Abstract
LBD (LATERAL ORGAN BOUNDARIES DOMAIN) genes are essential to the developmental programs of many fundamental plant organs and function in some of the basic metabolic pathways of plants. However, our historical perspective on the roles of LBD genes during plant evolution has, heretofore, been fragmentary. Here, we show that the LBD gene family underwent an initial radiation that established five gene lineages in the ancestral genome of most charophyte algae and land plants. By inference, the LBD gene family originated after the emergence of the green plants (Viridiplantae), but prior to the diversification of most extant streptophytes. After this initial radiation, we find limited instances of gene family diversification in land plants until successive rounds of expansion in the ancestors of seed plants and flowering plants. The most dynamic phases of LBD gene evolution, therefore, trace to the aquatic ancestors of embryophytes followed by relatively recent lineage-specific expansions on land.
Keywords: LBD genes, Viridiplantae, Charophyte algae, Phylogenomics
LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD) genes encode a plant-specific family of transcriptional regulators with roles in the developmental programs of several fundamental plant organs, including roots, leaves, and flowers, and the regulation of nitrogen metabolism (Majer and Hochholdinger 2011). The defining feature of LBD genes is an N-terminal LBD composed of a zinc finger-like motif required for DNA-binding, a conserved glycine residue, and a leucine-zipper-like sequence required for protein–protein interactions (Shuai et al. 2002; Husbands et al. 2007; Matsumura et al. 2009). Two types of LBD genes are recognized on the basis of LBD structure: In Class I LBD genes the LBD includes both zinc finger-like and leucine zipper-like motifs, whereas only the zinc finger-like motif is present in Class II LBD genes (Shuai et al. 2002). LBD genes involved in developmental processes are of the Class I type, whereas Class II genes are involved in nitrogen metabolism (Majer and Hochholdinger 2011).
Previous phylogenetic analyses support subdivision of the LBD gene family into Class I and II lineages and largely congruent subdivisions of the larger Class I clade (Iwakawa et al. 2002; Majer and Hochholdinger 2011; Coudert et al. 2013). Yet, due to the limited genomic information available to prior researchers, the origin and diversification history of LBD genes have not been clarified. For example, the single LBD gene detected in an early transcriptome database for the moss Physcomitrella patens (Yang et al. 2006) has expanded to 26 loci with whole-genome sequencing (Coudert et al. 2013). Likewise perhaps, two partial sequences with similarities to the LBD were detected in the transcriptomes of two charophyte algae (Coleochaete orbicularis and Spirogyra pratensis), potentially extending the origin of the LBD gene family to an earlier period of plant evolution (Coudert et al. 2013). However, as these sequences were not included in phylogenetic analyses (Coudert et al. 2013), the evolutionary significance of charophyte algae in LBD gene phylogeny is unresolved. Also, to date, LDB genes of monilophytes and gymnosperms have never been previously investigated, obscuring the diversification history of the gene family during land plant evolution. Thus, despite their prominence in plant developmental programs, our historical perspective of LBD genes is fragmentary.
We have gathered LBD sequences that sample extant green plant diversity much more thoroughly than previously possible. Our analyses include previously underrepresented groups, such as algae, monilophytes, gymnosperms, and basal angiosperms. Here, we use these newly assimilated data to conduct phylogenetic analyses that elucidate the origin of the gene family and reconstruct its diversification history during green plant evolution.
Results and Discussion
We obtained 2,762 LBD sequences from 307 species representing the major plant lineages as follows: Angiosperms (38 species:1,213 sequences), gymnosperms (82:750), monilophytes (61:240), lycophytes (22:124), liverworts (23:77), hornworts (2:7), mosses (39:233), and charophyte algae (40:118).
Origin and Early Evolution of the LBD Gene Family
Among charophyte algae, LBD sequences were only found in species representing the orders Coleochaetales, Klebsormidiales, and Zygnematales. Although no sequences were found in members of the Charales, Chlorokybales, and Mesostigmatales, which occupy relatively basal branches in streptophyte phylogeny (Timme et al. 2012; Ruhfel et al. 2014; Wickett et al. 2014) and represented by a single sample in the 1,000 plants (1KP) data set, we cannot rule out the possibility that LBD genes exist in these taxa. On the other hand, no LBD homologs were detected in multiple species of chlorophyte algae and other non-Viridiplantae lineages in the 1KP transcriptome data sets and publically available genome sequences. The phylogenetic distribution of LBD genes, therefore, indicates that they constitute a green plant-specific gene family that originated sometime during the early evolutionary history of charophyte algae.
