Abstract
Medicago truncatula is a model legume species used to investigate plant–microorganism interactions, notably root symbioses. Massive population genomic and transcriptomic data now available for this species open the way for a comprehensive investigation of genomic variations associated with adaptation of M. truncatula to its environment. Here we performed a fine-scale genome scan of selective sweep signatures in M. truncatula using more than 15 million single nucleotide polymorphisms identified on 283 accessions from two populations (Circum and Far West), and exploited annotation and published transcriptomic data to identify biological processes associated with molecular adaptation. We identified 58 swept genomic regions with a 15 kb average length and comprising 3.3 gene models on average. The unimodal sweep state probability distribution in these regions enabled us to focus on the best single candidate gene per region. We detected two unambiguous species-wide selective sweeps, one of which appears to underlie morphological adaptation. Population genomic analyses of the remaining 56 sweep signatures indicate that sweeps identified in the Far West population are less population-specific and probably more ancient than those identified in the Circum population. Functional annotation revealed a predominance of immunity-related adaptations in the Circum population. Transcriptomic data from accessions of the Far West population allowed inference of four clusters of coregulated genes putatively involved in the adaptive control of symbiotic carbon flow and nodule senescence, as well as in other root adaptations upon infection with soil microorganisms. We demonstrate that molecular adaptations in M. truncatula were primarily triggered by selective pressures from root-associated microorganisms.
Keywords: selective sweep, SNP, linkage disequilibrium, Medicago truncatula, transcriptomics, root, nodule, next generation sequencing
Introduction
Exploring the fine scale genomic signatures of natural selection is now possible thanks to massive species-scale single nucleotide polymorphism (SNPs) discovery through next generation sequencing. In a population, a selective sweep is the local genomic signature of fixation or near fixation of highly favorable, adaptive mutations together with linked neutral mutations (Nielsen 2005). In this context, statistical population genetics methods have been developed in the past decade to identify candidate genomic regions and genes targeted by selective sweeps in a population using SNPs data. These methods can be classified into two groups: Methods based on 1) the length and structure of haplotypes, measured by extended haplotype homozygosity and derived statistics (Sabeti et al. 2002; Voight et al. 2006) and 2) the properties of the site frequency spectrum (Kim and Stephan 2002; Jensen et al. 2005; Nielsen et al. 2005; Boitard et al. 2009). In parallel, genome annotation projects allow for functional predictions about observed gene models. Therefore, selective sweep genome scans combined with functional annotation should evidence for biological functions and pathways targeted by positive selection.
The species Medicago truncatula is a plant genomic model for the study not only of root symbioses with N-fixing bacteria and arbuscular mycorrhizae (Jones et al. 2007; Parniske 2008; Young et al. 2011) but also of root pathogen infections (Vailleau et al. 2007; Djébali et al. 2009; Samac et al. 2011; Ben et al. 2013). To perform Genome Wide Association Studies and other genome scans based on polymorphism data, more than 280 accessions representative of M. truncatula natural genetic variation (Ronfort et al. 2006) were sequenced using Illumina technology by the Medicago truncatula HapMap Project (medicagohapmap.org/, last accessed April 23, 2015), thereby producing high-resolution SNP polymorphism data (Branca et al. 2011; Stanton-Geddes et al. 2013). At the same time, a high-quality M. truncatula genome of the reference genotype (A17) was published (Young et al. 2011). From these data a first genome-wide analysis of selection was performed on more than 20,000 annotated genes for 56 M. truncatula accessions, using dN:dS, MK, and DTH summary statistics (Paape et al. 2013). Based on this early study, approximately 1% of M. truncatula genes harbor signature of positive selection, whereas ≈75% of the genes are apparently subject to strong purifying selection.
