fingers as well, which are required for their enzymatic activities (Klose et al., 2007; Yamane et al., 2006). Therefore, we hypothesized that cadmium could compete with zinc in the zinc fingers of histone demethylases, resulting in alteration of the conformation of zinc fingers and finally inhibiting the activities of histone demethylases. Second, cadmium has been reported to induce generation of reactive oxygen species both in vivo and in vitro (Joseph, 2009). As a consequence, cadmium would reduce the antioxidant ascorbate, an essential cofactor for the catalytic activity of the Jmjc-domain-containing demethylases and result in oxidation of ferrous iron to ferric iron to inactivate the enzyme. It has been reported that the levels of ascorbate were significantly reduced in the lungs of cadmium-exposed mice (Luchese et al., 2007). Therefore, cadmium may also inhibit the activities of H3K4 and H3K9 demethylases via depletion of ascorbate. In addition to investigate the activities of histone demethylases, we found that there was no significant change in the protein levels of KDM5A and KDM3A after exposure to cadmium. Overall, these results suggested that cadmium-induced elevations of H3K4me3 and H3K9me2 were modulated by inhibiting the histone demethylases, but not through modulating the protein levels of KDM5A and KDM3A.
In vitro morphological transformation of cells has been widely employed to study the potential carcinogenicity of chemicals (Barrett et al., 1984). Cadmium is an established human lung carcinogen and its transformative capacity has been confirmed in different human bronchial epithelial cell lines. It has been reported that 16HBE cells were malignantly transformed after exposure to 5−15 μM cadmium for 3−4 months (Lei et al., 2008). Jing et al. (2012) found that exposure of 5.0 μM cadmium for 26 weeks malignantly transformed BEAS-2B cells. In another study by Son et al. (2012), BEAS-2B cells exposed to 0.5, 1.0, and 2.0 μM cadmium for 2 months were malignantly transformed. In our study, BEAS-2B cells were transformed after 20 weeks of exposure to 2.0 μM cadmium, which was inconsistent with the study as reported by Son et al. The following reasons may explain the discrepancy. First, the densities of cells plating in soft agar were different. BEAS-2B cells were plated at 1 × 104 in their study, whereas the number of the cells plating in soft agar was only half (5 × 103) in our study. Second, the cells were grown in soft agar for 2 months in the study by Son et al., whereas we cultured the cells in soft agar for 3 weeks before colonies were counted. It’s worth noting that colonies were formed in control in this study, which was in accordance with the studies by Sun et al. (2011), Wang et al. (2011), and Stueckle et al. (2012). American Type Culture Collection has claimed that BEAS-2B cells do form colonies in semisolid medium, but are not tumorigenic in immunosuppressed mice. It has been proposed by Stueckle et al. (2012) that formation of colonies in control cells may be partially due to the existence of a mutated p53 in BEAS-2B cells, which is induced by SV40 large T-antigen that immortalizes the cells.
It was of interest that during transformation of BEAS-2B cells induced by cadmium, global H3K4me3 and H3K9me2 were significantly increased at 4 weeks, while no significant change was observed at 8, 12, 16, and 20 weeks compared with those of untreated cells. The result indicated that the increases of global H3K4me3 and H3K9me2 only occurred at an early stage of cadmium-induced cell transformation. The increases were induced by inhibiting the activities of histone demethylases, which has been confirmed by histone demethylation assay. However, at the mid or late stages of transformation, cadmium may trigger other molecular pathways that affect the methylating process as well, so that the balance of methyl addition and removal could be maintained. In addition, we found that there was no significant change in the protein levels of KDM5A and KDM3A following exposure to cadmium for 4 and 8 weeks, indicating that cadmium-induced increases of global H3K4me3 and H3K9me2 at the early stage of transformation of BEAS-2B cells were not mediated by altering the protein levels of the histone demethylases KDM5A and KDM3A.
In conclusion, our results showed that acute exposure to cadmium increased global H3K4me3 and H3K9me2 in BEAS-2B cells by inhibiting the activities of H3K4 and H3K9 demethylases, respectively, which would ultimately affect the expression of genes or noncoding RNA. The increases remained after the removal of cadmium. Moreover, the increases of global H3K4me3 and H3K9me2 only occurred at an early stage of cadmium-induced cell transformation, which suggested that aberrant histone methylation may be involved in the initiation step of cell transformation. Further study may focus on the expression of specific genes or noncoding RNA in relationship with cadmium-induced alterations of histone methylation, thus providing more insights into the mechanisms of cadmium-induced carcinogenesis.
ACKNOWLEDGMENT
We thank Dr Chuanshu Huang (New York University) for his generous gift of BEAS-2B cells.