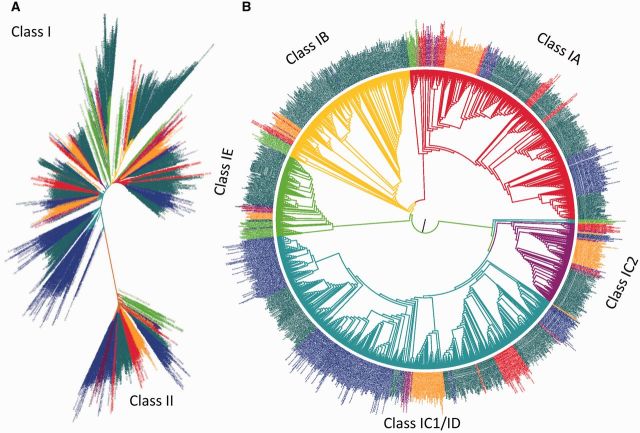
These newly acquired gene sequences allowed us to perform the first detailed phylogenetic analysis of the earliest phases of LBD gene family evolution. The maximum-likelihood (ML) tree (fig. 1A) resolves two major gene lineages that correspond with the traditional distinction of Class I and Class II genes. The Class I branch of the phylogenetic tree may be rerooted to show four gene lineages that correspond well with previously proposed groups of Class I genes (fig. 1B). Three of these correspond with the IA, IB, and IE Classes, whereas the fourth includes both Classes IC and ID as defined by Yang et al. (2006) and Coudert et al. (2013). Importantly, charophyte algae are placed basally with respect to land plants in the Class II and all four of the Class I gene lineages (fig. 1 and supplementary figs. S1–S5, Supplementary Material online), indicating that all five originated before the diversification of the streptophytes (charophyte algae and land plants). Accordingly, the initial radiation of the LBD gene family established five major branches of the gene family prior to the transition of plants from aquatic to terrestrial habitats.
Fig. 1.
ML phylogeny of LBD proteins. (A) Phylogram of global relationships among LBD genes shows that a long branch separates Class II proteins from the larger Class I clade. (B) Class I lineage of LBD proteins rooted along the branch separating Classes IA, IB, and IE, from Class IC/ID. Colored taxon names indicate the presence of charophyte algae (green), bryophytes (orange), lycophytes (purple), monilophytes (red), gymnosperms (blue), and angiosperms (teal), in all five of the main sublineages of the gene family.
Evolution of LBD Genes during Land Plant Diversification
Our expanded sampling also offers new insights into gene family evolution after the colonization of land approximately 450 Ma to the radiation of flowering plants during the past approximately 130 My. To explore this vast period of evolutionary history, we here evaluate LBD gene expansion(s) in the context of each of the major land plant lineages.
Bryophytes
Mosses, liverworts, and hornworts (collectively bryophytes, typically reconstructed as a paraphyletic grade) average 6.0, 3.3, and 3.5 LBD genes per species, respectively, distributed among six main clades (or grades): One each in the Class IA, IB, IE, and Class II gene lineages, and two in the Class IC/ID gene lineage (supplementary figs. S1–S5, Supplementary Material online). Notably, the latter two are placed in either of two reciprocally sister gene lineages that include all other land plants (lycophytes, monilophytes, gymnosperms, and flowering plants). This phylogenetic structure indicates that an ancient gene duplication event affected the ancestral Class IC/ID locus of the embryophytes (land plants). One of the descendant clades includes the Arabidopsis LBD13 and LBD15 genes (supplementary fig. S3, Supplementary Material online) and therefore corresponds with the Class IC2 group of Coudert et al. (2013), whereas the second is composed of their remaining Class IC/ID groups (i.e., Class IC1, ID1, and ID2).Of the five LBD gene lineages established in charophyte algae only the Class IC/ID lineage shows evidence of diversification prior to the radiation of extant land plants.