In this work, we have chosen to focus on the identification of selective sweeps in M. truncatula using a hidden Markov model (HMM) algorithm (sweep HMM) which makes use of the full site frequency spectrum and the spatial pattern of diversity along chromosomes, rather than classical summary statistics of genetic diversity (Boitard et al. 2009). We performed a whole-genome scan with ultrahigh SNP density, namely with 1 SNP each 30 bp on average, with more than 15 million SNPs identified in a collection of 283 M. truncatula accessions with the Mt3.5 reference genome version. To reduce false-positive signals, we took into account the M. truncatula population structure into two groups previously identified with genetic data (Ronfort et al. 2006; De Mita et al. 2011; Bonhomme et al. 2014). The “Far West” (FW) population is geographically restricted to the extreme west of Mediterranean basin (Spain, Portugal, Morocco, and west Algeria), whereas the other population called “Circum” (C) has a large geographic distribution throughout the Mediterranean basin with only few occurrences in the extreme west. The historical divergence of these two populations is unclear, but it seems that M. truncatula has undergone demographic expansion (Branca et al. 2011) from glacial refuges located both West and East of the Mediterranean basin (Ronfort et al. 2006; De Mita et al. 2011). However, the geographic regions spanned by these two populations do not show contrasted abiotic environments (Thompson 2005; De Mita et al. 2011). Our genome scan for selective sweeps, combined with gene annotation as well as gene expression data analyses, identified a set of genes under strong positive selection which clearly demonstrates the molecular adaptation of M. truncatula especially in the context of biotic interactions.
Results and Discussion
Population Genomic Features of M. truncatula Sweeps
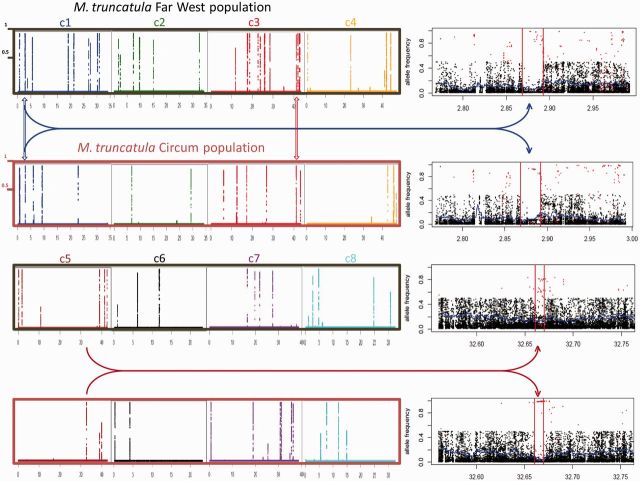
Selective sweep inference was performed on two M. truncatula populations (FW and C) using the sweep HMM algorithm (Boitard et al. 2009). We detected 58 regions in the M. truncatula genome showing specific features of selective sweeps such as lower minor allele frequencies (MAF) and recurrent high frequencies of the derived allele compared with the ancestral allele (fig. 1; supplementary data S1, Supplementary Material online). Concomitantly, these regions also showed a severe loss of genetic diversity: The average expected heterozygosity—He—was significantly lower in swept regions (0.09 and 0.06 for FW and C, respectively) than in genome-wide regions of the same size (0.16 and 0.14, for FW and C, respectively; t-test: P < 2.2 × 10−16 for both populations). The posterior probability of a given SNP to be in sweep state in these regions exceeds 0.99 (fig. 1 and supplementary fig. S1, Supplementary Material online). These regions are defined by 32,049 SNPs (i.e., 6,266, 1,430, 9,448, 2,271, 5,855, 1,724, 4,111, and 944 SNPs on chromosomes 1–8, respectively). It should be noted that the power to detect a selective sweep is linked to the proportion of missing data (i.e., coverage) in a given region, as chances to detect rare alleles are higher for low proportions of missing data (high coverage). We indeed found that all swept regions identified with sweep HMM algorithm showed a higher coverage (73.3 and 147.9 genotypes called on average for FW and C, respectively) than the genome-wide average (63.5 and 122.8; t-tests: P = 3.4 × 10−16 and 2 × 10−16 for FW and C population, respectively). Among the 58 candidate regions, two were common to both FW and C populations, on chromosomes 1 and 3, whereas 34 and 22 sweeps were detected specifically in FW and C, respectively (fig. 1). Assuming a genome size of 400 Mb for M. truncatula, one selective sweep signature was detected each approximately 7 Mb of M. truncatula genome, on average. All predicted gene models in sweep windows were extracted according to the Mt3.5 M. truncatula genome version, leading to 190 candidate genes—including 29 predicted transposable elements—potentially targeted by selective sweeps in M. truncatula (41, 11, 50, 17, 26, 17, 24, and 4 genes on chromosomes 1–8, respectively; supplementary data S2, Supplementary Material online). In the following sections, the recently released Mt4 genome version of this gene list was used as reference (and presumably improved) annotation.