FUNDING
The National Natural Science Foundation of China (Nos. 81202237 and 81172700); the Specialized Research Fund for the Doctoral Program of Higher Education (No. 20110142120027); and Hubei Health Department (No. QJX2012-01).
REFERENCES
- Arita A., Costa M. (2009). Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics 1, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Hesterberg T. W., Thomassen D. G. (1984). Use of cell transformation systems for carcinogenicity testing and mechanistic studies of carcinogenesis. Pharmacol. Rev. 36, 53S–70S. [PubMed] [Google Scholar]
- Benbrahim-Tallaa L., Waterland R. A., Dill A. L., Webber M. M., Waalkes M. P. (2007). Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ. Health Perspect. 115, 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyersmann D., Hartwig A. (1994). Genotoxic effects of metal compounds. Arch. Toxicol. Suppl. 16, 192–198. [DOI] [PubMed] [Google Scholar]
- Beyersmann D., Hechtenberg S. (1997). Cadmium, gene regulation, and cellular signalling in mammalian cells. Toxicol. Appl. Pharmacol. 144, 247–261. [DOI] [PubMed] [Google Scholar]
- Bolognesi C., Landini E., Roggieri P., Fabbri R., Viarengo A. (1999). Genotoxicity biomarkers in the assessment of heavy metal effects in mussels: experimental studies. Environ. Mol. Mutagen. 33, 287–292. [PubMed] [Google Scholar]
- Cedar H., Bergman Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304. [DOI] [PubMed] [Google Scholar]
- Chen H., Ke Q., Kluz T., Yan Y., Costa M. (2006). Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell Biol. 26, 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. J., Xu Y. M., Du J. Y., Huang D. Y., Lau A. T. (2014). Cadmium induces cytotoxicity in human bronchial epithelial cells through upregulation of eIF5A1 and NF-kappaB. Biochem. Biophys. Res. Commun. 445, 95–99. [DOI] [PubMed] [Google Scholar]
- Das P., Samantaray S., Rout G. R. (1997). Studies on cadmium toxicity in plants: a review. Environ. Pollut. 98, 29–36. [DOI] [PubMed] [Google Scholar]
- Forti E., Bulgheroni A., Cetin Y., Hartung T., Jennings P., Pfaller W., Prieto P. (2010). Characterisation of cadmium chloride induced molecular and functional alterations in airway epithelial cells. Cell. Physiol. Biochem. 25, 159–168. [DOI] [PubMed] [Google Scholar]
- IARC. (2012). International Agency for Research on Cancer. A review of human carcinogens: arsenic, metals, fibres, and dusts. IARC monographs on the evaluation of carcinogenic risks to humans, 100c. Lyon, France, pp. 121–141. [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Xu L., Song S., Zhu C., Wu Q., Zhang L., Wu L. (2008). Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 244, 49–55. [DOI] [PubMed] [Google Scholar]
- Jin Y. H., Clark A. B., Slebos R. J., Al-Refai H., Taylor J. A., Kunkel T. A., Resnick M. A., Gordenin D. A. (2003). Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34, 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Liu L. Z., Jiang Y., Zhu Y., Guo N. L., Barnett J., Rojanasakul Y., Agani F., Jiang B. H. (2012). Cadmium increases HIF-1 and VEGF expression through ros, erk, and akt signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol. Sci. 125, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P. (2009). Mechanisms of cadmium carcinogenesis. Toxicol. Appl. Pharmacol. 238, 272–279. [DOI] [PubMed] [Google Scholar]
- Kawamoto K., Okino S. T., Place R. F., Urakami S., Hirata H., Kikuno N., Kawakami T., Tanaka Y., Pookot D., Chen Z., et al. (2007). Epigenetic modifications of RASSF1A gene through chromatin remodeling in prostate cancer. Clin. Cancer Res. 13, 2541–2548. [DOI] [PubMed] [Google Scholar]
- Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D. G., Zhang Y., Kaelin W. G., Jr (2007). The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900. [DOI] [PubMed] [Google Scholar]
- Kothinti R. K., Blodgett A. B., Petering D. H., Tabatabai N. M. (2010). Cadmium down-regulation of kidney Sp1 binding to mouse SGLT1 and SGLT2 gene promoters: possible reaction of cadmium with the zinc finger domain of Sp1. Toxicol. Appl. Pharmacol. 244, 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O. H., Park J. L., Kim M., Kim J. H., Lee H. C., Kim H. J., Noh S. M., Song K. S., Yoo H. S., Paik S. G., et al. (2011). Aberrant up-regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem. Biophys. Res. Commun. 406, 539–545. [DOI] [PubMed] [Google Scholar]
- Lei Y. X., Wei L., Wang M., Wu G. R., Li M. (2008). Malignant transformation and abnormal expression of eukaryotic initiation factor in bronchial epithelial cells induced by cadmium chloride. Biomed. Environ. Sci. 21, 332–338. [DOI] [PubMed] [Google Scholar]