Lycophytes
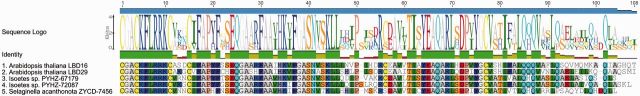
On average we found 5.6 genes per lycophyte species, and as in the bryophytes, these genes form single clades within the major LBD gene lineages, including the two sublineages of Class IC/ID genes. Notably, as also reported by Coudert et al. (2013), we confirm that the genome sequence of the lycophyte Selaginella moellendorphii does not contain a Class IB gene (supplementary fig. S2, Supplementary Material online). As the Class IB gene lineage includes all LBD genes with known roles in root development, including LBD16 and LBD29 of Arabidopsis, its absence in Selaginella was noted by Coudert et al. (2013) to propose that the independently evolved roots of lycophytes (Pires and Dolan 2012) may also have distinct genetic programs relative to other vascular plants. However, we have identified genes from Selaginella acanthonota and a species of Isoetes that possess the signature motif of Class IB proteins, “CGACKFLRRKC” (fig. 2; see also Yang et al. 2006), and which are placed in the Class IB clade with strong bootstrap support (supplementary fig. S2, Supplementary Material online). These lycophyte genes were obtained from transcriptome sequencing of vegetative shoot tissues, therefore whether they are also expressed in roots is still unknown. Nevertheless, Class IB LBD genes have not been entirely lost during lycophyte evolution, and are available to function in root development as they do in other vascular plants.
Fig. 2.
Alignment of Arabidopsis, Selaginella, and Isoetes Class IB LBD proteins. Note that the characteristic motif of the angiosperm Class IB sequences (“CGACKFLRRKC,” residues 1–11) is conserved in the lycophyte species.
Monilophytes
The monilophytes average 3.9 genes per species, fewer than the number of major LBD gene lineages, but are represented in all lineages, including the two Class IC/ID lineages descended from the embryophyte duplication discussed above. The lower average gene complement of LBD genes in monilophytes relative to mosses and lycophytes, therefore, may represent insufficient sampling depth of monilophyte transcriptomes rather than extinction of LBD gene lineages. In general, monilophytes form single clades in the six land plant LBD genes lineages, but a few smaller clades of monilophyte sequences are scattered among seed plant clades in the Class IA (supplementary fig. S1, Supplementary Material online) and Class II (supplementary fig. S5, Supplementary Material online) gene lineages. As these do not represent duplicate sets of the main monilophyte clades, it is unlikely that they represent ancient gene family expansions along the ancestral lines below extant monilophytes or euphyllophytes (monilophytes and seed plants).
Spermatophytes (Gymnosperms and Angiosperms)
Gymnosperms average 9.2 LBD genes per species, suggesting an approximately 2-fold increase over nonseed plants, with expansions into three major clades in the Class II lineage, four in the Class IC/ID lineage, and two in the Class IA lineage. These gymnosperm lineages are each associated with clades of flowering plant genes, indicating substantial gene family expansion prior to the radiation of all living seed plants. Yet, angiosperms possess an average of 31.9 LBD genes, suggesting a 3-fold increase over gymnosperms. This increase may be explained in part by the more complete sampling of LBD genes in sequenced angiosperm genomes, but phylogenetic reconstruction also implicates expansions during flowering plant diversification. We found 16 LBD genes in the Amborella genome, which is approximately twice as many as the average gymnosperm and, may be associated with the ancient whole-genome duplication (WGD) in the ancestor of all flowering plants (Jiao et al. 2011; Amborella Genome Project 2013). Further, 42 genes have been reported for Arabidopsis (Iwakawa et al. 2002), and similar numbers have been reported for Vitis and Populus (Coudert et al. 2013). The 3-fold increase relative to Amborella may relate to the multiple WGD events that predate the origin of core eudicots (Jiao et al. 2012). Similarly, multiple WGD events prior to the origin of the grasses (Paterson et al. 2012) may have led to the accrual of 35 (Yang et al. 2006) and 43 (Majer and Hochholdinger 2011) LBD genes in rice and maize, respectively. In addition to the above WGD events, a remarkable instance of LBD gene family amplification prior to the radiation of the angiosperms is seen in the Class IE lineage. Here, the gymnosperms (Zamia and Astrotaxus) are sister to four angiosperm-wide gene lineages—three with a basally positioned Amborella gene and one with Amborella placed outside the main angiosperm group (supplementary fig. S4, Supplementary Material online). The only functionally characterized Class IE gene, POLLEN CAR (AtLBD27), is expressed specifically in microspores (Oh et al. 2010).
