Abstract
Several hypotheses have been proposed regarding the origin and evolution of the secretin family of peptides and receptors. However, identification of homologous ligand–receptor pairs in invertebrates and vertebrates is difficult because of the low levels of sequence identity between orthologs of distant species. In this study, five receptors structurally related to the vertebrate class B1 G protein-coupled receptor (GPCR) family were characterized from amphioxus (Branchiostoma floridae). Phylogenetic analysis showed that they clustered with vertebrate parathyroid hormone receptors (PTHR) and pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon receptors. These PTHR-like receptors shared synteny with several PTH and PACAP/glucagon receptors identified in spotted gar, Xenopus, and human, indicating that amphioxus preserves the ancestral chordate genomic organization of these receptor subfamilies. According to recent data by Mirabeau and Joly, amphioxus also expresses putative peptide ligands including homologs of PTH (bfPTH1 and 2) and PACAP/GLUC-like peptides (bfPACAP/GLUCs) that may interact with these receptors. Functional analyses showed that bfPTH1 and bfPTH2 activated one of the amphioxus receptors (bf98C) whereas bfPACAP/GLUCs strongly interacted with bf95. In summary, our data confirm the presence of PTH and PACAP/GLUC ligand–receptor pairs in amphioxus, demonstrating that functional homologs of vertebrate PTH and PACAP/glucagon GPCR subfamilies arose before the cephalochordate divergence from the ancestor of tunicates and vertebrates.
Keywords: amphioxus, GPCR, PACAP/glucagon, PTH
Introduction
Early in vertebrate evolution, after the split with urochordates, there were two rounds of whole-genome duplication (WGD), with an additional third round of WGD specific to the teleost lineage (Jaillon et al. 2004; Putnam et al. 2008; Santini et al. 2009). Subsequently, duplicate genes originating from the ancestral chordate complement are thought to have facilitated development of vertebrate innovations (Holland et al. 1994; Dehal and Boore 2005; Carroll 2008; Putnam et al. 2008; Holland 2013). Since amphioxi (cephalochordates) and tunicates (urochordates) diverged from the vertebrate lineage before the WGD events, they have been extensively used as models for investigating the evolutionary origin of vertebrates. Although tunicates share a more recent common ancestor with vertebrates than amphioxus (Delsuc et al. 2006; Putnam et al. 2008), amphioxus is considered to resemble ancestral chordates more closely, because tunicates have evolved quickly (Tsagkogeorga et al. 2012; Holland 2013).
G protein-coupled receptors (GPCR) constitute the largest protein family bridging extracellular stimuli with intracellular responses (Fredriksson et al. 2005). The vertebrate GPCR gene expansions may facilitate survival, as these receptors provide more sensory information from interactions with the environment and allow for more complex homeostatic regulation (Strotmann et al. 2011). Members of the class B1 GPCR, also known as the secretin receptor family, are distinguishable from other GPCR classes because of the highly conserved sequences in the seven transmembrane domains and relatively long N-termini (∼120 amino acids) with disulfide bridges critical for ligand binding (Harmar 2001; Couvineau et al. 2004; Harmar et al. 2012). Receptors of the bilaterian-specific GPCR family interact with a family of physiologically important peptide hormones were proposed to descend from adhesion receptors, prior to protostome–deuterostome divergence (Cardoso et al. 2006; Nordstrom et al. 2009). In humans, 15 class B1 receptors have been divided into five subfamilies based on structure: 1) corticotrophin-releasing hormone receptors (CRHR1 and CRHR2), 2) calcitonin receptors (CALCR and CALCR-like), 3) parathyroid hormone receptors (PTHR and PTH2R), 4) glucagon receptors (GCGR, glucagon-like peptide 1 receptor [GLP1R], GLP2R, glucagon-related peptide receptor [GCRPR], and gastric inhibitory polypeptide receptor), and 5) pituitary adenylate cyclase-activating polypeptide (PACAP) receptor ([PAC1R], vasoactive intestinal peptide/PACAP receptors [VPAC1R and VPAC2R], growth hormone-releasing hormone receptor [GHRHR], and secretin receptor [SCTR]; Hwang et al. 2013). Among the five subfamilies, only homologs of CRHR, CALCR, and PTHR have been identified in protostomes (Cardoso et al. 2006). Two additional groups of the family, Cluster A and pigment-dispersing factor receptor (PDFR)/PDFR-related, are proposed to be lost in vertebrates (Cardoso et al. 2014). In the amphioxus genome, orthologous genes expressing CRHR, CLACR, and the PTHR families are found, but not orthologs of glucagon and PACAP receptor subfamilies (Nordstrom et al. 2008; Hwang et al. 2013; Mirabeau and Joly 2013). Putative GCGR-like genes have been detected in the tunicate genome (Cardoso et al. 2006). Thus, after the divergence of amphioxus from the stem chordate lineage, the WGD events, and local gene duplications resulted in at least eight new class B1 GPCR members in vertebrates (Ng et al. 2010, 2012; Hwang et al. 2013).
Previous phylogenetic analyses have provided clues on the possible evolutionary course of these receptors and peptides (Cardoso et al. 2006, 2010, 2014; Ng et al. 2012; Pinheiro et al. 2012; Hwang et al. 2013). Recent genome sequencing has facilitated reexamination of the evolution of class B1 GPCRs and their ligands in cephalochordates. Recently, putative secretin-like peptides were identified from online genome data from amphioxus (Mirabeau and Joly 2013). In this study, the functional properties of several putative class B1 receptors of the amphioxus (Branchiostoma floridae) were characterized. This data, in combination with phylogenetic and chromosome synteny analyses, show that the amphioxus receptors bf98C and bf95 are functional homologs of the vertebrate PTH and PACAP/glucagon receptors, respectively, and indicate that PTH and PACAP/glucagon ligand–receptor pairs evolved before the cephalochordate split from the lineage that led to vertebrates.