Fig. 1.
Genome scan of selective sweeps in the two Medicago truncatula populations. The left part of the figure displays the posterior probability (y axis) to be in sweep state along each of the eight chromosomes (c1–c4 and c5–c8), for each population (FW in black and C in brown). The x axis indicates the chromosomal position, in Mb. The right part of the figure corresponds to zooms of the corresponding genomic positions indicated by arrows, in each population. These graphics represent the population frequency of 1) the minor allele for SNPs with unknown ancestral allele (black) and 2) the derived allele for SNPs with known ancestral allele (red). Red bars indicate the selective sweep window. The expected heterozygosity (He) is shown by the blue curve. The two uppermost graphics in the right part highlight a common selective sweep window among populations whereas the two lowermost graphics highlight a selective sweep identified only in one population (here in the C population).
The average number of gene models per 10 kb of swept region (2.5 ± SD = 1.1) was significantly higher (t-test, P = 6.9 × 10−7) than the genome-wide average of 1.7 genes per 10 kb estimated in Young et al. (2011). The average number of gene models in a swept region was 3.3 (±SD = 1.8) for the whole species sample. This number was higher in the FW population (3.6 ± SD = 2) than in the C population (2.9 ± SD = 1.4), but the difference between the two populations was not significant (Wilcoxon Rank Sum test, P = 0.22). The average length of a swept region was 14.9 kb ( ± SD = 9.5 kb) for the whole species sample. Again, it was higher in the FW population (16 kb ± SD = 10.8) than in the C population (13.2 ± SD = 7.1), but the difference was not significant (Wilcoxon Rank Sum test, P = 0.61). These findings suggest that swept regions in M. truncatula are enriched in coding sequences by reference to a genome-wide average, and that they show similar gene density and genomic length across populations. Interestingly, the estimated average sweep length of ≈15–16 kb was higher, though of the same order, as the average genomic linkage disequilibrium (LD) decay of ≈10 kb estimated for M. truncatula genome (Branca et al. 2011). This illustrates the erosion of a selective sweep signature beyond such physical distances.
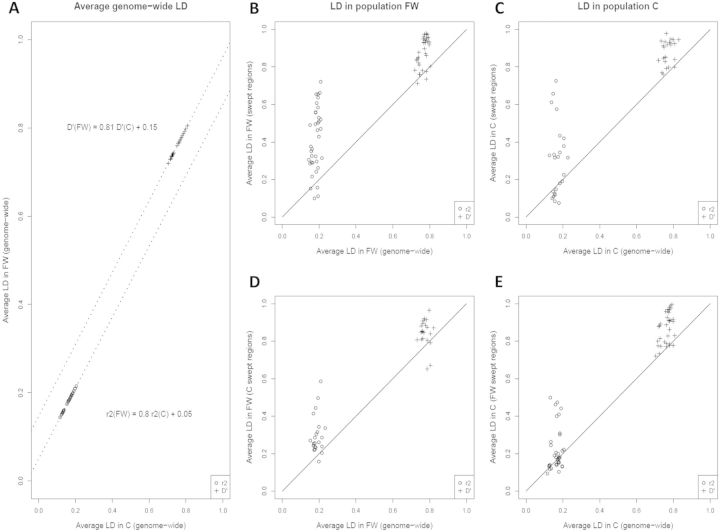
Although LD is mostly influenced by genome-wide neutral processes such as recombination rate and genetic drift, selective sweeps can also strongly affect local LD, depending on selection intensity. We used the r2 and D′ measures to estimate LD in swept regions and in genome-wide regions of the same lengths. First, we compared genome-wide LD between FW and C populations for regions of the same lengths (fig. 2A), and found almost complete positive correlation between average LD in the two populations (r = 0.9999822 and 0.9999854 with r2 and D′ measures, respectively). Linear regressions of average r2 and D′ values of population FW against population C estimated slopes below 1 (0.80 and 0.81 for r2 and D′, respectively; fig. 2A) indicating that effective population size (Ne) in FW is 1.25 (1/0.8)-fold larger than effective population size in C. This ratio is consistent with that estimated from the number of SNPs in