- Luchese C., Brandao R., de Oliveira R., Nogueira C. W., Santos F. W. (2007). Efficacy of diphenyl diselenide against cerebral and pulmonary damage induced by cadmium in mice. Toxicol. Lett. 173, 181–190. [DOI] [PubMed] [Google Scholar]
- Nawrot T. S., Staessen J. A., Roels H. A., Munters E., Cuypers A., Richart T., Ruttens A., Smeets K., Clijsters H., Vangronsveld J. (2010). Cadmium exposure in the population: from health risks to strategies of prevention. Biometals 23, 769–782. [DOI] [PubMed] [Google Scholar]
- Person R. J., Tokar E. J., Xu Y., Orihuela R., Ngalame N. N., Waalkes M. P. (2013). Chronic cadmium exposure in vitro induces cancer cell characteristics in human lung cells. Toxicol. Appl. Pharmacol. 273, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen P. B., Staller P. (2014). The KDM5 family of histone demethylases as targets in oncology drug discovery. Epigenomics 6, 277–286. [DOI] [PubMed] [Google Scholar]
- Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., Allis C. D. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C. T., Schreiber S. L., Mellor J., Kouzarides T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. [DOI] [PubMed] [Google Scholar]
- Shin S., Freedman V. H., Rissert R., Pollack R. (1975). Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Cell Biol. 72, 4435–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somji S., Garrett S. H., Toni C., Zhou X. D., Zheng Y., Ajjimaporn A., Sens M. A., Sens D. A. (2011). Differences in the epigenetic regulation of MT-3 gene expression between parental and Cd+2 or As+3 transformed human urothelial cells. Cancer Cell Int. 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son Y., Wang L., Poyil P., Budhraja A., Hitron J. A., Zhang Z., Lee J., Shi X. (2012). Cadmium induces carcinogenesis in BEAS-2B cells through ROS-dependent activation of PI3K/AKT/GSK-3β/β-catenin signaling. Toxicol. Appl. Pharmacol. 264, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueckle T. A., Lu Y., Davis M. E., Wang L., Jiang B. H., Holaskova I., Schafer R., Barnett J. B., Rojanasakul Y. (2012). Chronic occupational exposure to arsenic induces carcinogenic gene signaling networks and neoplastic transformation in human lung epithelial cells. Toxicol. Appl. Pharmacol. 261, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Clancy H. A., Kluz T., Zavadil J., Costa M. (2011). Comparison of gene expression profiles in chromate transformed BEAS-2B cells. PLoS ONE 6, e17982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Zhou X., Chen H., Li Q., Costa M. (2009). Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol. Appl. Pharmacol. 237, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M., Achanzar W. E., Qu W., Li G., Waalkes M. P. (2003). Effects of cadmium on DNA-(cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 286, 355–365. [DOI] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006). Histone demethylation by a family of jmjc domain-containing proteins. Nature 439, 811–816. [DOI] [PubMed] [Google Scholar]
- Verougstraete V., Lison D., Hotza P. (2002). A systematic review of cytogenetic studies conducted in human populations exposed to cadmium compounds. Mutat. Res. 511, 15–43. [DOI] [PubMed] [Google Scholar]
- Waalkes M. P. (2003). Cadmium carcinogenesis. Mutat. Res. 533, 107–120. [DOI] [PubMed] [Google Scholar]
- Waalkes M. P., Rehm S., Riggs C. W., Bare R. M., Devor D. E., Poirier L. A., Wenk M. L., Henneman J. R., Balaschak M. S. (1988). Cadmium carcinogenesis in male wistar [Crl:(WI)BR] rats: dose-response analysis of tumor induction in the prostate and testes and at the injection site. Cancer Res. 48, 4656–4663. [PubMed] [Google Scholar]
- Wang L., Luanpitpong S., Castranova V., Tse W., Lu Y., Pongrakhananon V., Rojanasakul Y. (2011). Carbon nanotubes induce malignant transformation and tumorigenesis of human lung epithelial cells. Nano Lett. 11, 2796–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006). JHDM2A, a jmjc-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495. [DOI] [PubMed] [Google Scholar]
- Zhou Z. H., Lei Y. X., Wang C. X. (2012). Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol. Sci. 125, 412–417. [DOI] [PubMed] [Google Scholar]
- Zhou X., Li Q., Arita A., Sun H., Costa M. (2009). Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol. Appl. Pharmacol. 236, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Sun H., Chen H., Zavadil J., Kluz T., Arita A., Costa M. (2010). Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 70, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Sun H., Ellen T. P., Chen H., Costa M. (2008). Arsenite alters global histone H3 methylation. Carcinogenesis 29, 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]