Results
Identification of Putative Class B1 GPCRs and Corresponding Peptides in Amphioxus (B. floridae)
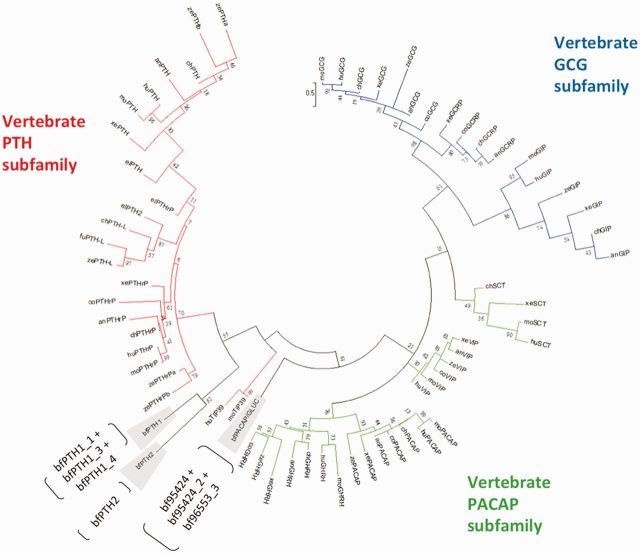
Six putative PTHR-like sequences were identified from the online B. floridae (bf) genome (http://genome.jgi-psf.org/pages/blast.jsf?db=Brafl1). Among these, five full-length cDNA sequences were cloned from sexually mature B. floridae and designated bf98A, bf98B, bf98C, bf95, and bf173, according to their locations on the scaffolds (supplementary fig. S1A–E, Supplementary Material online). These receptors possessed characteristics of class B1 GPCR, with seven conserved transmembrane domains and six N-terminal cysteine residues in the peptide binding domain, except bf173, which had a relatively shorter N-terminus (∼75 amino acids) with no signal peptide and lacked one of the conserved cysteine residues (supplementary fig. S2, Supplementary Material online). Phylogenetic analysis (fig. 1) placed all five putative bfPTHR-like receptors at the base of the clade comprising the vertebrate PTHR subfamilies, with tunicate PTHR-like sister to the vertebrate orthologs. Among the five bfPTHR-like sequences, bf98A, bf98B, and bf98C formed a cluster (with 95% support value) immediately basal to the vertebrate/tunicate PTHR subfamily. It is worth noting that the nodes grouping bf95 and bf173 with the remaining PTHRs had low bootstrap support (54% and 48% in fig. 1, respectively). To further evaluate the tree topology, the phylogenetic tree with different parameter (JTT +I +G without F, the second best-fit model from Protest) was calculated (supplementary fig. S7, Supplementary Material online). The overall topology is the same and the bootstrap value of the nodes that separating bf95 and bf173 are only slightly different (49% and 50%). In addition, an alternative phylogenetic method, Bayesian inference (Ronquist et al. 2012), was applied to the same alignment (supplementary fig. S8, Supplementary Material online). These new analyses support the trees that were generated by the maximum likelihood method in most of the nodes, except that bf95 is now placed at the base of the clade comprising the vertebrate PACAP/glucagon receptor. This indicates the identification of bf95 is not reliable by phylogenetic analysis and further evidence is required to prove the true identity of this receptor.
Fig. 1.
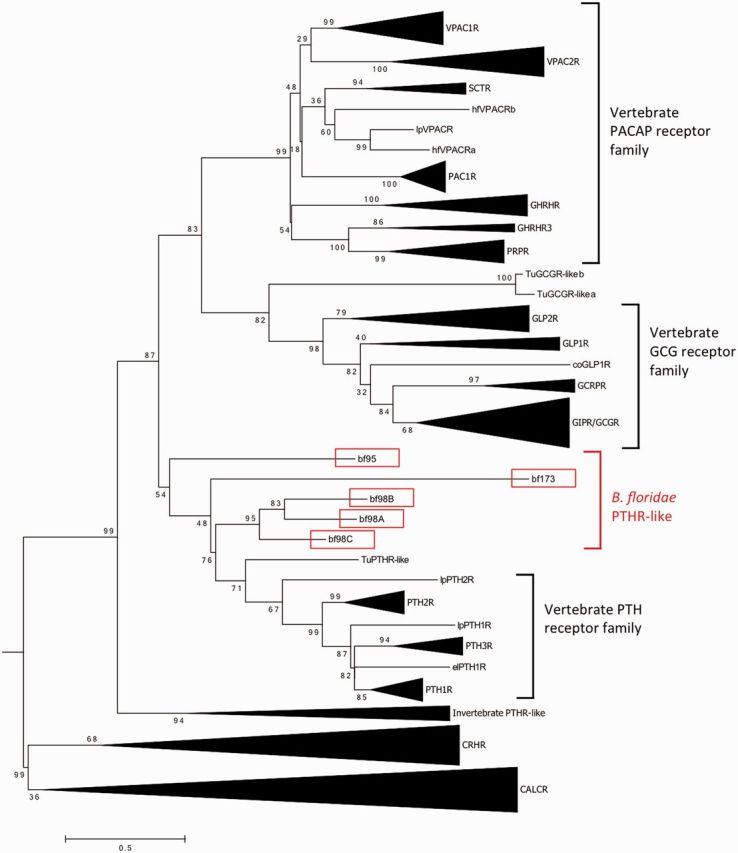
Phylogenetic tree of bfPTHR-like and vertebrate PTHR, PACAP and Glucagon receptor families. The maximum likelihood tree was generated using Mega 6.06. The trees were calculated by maximum likelihood method with the JTT +I, +G, +F, model. One thousand bootstrap simulations were used to test the reliability of branching. The bfPTHR-like genes are highlighted in red boxes. CALCR and CRHR subfamilies were used as outgroups. Accession numbers of the sequences used are listed in supplementary table S3, Supplementary Material online. The uncompressed version of this phylogenetic tree is showed in supplementary figure S6, Supplementary Material online. Species abbreviation: coelacanth (co), elephant shark (el), lamprey (lp), tunicate (Tu) and Branchiostoma floridae (bf). Number of sites used in this phylogenetic reconstruction is 277.
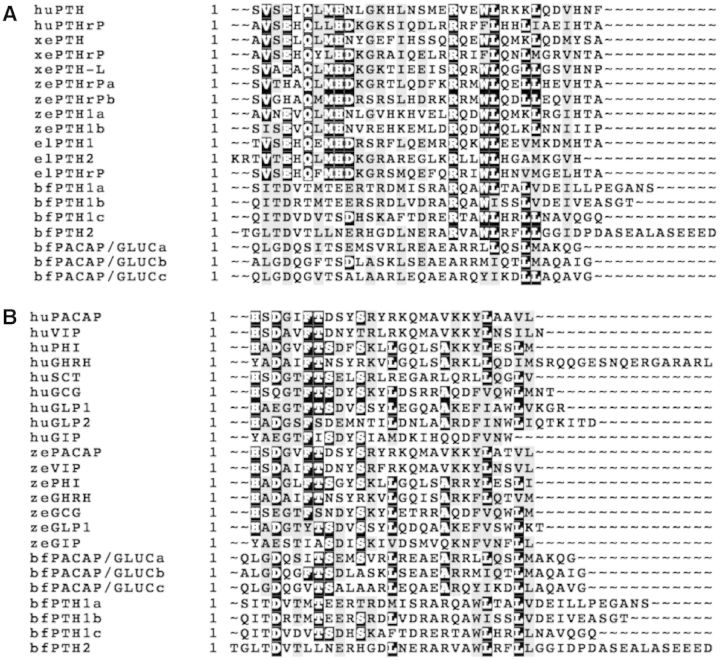
Three putative secretin-like peptide precursors (bfPTH1, bfPTH2, and bfPACAP/GLUC) were uncovered from online genome data (Mirabeau and Joly 2013; supplementary fig. S3A–C, Supplementary Material online). Sequence of the predicted bfPTH1 precursor is encoding three putative PTH-like peptides (bfPTH1a, bfPTH1b, and bfPTH1c). The bfPTH2 gene is encoding one putative PTH-like peptide (bfPTH2). A precursor gene encodes three putative PACAP/GLUC-like peptides (bfPACAP/GLUCa, bfPACAP/GLUCc, and bfPACAP/GLUCb). An alignment of the predicted mature peptide sequences with those in the same family from different species is shown in figure 2. As shown in the alignment of PTH and PTHrP (fig. 2A), two highly conserved regions are found in the vertebrate PTHs and PTHrPs. One of the regions is located near the N-terminus with VxExQxMHD sequence, whereas the other is close to the C-terminus with RxxWLxxLL sequence. In amphioxus peptides, the C-terminal RxxWLxxLL sequence is highly conserved in bfPTHs. But this sequence is not conserved in bfPACAP/GLUCs. In the alignment between amphioxus peptides with vertebrate secretin peptide family (fig. 2B), several residues are identical across species and they are evenly distributed along the peptides. The number of conserved residues in bfPACAP/GLUCs (with 6–7 identical residues) is slightly more than bfPTHs (with 4–5 identical residues). In summary, the alignments indicate that the bfPTHs are structurally more similar to vertebrate PTHs and PTHrPs when compare to bfPACAP/GLUCs, especially at the region near the C-termini. The bfPACAP/GLUCs is slightly more conserved than bfPTHs when compare to vertebrate secretin peptide family.