each population (1.35; see Materials and Methods for Watterson’s theta) or the proportion of pairwise differences between individuals in each population (see the average genome-wide He in fig. 3). The contrasted intercepts of these linear regressions (0.05 and 0.15 for r2 and D′, respectively) most likely result from a coverage difference (i.e., a different proportion of genotypes called for a given SNP) between the two populations, which has more impact on D′ than on r2. Indeed, D′ is expected to show higher values when rare alleles are missing. Second, comparisons of average LD in swept and genome-wide regions of the same length clearly showed that LD was higher in swept regions, both in the FW population (t-tests; P = 8.26 × 10−10 and 1.69 × 10−10 for r2 and D′, respectively; fig. 2B) and in the C population (t-tests; P = 0.0014 and 5.49 × 10−9 for r2 and D′, respectively; fig. 2C). These results are consistent with the fact that selective sweeps generally tend to increase LD (Kim and Nielsen 2004) although different and sometimes opposite effects might be observed depending on the phase of the sweep and the position of the selected locus in the sweep region (McVean 2007). Finally, to evaluate the level of population specificity of the sweeps identified, we compared within each population the genome-wide LD with LD in regions swept in the other population (fig. 2D and E). In the FW population we found that LD was higher in the regions that have swept in the C population than in genome-wide regions (t-tests; P = 2.55 × 10−5 and 1.4 × 10−4 for r2 and D′, respectively; fig. 2D). The same was found in the C population (t-tests; P = 0.0042 and 5.48 × 10−9 for r2 and D′, respectively; fig. 2D). These results clearly indicate that sweeps in a given population tend to leave an LD signature in the other population; in other words, sweeps identified in each population tend to be shared among populations and are thus not specific.
Fig. 2.
Levels of LD in swept and genome-wide regions in Medicago truncatula populations. We estimated average r2 and D′ measures of LD in swept regions and genome-wide regions of the same length, in both M. truncatula populations. (A) Correlation of average genome-wide LD between the FW and C populations (the dotted lines show the linear regressions). (B) comparison of average LD in swept and genome-wide regions of the same length in the FW population, and in the C population (C). (D) For the FW population, comparison of genome-wide LD with LD in regions swept in the C population. (E) For the C population, comparison of genome-wide LD with LD in regions swept in the FW population. In (B)–(E), the solid line indicates the section of equal average LD.
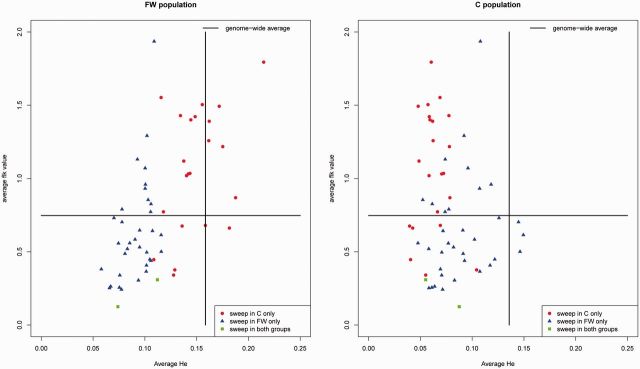
Fig. 3.
Levels of genetic diversity (He) and genetic differentiation (FLK) in swept and genome-wide regions in Medicago truncatula populations. We estimated average He and FLK (Bonhomme et al. 2010) values in swept regions identified in each population. For each population, we plotted FLK against He values for each swept region, by distinguishing swept regions identified in FW, C, and in both populations (see the color code in the figure). Solid lines indicate the genome-wide averages of He and FLK, in each population.