Conclusion
LBD genes originated and diversified during the early evolution of charophyte algae. This initial diversification established five gene lineages in the ancestor of the streptophytes. A subsequent expansion in one of these lineages, Class C/D, led to a total of six LBD gene lineages in the land plants. Lineage-specific expansions can be found in almost all major plant groups, but the gene family has expanded most appreciably in the seed plants, especially in the flowering plants. We also find that the Class IB gene lineage that has been associated with root development (see Coudert et al. 2013) is present in at least some lycophytes. Class IB LBD genes may therefore represent a conserved component of root development during plant evolution.
Materials and Methods
LBD homologs were identified through BLASTp searches using Arabidopsis LBD proteins to query protein databases provided by the Joint Genome Institute (JGI, http://www.phytozome.net/, last accessed on April 6, 2015), the Amborella genome project (http://www.amborella.org/, last accessed on April 6, 2015), and the 1KP Consortium (https://www.bioinfodata.org/Blast4OneKP/, last accessed on April 6, 2015). In addition, tBLASTn searches were run locally on transcriptome assemblies downloaded from the Ancestral Angiosperm Genome Project (http://ancangio.uga.edu/, last accessed on April 6, 2015), and resulting hits translated using the ORF finder function of the Geneious package (version 6.4; created by Biomatters, available from http://www.geneious.com/, last accessed on April 6, 2015). BLASTp and tBLASTn searches were run under default parameters, adjusting E value to 1 if necessary. Amino acid alignments were constructed using the FFT-NS-1 algorithm under default settings of MAFFT (Katoh et al. 2002) implemented in Geneious. Alignments were trimmed to the LBD region and the optimal protein substitution model (JTT [Jones, Taylor, and Thorton]) determined using a perl script available at http://sco.h-its.org/exelixis/web/software/raxml/, last accessed on April 6, 2015. ML topology searches were performed with RAxML version 8.0.0 (Stamatakis 2014) using 100 separate heuristic searches from random starting trees, whereas bootstrap analyses were based on 1,000 pseudoreplicates. Phylogenetic trees were examined and manipulated with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed on April 6, 2015).
Supplementary Material
Sequence alignment, accession data, and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the China Scholarship Council (CSC, No. 2011853505 to F.H.) and the National Science Foundation (NSF IOS-0922742). The authors are grateful to Gane Ka-Shu Wong (Department of Biological Sciences, University of Alberta) for access to the 1KP transcriptome data sets. F.H. is an equal contributor to aspects of this project.
References
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Dievart A, Droc G, Gantet P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol Biol Evol. 2013;30:569–572. doi: 10.1093/molbev/mss250. [DOI] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers J, McKain M, McNeal J, Rolf M, Ruzicka D, Wafula E, Wickett N, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J Cell Mol Biol. 2009;58:525–537. doi: 10.1111/j.1365-313X.2009.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Park KS, Twell D, Park SK. The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 2010;64:839–850. doi: 10.1111/j.1365-313X.2010.04374.x. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Wang X, Li J, Tang H. Ancient and recent polyploidy in monocots. In: Soltis PS, Soltis DE, editors. Polyploidy and genome evolution. Berlin Heidelberg: Springer; 2012. pp. 93–108. [Google Scholar]
- Pires ND, Dolan L. Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond B Biol Sci. 2012;367:508–518. doi: 10.1098/rstb.2011.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;7:e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006;39:248–262. doi: 10.1016/j.ympev.2005.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.