Fig. 2.
Alignment of (A) bfPTH-like and bfPACAP/GLUCs with vertebrate PTH and (B) bfPTH-like and bfPACAP/GLUsC with human and zebrafish secretin peptide family. The alignment was generated using the ClustalW and displayed by BOXSHADE software (version 3.2, by ExPASy Bioinformatics Resource Portal, http://www.ch.embnet.org/). Black background indicates conserved residues, while the gray background indicates similar residues. bf, Branchiostoma florida; ef, elephant shark; hu, human; xe, Xenopus; ze, zebrafish.
Phylogenetic analysis of these propeptide sequences (fig. 3) led to grouping of bfPTH1 and bfPTH2 with members of the vertebrate PTH/PTHrP family and bfPTH1 and bfPTH2 clustered together to form a basal clade. The bfPACAP/GLUC propeptides located at the base of the clade comprising the vertebrate PACAP and glucagon propeptides. Similar to the receptor, low support values were found in some critical nodes. But this tree topology can be reproduced in other phylogenetic analyses using other parameters (JTT +G +F without I, second best-fit model by Protest, supplementary fig. S9, Supplementary Material online) and in Bayesian inference method (supplementary fig. S10, Supplementary Material online). These data, therefore, suggest that homologous PTH and PACAP/GLUC ligands are found in cephalochordates.
Fig. 3.
Phylogenetic analysis of bfPTH-like and bfPACAP/GLUCs peptide precursors with vertebrate PTH, GCG and PACAP families. Full-length precursors amino acid sequences from vertebrates anole lizard (an), chicken (ch), elephant shark (el), human (hu), mouse (mo), Xenopus (xe), zebrafish (ze), and invertebrate Branchiostoma floridae (bf) were used to generate a maximum likelihood tree using Mega 6.06 (with JTT model and combined with +I, +G, +F). One thousand bootstrap simulations were used to test the reliability of branching. Vertebrate PACAP, GCG, and PTH subfamilies are highlighted in different colors. Predicted mature peptides within bfPTH1, bfPTH2, and bfPACAP/GLUCs precursors are labeled next to the gene names. Number of sites used in this phylogenetic reconstruction is 57.
Evidence of Functional Pairing of Amphioxus PTH-Like Peptides and Receptors via the cAMP Assay
Activation of cAMP production is a common pathway utilized by all class B1 GPCRs. In this study, all cloned amphioxus receptors were treated with a wide range of synthetic amphioxus and vertebrate PTH-like and PACAP/GLUC peptides for measurement of cAMP production. In the peptide screening, only two receptors, bf98C and bf95, showed strong responses. Interestingly, bf95 was activated by all three bfPACAP/GLUCs, but not other ligands, including vertebrate PACAP/GLUC peptides (fig. 4B). bf98C was activated strongly upon treatment with all amphioxus and lamprey PTH-like (lpPTH, lpPTHrPa, and lpPTHrPb), as well as human PTHrP, and weakly by human PTH, lamprey/zebrafish PACAP, and goldfish vasoactive intestinal petpide (VIP) (fig. 4A). None of the tested peptides was able to stimulate cAMP production in bf98A, bf98B, and bf173-transfected cells (fig. 4C–E), except human PTH, which weakly stimulated cAMP synthesis in bf98B-expressing cells (fig. 4D). The cells transfected with empty pcDNA3.1 were served as the negative control and none of the peptides can stimulate cAMP production in these cells (fig. 4F). To show similar transfection efficiencies, real-time polymerase chain reaction (PCR) was performed using first strand cDNA generated from receptor-transfected Chinese hamster ovary (CHO)-K1 cells. Real-time PCR revealed that all receptors are expressed in similar levels (supplementary fig. S4, Supplementary Material online). This control validates that the negative responses of bf98b and bf173 are not due to problems related to transfection efficiency or transcript level of the receptors.
Fig. 4.
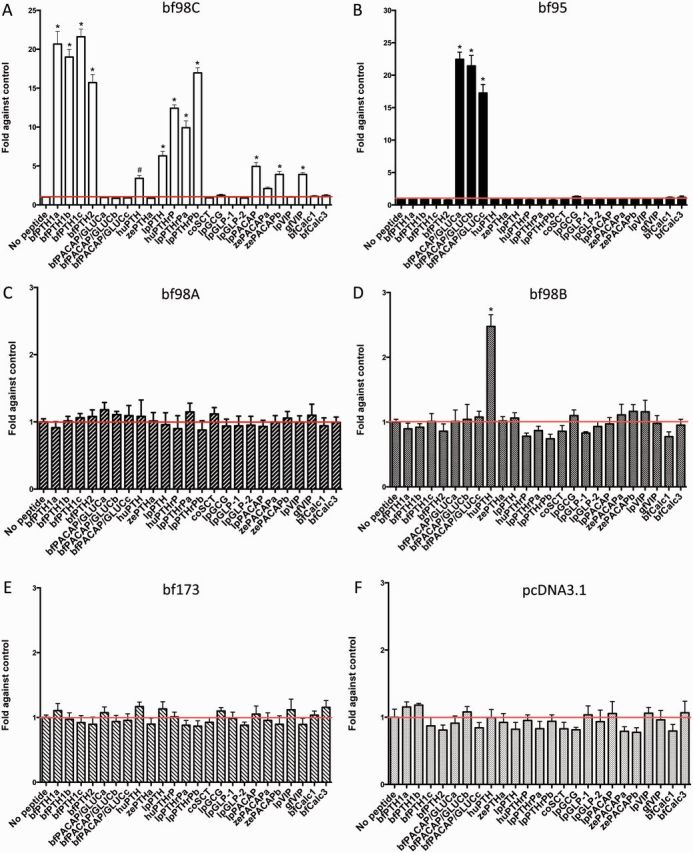
cAMP response of bfPTHR-like receptors to various peptides. Intracellular cAMP accumulation in response to 100 nM amphioxus and vertebrate superfamily peptides was tested on CHO-K1 cells transiently transfected with (A) bf98C, (B) bf95, (C) bf98A, (D) bf98B, (E) bf173, and (F) pcDNA3.1, negative control. Peptide species: Branchiostoma floridae (bf), coelacanth (co), goldfish (gf), human (hu), sea lamprey (lp), and zebrafish (ze). Fold changes in cAMP were obtained by comparing with the control (no peptide treatment). The red lines represent the control value. Data are means ± SEM from at least three experiments performed in duplicate. All data were analyzed by one-way analysis of variance (ANOVA) and followed by Dunnett’s test using the computer software PRISM (GraphPad Software Inc., San Diego, CA). *, P ≤ 0.001 and #, P ≤ 0.05.
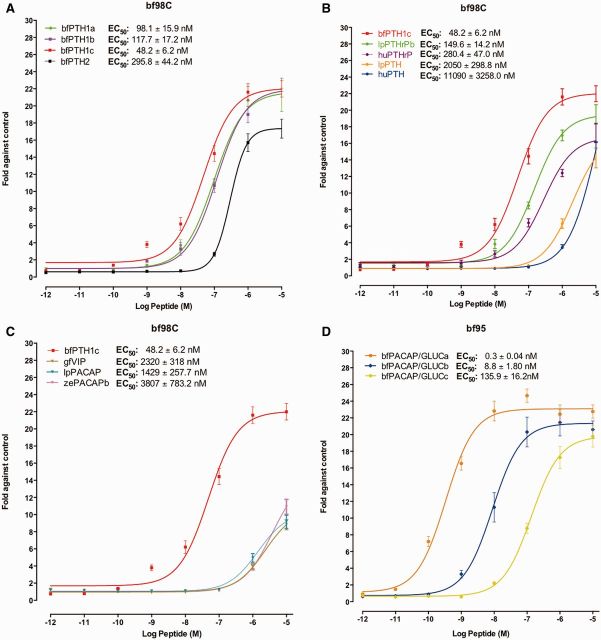
Peptides displaying positive results were further examined with graded doses of peptides to obtain dose-dependent curves (fig. 5). For bf98C, all amphioxus synthetic endogenous ligands (bfPTH1a, bfPTH1b, and bfPTH1c; fig. 5A) derived from the bfPTH1 precursor were able to trigger dose-dependent cAMP production with the affinity order: bfPTH1c > bfPTH1a ≈ bfPTH1b > bfPTH2. Regarding vertebrate PTH peptides (fig. 5B), lamprey PTHrPb and human PTHrP were able to stimulate cAMP production in bf98C receptor-transfected cells, whereas lamprey PTH and human PTH had lower affinity. On the other hand, lamprey PACAP, zebrafish PACAPb, and goldfish VIP were very weak agonists for bf98C (fig. 5C).
Fig. 5.
cAMP response of bf98C and bf95 to graded concentrations of peptides. Effects of (A) bfPTH-like, (B) vertebrate PTH and PTHrP, and (C) vertebrate PACAP or VIP on graded peptide concentrations assessed as intracellular cAMP accumulation in bf98C-expressing cells. (D) Measurement of intracellular cAMP elevation in CHO-K1 cells transiently expressing bf95 in response to graded concentrations of endogenous ligands, bfPACAP/GLUC peptides (bfPACAP/GLUCa, bfPACAP/GLUCb, and bfPACAP/GLUCc). Data are means ± SEM from at least three experiments performed in triplicate.
For receptor bf95, the synthetic bfPACAP/GLUCa peptide was the strongest agonist in the cAMP assay (fig. 5D). The other ligands expressed from the same precursor gene, bfPACAP/GLUCb and bfPACAP/GLUCc, exhibited significantly lower affinity. As these bfPACAP/GLUC peptides are structurally similar to vertebrate PACAP/GLUC peptides, our results indicate that bf95 is a homolog of vertebrate PACAP/GLUC receptors. Thus, functional and bioinformatics data from this study have provided new evidence that amphioxus bf98C and bf95 receptors are functional homologs of the vertebrate PTHrP/PTH and PACAP/GLUC receptors, respectively.
For other amphioxus receptors, especially bf98A and bf173, they are not reactive to any of the peptides tested. We cannot exclude the possibility that the lack of activity is due to ineffective functional coupling of these amphioxus GPCRs with hamster Gs protein in CHO cells, although there were several recent reports indicating the capability of amphioxus GPCRs in activating Gs and Gq in mammalian cell lines (Bayliss and Evans 2013a, 2013b; Xu et al. 2015).
To further explore the cross-species reactivity, we had tested the effects of bfPTHs and bfPACAP/GLUCs on human receptors by cAMP assay. Interestingly, as shown in our data in supplementary figure S5, Supplementary Material online, both bfPTH1a and bfPTH2 are able to active the human PTH2R, but not to human PTH1R (supplementary fig. S5A and B, Supplementary Material online). The bfPTH1a peptide is a stronger agonist when compare to bfPTH2 (supplementary fig. S5F, Supplementary Material online). This functional study provided an addition evidence to support the idea that bfPTH1 and bfPTH2 are orthologs of the human PTH. This result also indicates the binding mechanism between PTH2R and PTH was well conserved throughout evolution from cephalochordates to mammals. For the human PACAP/GLUC receptors, we have tested PAC1R, VPAC2R, and GCGR (supplementary fig. S5C–E, Supplementary Material online), and none of the bfPTHs and bfPACAP/GLUCs is able to activate them.
Synteny Analysis of bfPTH-Like Paralogs in Amphioxus, Spotted Gar, Xenopus, and Human
The amphioxus genome exhibits synteny with vertebrates (Putnam et al. 2008). Accordingly, this animal provides an excellent model to investigate the evolutionary genesis of vertebrates (Holland and Sower 2010). Other than Xenopus and human as the representatives of amphibians and mammals in this synteny test, a basal actinopterygian, the spotted gar (Lepisosteus oculatus) was used since gars did not undergo the teleost-specific WGD (Amores et al. 2011).
In the synteny analysis (fig. 6), genes expressing four class B1 GPCR members (PTHR, CALCR, corticotropin receptor [CRHR], and PACAP/GLUC receptor) were located in three syntenic loci within the gnathostome genomes. The first locus (Locus 1) contained the genes expressing PTHR together with those expressing GHRHR, PAC1R, PRPR, VPAC1R, VPACR2, CRHR2, and CALCR. Theses GPCRs were located at a single locus in spotted gar (Ch. LG9) and Xenopus (scaffold NW_004668239). The gene environment of this locus was highly conserved within the vertebrates. The corresponding locus in humans was separated into three (Ch. 3, Chr. 7, and Ch. 10). Totally, 14 genes (excluded GPCR) were conserved in spotted gar, Xenoups and human. The second syntenic locus (Locus 2) included genes expressing CRHR1, PTH3R, and GHRHR3. A number of genes within this locus were conserved in the vertebrates including vat1, ddx42, dcaf7, dcakd, grb7, med1, pcgf2, psmb3, and pip4k2. In mammals, loss of the genes expressing PTH3R and GHRHR3 have been reported (Pinheiro et al. 2012; Tam et al. 2013). gcgr and glp2r were located within this locus in human, but they were located in different chromosomes or scaffolds in spotted gar and Xenopus. In spotted gar, the ddx5 gene near gcgr and glp2r was identified in human Ch. 17. In Xenopus, gcgr was located in scaffold NW_004668240 and colocalized with the glb1-like gene. Based on these findings, we propose that gcgr and glp2r may share the same ancestral locus with CRHRH1, PTH3R, and GHRHR3 before vertebrate evolution. PTH2R, SCTR, and CALCRL were located in the third conserved locus (Locus 3), except in Xenopus, where SCTR was not included in scaffold NW_004668244. Within this locus, eight genes were conserved across the vertebrates.
Fig. 6.
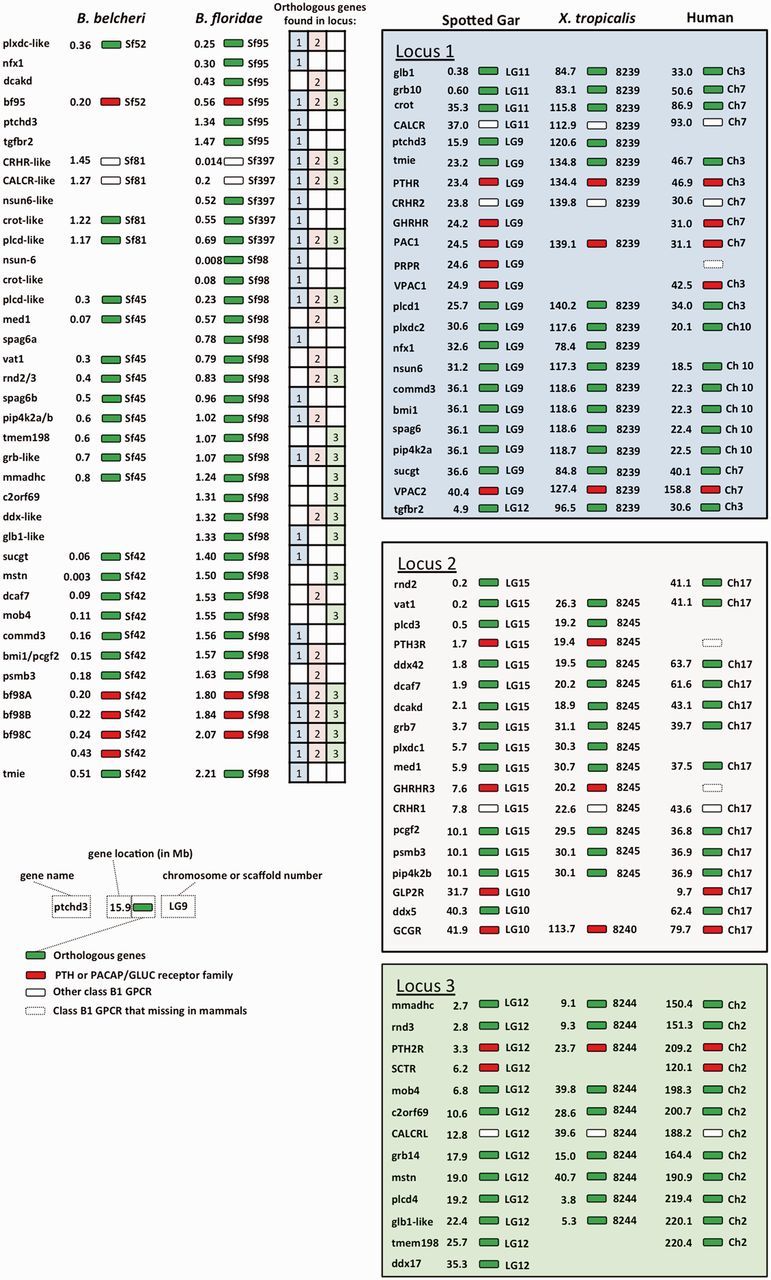
Synteny of PACAP/GCG and PTH peptide and receptor subfamily in amphioxus with spotted gar (Lepisosteus oculatus), Xenopus, and human. Chromosome numbers are on the right side of the indicated gene, and the gene locations (megabase) on the chromosome are shown on the left hand side. In vertebrates, paralogs (or orthologs) of each gene on different paralogons were aligned on the same row. Orthologous genes are labeled with green colors. PTH/PTHrP and PACPA/GLUC receptor family, including the putative amphioxus PTHR-like and PACAP/GLUC receptor, are highlighted in red. White boxes represent other class B1 GPCR. The GPCRs that missing in mammals are represented by white boxes with dash line. The table summarized all the connection of the orthologous genes between amphioxi and vertebrates. The strong connection between amphioxi and vertebrates syntenic loci implies that ancestral class B1 GPCRs were probably located on the same locus before the emergence of amphioxus.
In amphioxus, two functional receptors, bfPTHR-like (bf98C) and bfPACAP/GLUC receptor (bf95), were located on scaffolds 98 and 95. Many of the genes within these two scaffolds were found in the previously discussed syntenic loci in the vertebrates. In particular, orthologs of plcd-like and grb-like genes were identified in all the conserved loci. This evidence strongly suggests that the gene environment of the conserved loci in spotted gar, Xenopus, and human are inherited from ancestral locus/loci similar to the combined amphioxus scaffolds 98 and 95. In addition, the transmembrane inner ear gene was always positioned next to PTH1R in all three species and bf98C, further suggesting that bf98C is the homolog of vertebrate PTHR. Amphioxus CRHR-like and CALCR-like receptor genes were located on scaffold 397. Interestingly, scaffolds 397 and 98 shared a long segment with similar gene environment, which included three genes, nsun-6 and crot-like, and plcd-like, indicating the two scaffolds are potentially part of a single chromosome in amphioxus. In the recently released Chinese amphioxus (B. belcheri) genome, the PACAP/GLUC receptor, CRHR-like, CALCR-like, and PTHR-like receptor genes were located on scaffolds 52, 81, 45, and 42, respectively. These scaffolds contained gene environments highly conserved with those of B. floridae. These data further suggest that CRHR and CALCR are colocalized with PTHR and PACAP/GLUC receptor on the same locus in amphioxus. The table inside figure 6 provides a summary of the gene synteny of this amphioxus locus with the above mentioned vertebrates syntenic loci. Our result suggests that for many genes that are located on this amphioxus locus, their orthologs are also found in the vertebrate loci that contain the class B1 GPCRs. For example, excluding the class B1 GPCRs, the orthologus genes corresponding to 19 other genes in the amphioxus locus are also present in syntenic Locus 1, whereas 13 and 11 orthologus genes are observed in Locus 2 and Locus 3, respectively. In summary, the synteny analysis collectively suggests that class B1 GPCRs are located in the same locus in cephalochordate. In addition, the vertebrate class B1 GPCR family may have originated from an ancestral locus similar to the one found in the amphioxus.
Discussion
Coevolution of the PACAP/GLUC Ligand–Receptor Pair in Invertebrate to Vertebrate
Due to similarities in tissue expression, function, receptor interactions, gene organization, and mature peptide sequences, members of the vertebrate PACAP/GLUC subfamily have long been proposed to be originated from exon duplication of ancestral peptides following local gene duplication and 2 round WGD (Sherwood et al. 2000; Ng et al. 2012; Hwang et al. 2013). Evolution of this subfamily has resulted in seven genes after vertebrate expansion. However, even with considerable research efforts on the origin of the PACAP/GLUC subfamilies using invertebrates, no functional data are currently available from these animals.
At present, functional data on the PACAP receptor subfamily from the most anciently diverged lineage comes from the cyclostomes (jawless fishes; Ng et al. 2012), while functional receptors for the glucagon subfamily are found only in the teleost (Hwang et al. 2014). Our study describes the characterization of a more anciently diverged form that is the first invertebrate PACAP/GLUC receptor characterized. In view of the presence of the functional PACAP receptor in Japanese lamprey, the PACAP/GLUC receptor duplication presumably happened after the divergence of amphoxi but before the divergence of tunicates from the chordate stem lineage.
The maximum likelihood phylogenetic analysis, together with other studies, has placed the bfPTHR-like to the base of PTH receptor subfamily with low bootstrap value (fig. 1 and supplementary figs. S7 and S8, Supplementary Material online). Also, in Bayesian inference method, the bf95 is placed at the base of PACAP/glucagon receptor subfamily. This is a clear case where phylogenetic and bioinformatics studies could not provide sufficient detail information regarding the true identity of receptors. Our study suggests that bfPACAP/GLUCs, rather than bfPTH-like peptides, are the endogenous agonists of bf95. Thus, the bfPACAP/GLUC and bf95 ligand–receptor pair represents the most anciently diverged example of the PACAP/GLUC family in chordates.
By chromosome synteny analysis, the bfPTHR-like gene locus was found to share similar gene environment with PTH/PTHrP receptors in spotted gar, Xenopus and humans (fig. 6). According to the chromosome synteny pattern, scaffolds 98 and 95 appear to be close neighbors on the same chromosome in amphioxus. Therefore, we propose that ancestral chordate class B1 GPCRs were located on the same chromosomal locus, prior to the divergence of amphioxus. This observation in amphioxus is consistent with data from a large-scale genome comparison study (Hwang et al. 2013) that also hypothesized that all pre-2R ancestral members of class B1 GPCRs resided on the same ancestral chromosomal locus.
The predicted precursors of bfPACAP/GLUC genes were phylogenetically assigned to the cluster of the PACAP/GLUC peptide subfamily (fig. 3; Hwang et al. 2013; Mirabeau and Joly 2013). These bfPACAP/GLUC peptides share relatively low levels of similarity with members of the vertebrate PACAP/GLUC subfamilies, but nevertheless, several residues involved in receptor interactions of the two subfamilies are well conserved (fig. 2), including Phe6 and Tyr22 of VIP and PACAP in bfPACAP/GLUCb, and Thr/Ser7, Ile/Val23, and Leu26 of GLP1, GLP2 and glucagon in all bfPACAP/GLUCs, with bfPACAP/GLUCa containing Leu instead of Ile/Val23 (Sun et al. 2007; Bourgault et al. 2009; Underwood et al. 2010; Dejda et al. 2011; Moon, Kim, et al. 2012; Moon, Park, et al. 2012). Thus, bf95-bfPACAP/GLUCs pairing may have preserved some of the ancestral character for this family prior to the gene duplication that led to the PACAP and glucagon subfamilies. Even so, the tested lamprey peptides were unable to trigger an increase in intracellular cAMP synthesis in bf95-transfected cells, possibly due to the relatively low sequence similarity between vertebrate and amphioxus PACAP/GLUC peptides particularly at the N-termini, which are critical for ligand–receptor binding. Consistent with this, all these amphioxus PACAP/GLUC peptides are unable to activate several of the human PACAP/GLUC receptors tested. Unlike PTH and PTH receptors, the lack of cross amphioxus–human species activities of PACAP/GLUC peptides indicates either the missing of critical residue(s) for the activation of human receptors in these amphioxus peptides, and/or the fact that human receptors have evolved a different binding pocket. It will be interesting to study the structural and functional coevolution of class B1 GPCRs and their ligands in representative species from amphioxus to human in the future.
PTH Subfamily Ligand–Receptor Interactions in Invertebrates
A recent bioinformatics study in the protostome lineage showed that nematodes and arthropods possess the PTHR-like receptor, suggesting the existence of an ancient form of PTHR before the protostome-deuterostome spilt (Cardoso et al. 2014). In total, six bfPTH-like receptors were identified from the B. floridae genome. An unusually large number of receptors (nine in total) belonging to CALCR and CRHR subfamilies are also present in the amphioxus genome (Hwang et al. 2013). This many receptors from the PTHR, CALCR, and CHRHR subfamilies have not been observed in other vertebrate and invertebrate species. As there is only one PTHR-like in tunicates, we believe that ancestral chordates have only a single PTHR-like while other receptors in this gene family were generated by gene duplication events specific to amphioxus. In amphioxus, bf98A, bf98B, bf98C, and bf173 are closely related and are clustered together to form a subbranch, whereas bf95 was phylogenetically separated from its paralogs. These receptors produced by gene duplications may have developed other functions (neofunctionalization) or lost their function (nonfunctionalization) in amphioxus. Here, our data suggest that only bf98C is a functional receptor for bfPTH-like peptides. Specifically, in the cAMP assay, bf98C was found to be responsive to all bfPTH-like peptides as well as lamprey and human PTH or PTHrP. The cross-reactivity of bf98C to vertebrate PTHrP and PTH orthologs indicates preservation of ligand–receptor structural constraints throughout the chordate lineage was under considerable selective pressure. The functional assay data are consistent with protein sequence analysis showing the requirement of several key residues (T33, Q37, and F184) for mammalian PTH/PTHrP-PTHR interactions (Juppner et al. 1994; Gardella and Juppner 2001) found solely at the N-terminus of bf98C. Surprisingly, bf98C was also activated by lpPACAP, zePACAP and gfVIP at high doses. This finding demonstrates some degree of structural similarity between PTH and PACAP/GLUC ligand–receptor pairs. Activation of bf98C by PACAP and VIP also suggests that the PACAP/GLUC receptors retain structural constraints found in the gene from which they are descended with PTHR. So far, such cross-subfamily reactivity has not been described in PTH receptors in other species. When we use amphioxus PTH peptides to stimulate human PTH receptors, we found that both bfPTH1a and bfPTH2 peptides are capable of activating the human PTH2R. This further supports the preservation of PTH ligand–receptor structural constraints throughout the chordate lineage. Consider the high sequence similarity shared by bfPTH1 peptides, it is surprising to us that both bfPTH1b and bfPTH1c are unable activate the human PTH2R. The sequence comparison of bfPTH1a peptide with 1b and 1c peptides should therefore provide insights into the structural determinants that may be needed for activating PTH2R. Further studies are required to clarify the binding mechanism of bfPTHs with human PTH2R.
Conclusions
Our data support the existence of functional homologs of vertebrate PTH and PACAP/GLUC ligand–receptor pairs prior to the cephalochordate–vertebrate split. The similarities between bf98C and bf95 indicate that they shared a common ancestor and were duplicated before the divergence of amphioxus. Based on the collective findings, we propose that the genes encoding the PTHR, VPACR, CALCR, and CRHR subfamilies potentially evolved from a single ancestral locus in the genome of the ancestral chordate, similar to that found in amphioxus.
Materials and Methods
Animal Collection, Total RNA Isolation, and First-Strand cDNA Synthesis
Amphioxus (B. floridae) were collected from Old Tampa Bay, Florida, as described previously (Yu and Holland 2009). Adult (sexually mature) amphioxus total RNA was isolated from excised tissues with TriPure reagent (Invitrogen, Carlsbad, CA), in keeping with previous publications (Lee et al. 2009). Total RNA (4 µg) and anchored-oligo (dT)18 primer were used to synthesize first-strand cDNA with the Transciptor First-Strand cDNA Synthesis Kit (Roche).
Peptides
Sea lamprey VIP and PACAP (predicted amino acid sequences from the P. marinus preassembled genome) peptides were synthesized by Professor Alain Fournier and Professor Hubert Vaudry from the Institut National de la Santé et de la Recherche Mé dicale U413 (European Institute for Peptide Research, University of Rouen, France). Amphioxus peptides (PTH and PACAP/GLUC peptides, supplementary table S1, Supplementary Material online) and other peptides were synthesized by Genscript Inc. (Piscataway, NJ). Peptide sequences were based on the prediction of Mirabeau and Joly (2013). All synthetic peptides were greater than 95.0% pure.
Molecular Cloning of PTH-Like Receptors from B. floridae
Using BLAST analysis (tBLASTx) and the genome data from B. floridae (acquired from JGI B. floridae database http://genome.jgi-psf.org/Brafl1/Brafl1.home.html), putative PTH-like receptors in amphioxus were predicted based on the seven transmembrane domain sequences from zebrafish and mammalian PTHRs and PACAP/GLUC receptors (accession numbers shown in supplementary table S3, Supplementary Material online). According to the predicted sequences, conserved transmembrane domain sequences were selected to design specific primers for amplifying partial sequences of the putative PTH-like receptors. Using the confirmed partial sequences, specific primers (supplementary table S2, Supplementary Material online) were designed for performing 5′- and 3′-rapid amplification of cDNA ends, according to the manufacturer’s protocol (Invitrogen). Full-length cDNA sequences were cloned and confirmed by PCR using primers targeting the 5′- and 3′-UTR regions. The coding region of bfPTHR-like was subcloned into pcDNA3.1(+) vector for functional characterization.
Transient Expression of bfPTHR-Like in CHO-K1 Cells and cAMP Assay
CHO-K1 cells were cultured in minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 U/ml Penicillin, and 100 µg/ml streptomycin (Invitrogen) in a 5% (v/v) CO2 humidified chamber at 37 °C. For the cAMP assay, 3 × 105 cells were seeded onto a six-well plate (35 mm/well; Costar, San Diego, CA) and allowed to grow for 24 h before transfection. In each well, 2 µg receptor-pcDNA3.1 construct was transfected into CHO-K1 cells with X-treme GENE HP DNA Transfection Reagent (Roche) in 1:3 ratio (w/v). Two days later, transfected CHO-K1 cells were treated with peptides for 45 min. Measurement of intracellular cAMP was performed using the LANCE cAMP assay kit (Perkin Elmer, Waltham, MA) with Victor x4 multilabel reader (Perkin-Elmer) following the manufacturer’s protocol. Results were presented as fold change against the basal intracellular cAMP level (no peptide control). CHO-K1 cells transfected with pcDNA3.1 vector were used as the negative control. To confirm the expression of the receptors, cells were harvested for total RNA extraction (TriPure reagent, Invitrogen) after transfection. Five micrograms of total RNA were used for first strand cDNA synthesis as described before. The transcript levels of the amphioxus receptors were measured real-time PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and the 7300 Real Time PCR System (Applied Biosystems).
Phylogenetic Analysis
All amino acid sequences for phylogenetic analysis were obtained from the Ensembl Genome database, NCBI GeneBank database, or JGI Genome project. Accession numbers of the sequences used in these analyses are listed in supplementary table S3, Supplementary Material online. For class B1 receptor and secreitn-like peptide phylogenetic trees, full-length amino acid sequences of the respective precursors were used. Alignment of sequences for both trees was performed using subsidiary MUSCLE function with the default setting in MEGA6.06 (Tamura et al. 2013). The best-fit models of the trees were selected by ProtTest 3.0 (Abascal et al. 2005). The trees were calculated by maximum likelihood method with the JTT model and combined with +I: invariable sites, +G: rate heterogeneity among sites, and with or without +F: observed amino acid frequencies. One thousand bootstrap and partial deletion with 95% site coverage cutoff selected in the gaps/missing data treatment were used. Numbers on the nodes of the trees indicate the percentage of bootstrap replicates in which the labeled branch was reproduced. An alternative method, Bayesian inference, was performed using MrBayes version 3.2 (Ronquist et al. 2012) with JTT substitution model plus empirical frequency along with gamma distribution with invariant sites. In the MrBayes analysis, two Markov chain Monte Carlo runs with four chains were executed for 1,500,000 (for receptor tree) and 1,000,000 generations (for peptide tree). Each chain resulted in 15,000 for receptor tree and 10,000 for peptide tree. Twenty-five percent of the samples from two runs were discarded as “burn-in” and a 50% majority-rule consensus tree was constructed.
Chromosomal Synteny Analysis
Proteins on amphioxi scaffolds were retrieved and used for BLAST analysis (BLASTP). Spotted gar (L. oculatus), Xenopus tropicalis, and human proteins with E values less than 1e−20, greater than 30% identity, and greater than 40% coverage in BLASTP results were classified as conserved. The corresponding genes were used in the synteny analysis and marked on the chromosome region with class B1 GPCRs.
Statistical Analysis
In the functional assay, results are presented as ±SEM of triplicate assays from at least three independent experiments. GraphPad Prism version 5.0 (GraphPad Software, Inc., San Diego, CA) was used to plot sigmoidal curves in the cAMP assay.
Supplementary Material
Supplementary figures S1–S10 and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Prof. Linda Z. Holland from University of California San Diego for the generous supplies of the amphioxus samples and reading this manuscript. The authors also thank Prof. Hubert Vaudry and Prof. Alain Fournier for synthesis of the sea lamprey peptides. The present study was supported by the Hong Kong Government RGC Grant HKU/CRF/11G to B.K.C.C., and 770212 and 17112014 to L.T.O.L.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss A, Evans PD. Characterisation of AmphiAmR4, an amphioxus (Branchiostoma floridae) alpha(2)-adrenergic-like G-protein-coupled receptor. Invert Neurosci. 2013a;13:71–84. doi: 10.1007/s10158-012-0145-6. [DOI] [PubMed] [Google Scholar]
- Bayliss AL, Evans PD. Characterisation of AmphiAmR11, an amphioxus (Branchiostoma floridae) D2-dopamine-like G protein-coupled receptor. PLoS One. 2013b;8:e80833. doi: 10.1371/journal.pone.0080833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgault S, Vaudry D, Segalas-Milazzo I, Guilhaudis L, Couvineau A, Laburthe M, Vaudry H, Fournier A. Molecular and conformational determinants of pituitary adenylate cyclase-activating polypeptide (PACAP) for activation of the PAC1 receptor. J Med Chem. 2009;52:3308–3316. doi: 10.1021/jm900291j. [DOI] [PubMed] [Google Scholar]
- Cardoso JC, Felix RC, Power DM. Nematode and arthropod genomes provide new insights into the evolution of class 2 B1 GPCRs. PLoS One. 2014;9:e92220. doi: 10.1371/journal.pone.0092220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM. Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol. 2006;6:108. doi: 10.1186/1471-2148-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso JC, Vieira FA, Gomes AS, Power DM. The serendipitous origin of chordate secretin peptide family members. BMC Evol Biol. 2010;10:135. doi: 10.1186/1471-2148-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Couvineau A, Rouyer-Fessard C, Laburthe M. Presence of a N-terminal signal peptide in class II G protein-coupled receptors: crucial role for expression of the human VPAC1 receptor. Regul Pept. 2004;123:181–185. doi: 10.1016/j.regpep.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejda A, Bourgault S, Doan ND, Letourneau M, Couvineau A, Vaudry H, Vaudry D, Fournier A. Identification by photoaffinity labeling of the extracellular N-terminal domain of PAC1 receptor as the major binding site for PACAP. Biochimie. 2011;93:669–677. doi: 10.1016/j.biochi.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Schioth HB. Expansion of the superfamily of G-protein-coupled receptors in chordates. Ann N Y Acad Sci. 2005;1040:89–94. doi: 10.1196/annals.1327.011. [DOI] [PubMed] [Google Scholar]
- Gardella TJ, Juppner H. Molecular properties of the PTH/PTHrP receptor. Trends Endocrinol Metab. 2001;12:210–217. doi: 10.1016/s1043-2760(01)00409-x. [DOI] [PubMed] [Google Scholar]
- Harmar AJ. Family-B G-protein-coupled receptors. Genome Biol. 2001;2(12) doi: 10.1186/gb-2001-2-12-reviews3013. :REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Fahrenkrug J, Gozes I, Laburthe M, May V, Pisegna JR, Vaudry D, Vaudry H, Waschek JA, Said SI. Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br J Pharmacol. 2012;166:4–17. doi: 10.1111/j.1476-5381.2012.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ. Evolution of new characters after whole genome duplications: insights from amphioxus. Semin Cell Dev Biol. 2013;24:101–109. doi: 10.1016/j.semcdb.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Sower SA. “Insights of early chordate genomics: endocrinology and development in amphioxus, tunicates and lampreys": introduction to the symposium. Integr Comp Biol. 2010;50:17–21. doi: 10.1093/icb/icq039. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994;(Supplement):125–133. [PubMed] [Google Scholar]
- Hwang JI, Moon MJ, Park S, Kim DK, Cho EB, Ha N, Son GH, Kim K, Vaudry H, Seong JY. Expansion of secretin-like G protein-coupled receptors and their peptide ligands via local duplications before and after two rounds of whole-genome duplication. Mol Biol Evol. 2013;30:1119–1130. doi: 10.1093/molbev/mst031. [DOI] [PubMed] [Google Scholar]
- Hwang JI, Yun S, Moon MJ, Park CR, Seong JY. Molecular evolution of GPCRs: GLP1/GLP1 receptors. J Mol Endocrinol. 2014;52:T15–27. doi: 10.1530/JME-13-0137. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Juppner H, Schipani E, Bringhurst FR, McClure I, Keutmann HT, Potts JT, Jr, Kronenberg HM, Abou-Samra AB, Segre GV, Gardella TJ. The extracellular amino-terminal region of the parathyroid hormone (PTH)/PTH-related peptide receptor determines the binding affinity for carboxyl-terminal fragments of PTH-(1-34) Endocrinology. 1994;134:879–884. doi: 10.1210/endo.134.2.8299582. [DOI] [PubMed] [Google Scholar]
- Lee LT, Tam JK, Chan DW, Chow BK. Molecular cloning and mRNA distribution of pituitary adenylate cyclase-activating polypeptide (PACAP)/PACAP-related peptide in the lungfish. Ann N Y Acad Sci. 2009;1163:209–214. doi: 10.1111/j.1749-6632.2008.03661.x. [DOI] [PubMed] [Google Scholar]
- Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A. 2013;110:E2028–E2037. doi: 10.1073/pnas.1219956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon MJ, Kim HY, Park S, Kim DK, Cho EB, Park CR, You DJ, Hwang JI, Kim K, Choe H, et al. Evolutionarily conserved residues at glucagon-like peptide-1 (GLP-1) receptor core confer ligand-induced receptor activation. J Biol Chem. 2012a;287:3873–3884. doi: 10.1074/jbc.M111.276808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon MJ, Park S, Kim DK, Cho EB, Hwang JI, Vaudry H, Seong JY. Structural and molecular conservation of glucagon-like Peptide-1 and its receptor confers selective ligand-receptor interaction. Front Endocrinol. 2012b;3:141. doi: 10.3389/fendo.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Chow BK, Kasamatsu J, Kasahara M, Lee LT. Agnathan VIP, PACAP and their receptors: ancestral origins of today's highly diversified forms. PLoS One. 2012;7:e44691. doi: 10.1371/journal.pone.0044691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Lee LT, Chow BK. Insights into the evolution of proglucagon-derived peptides and receptors in fish and amphibians. Ann N Y Acad Sci. 2010;1200:15–32. doi: 10.1111/j.1749-6632.2010.05505.x. [DOI] [PubMed] [Google Scholar]
- Nordstrom KJ, Fredriksson R, Schioth HB. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol. 2008;8:9. doi: 10.1186/1471-2148-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom KJ, Lagerstrom MC, Waller LM, Fredriksson R, Schioth HB. The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol Biol Evol. 2009;26:71–84. doi: 10.1093/molbev/msn228. [DOI] [PubMed] [Google Scholar]
- Pinheiro PL, Cardoso JC, Power DM, Canario AV. Functional characterization and evolution of PTH/PTHrP receptors: insights from the chicken. BMC Evol Biol. 2012;12:110. doi: 10.1186/1471-2148-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood NM, Krueckl SL, McRory JE. The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon superfamily. Endocr Rev. 2000;21:619–670. doi: 10.1210/edrv.21.6.0414. [DOI] [PubMed] [Google Scholar]
- Strotmann R, Schrock K, Boselt I, Staubert C, Russ A, Schoneberg T. Evolution of GPCR: change and continuity. Mol Cell Endocrinol. 2011;331:170–178. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Sun C, Song D, Davis-Taber RA, Barrett LW, Scott VE, Richardson PL, Pereda-Lopez A, Uchic ME, Solomon LR, Lake MR, et al. Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS. Proc Natl Acad Sci U S A. 2007;104:7875–7880. doi: 10.1073/pnas.0611397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JK, Chow BK, Lee LT. Structural and functional divergence of growth hormone-releasing hormone receptors in early sarcopterygians: lungfish and Xenopus. PLoS One. 2013;8:e53482. doi: 10.1371/journal.pone.0053482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkogeorga G, Cahais V, Galtier N. The population genomics of a fast evolver: high levels of diversity, functional constraint, and molecular adaptation in the tunicate Ciona intestinalis. Genome Biol Evol. 2012;4:740–749. doi: 10.1093/gbe/evs054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, Rudolph R, Reedtz-Runge S. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J Biol Chem. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Bergqvist CA, Sundstrom G, Lundell I, Vaudry H, Leprince J, Larhammar D. Characterization of peptide QRFP (26RFa) and its receptor from amphioxus, Branchiostoma floridae. Gen Comp Endocrinol. 2015;210:107–113. doi: 10.1016/j.ygcen.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Yu JK, Holland LZ. Extraction of DNA from adult amphioxus tissue. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5287. pdb prot5287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.