To further investigate this issue, we also explored genetic diversity and differentiation in swept and genome-wide regions for both populations using He and the FLK statistics. This latter statistic is related to Fst but accounts for the phylogenetic structure of populations and for genetic drift differences between populations, by estimating a population kinship matrix (Bonhomme et al. 2010). For each population, we plotted FLK against He values for each swept region, by distinguishing swept regions identified in FW, C, and in both populations (fig. 3). We observed that FLK values in swept regions of population C tend to be higher than the genome-wide average and show high He values in the FW population, suggesting that these regions are more population-specific. In contrast, FLK values in swept regions of population FW are not very high compared with the genome-wide average and show low He values in the C population, suggesting that these swept regions are less population-specific (fig. 3). One hypothesis that could explain this difference is that selective sweeps in FW are more ancient and thus more likely to be shared between populations, whereas selective sweeps in C could be more recent and thus limited to this population. The estimated effective population size differences (ratio of 1.25) between populations may be related to these contrasted patterns of sweep specificity. Indeed, the genomic signature of a sweep is related to the age of this sweep in Ne units, the time scale at which coalescence events occur. Thus, sweeps detected in FW and C are likely equally old in this rescaled unit, and consequently older in FW when measured in generations, due to the larger population size. However, no single significant FLK value was found in the sweeps regions, even the more specific ones detected in the C population (P values were all above 0.01). This confirms that the sweeps detected in this analysis were not due to divergent adaptation between populations, as suggested by the LD analysis.
Identification of Molecular and Biological Functions Underlying Sweeps
We observed that the posterior probability of sweep state along a swept region was unimodal and that the highest posterior probability was located in or close to the middle of each swept region (supplementary fig. S1, Supplementary Material online). This means that selective sweeps in M. truncatula seem to follow a classic model in which positive selection targets one causal gene variant (the adaptive gene) whereas noncausal variants at neighboring genes undergo genetic hitchhiking with symmetrical intensity. Selective sweep simulations have shown that the middle of the region with lowest nucleotide diversity is not always centered on the site targeted by selection, but this phenomenon is more pronounced in populations with small effective sizes (Kim and Stephan 2002). Given that M. truncatula populations have most probably experienced demographic expansion thereby involving high effective sizes, we chose to analyze the subset of 58 candidate gene models associated with all maximum posterior probabilities to be in sweep state. This subset of candidate genes included eight transposable elements, and the predicted functions of several annotated genes suggest roles in perception and adaptation to environment (receptor genes and signaling), epigenetic plasticity (DNA modification, DNA repair), transport, cell organization (cytoskeleton) but also primary metabolism (table 1 and supplementary data S3 for more details, Supplementary Material online). It is interesting to note that none of the identified swept regions contained tandem clusters of genes of the same family, which are highly frequent in M. truncatula genome (Ameline-Torregrosa et al. 2008; Young et al. 2011; Nallu et al. 2014; Trujillo et al. 2014). Consequently, we do not exclude the possibility that gene families and/or tandem clusters exist within or very close to some swept regions, but missed during the Mt4.0 annotation.
Table 1.
Medicago truncatula Best Candidate Gene Models for Selective Sweep (Mt4 genome version).
| Gene Model (Mt4) | Pop | Mt4 Gene Annotation | Functional Class Classification (Mercator) | Proposed Classification |
|---|---|---|---|---|
| Medtr1g104850 | FW | Serine/threonine kinase | Signaling.receptor kinases | Signaling |
| Medtr3g116250 | FW | Phosphatidylinositol 4-kinase alpha | Signaling.phosphinositides | Signaling |
| Medtr5g099170 | FW | Calcium-binding EF hand-like | Signaling.calcium | Signaling |
| Medtr7g088830 | FW | Cam interacting protein | Signaling.calcium | Signaling |
| Medtr7g112870 | C | Serine/threonine-protein phosphatase PP1 | Protein.postranslational modification | Signaling |
| Medtr5g005450 | FW | Cysteine-rich receptor-kinase-like | Signaling.receptor kinases.DUF 26 | Receptor |
| Medtr3g041560 | C | Leucine-rich receptor-like kinase | Stress.biotic.PR-proteins | Receptor |
| Medtr7g071940 | C | NBS-LRR type disease-resistance | Stress.biotic.PR-proteins | Receptor |
| Medtr3g114150 | FW/C | DNA topoisomerase I | DNA.synthesis / chromatin structure | DNA modification |
| Medtr2g008640 | FW | ATP-dependent_DNA_helicase_PIF1a | DNA.unspecified | DNA modification |
| Medtr4g123600 | C | DNA methyltransferase, putative | RNA.regulation of transcription | DNA modification |
| Medtr1g012370 | FW/C | AFG1-family ATPase | NA | DNA repair? (UV-response) |
| Medtr1g082570 | FW | DNA excision repair protein ERCC-1 | RNA.regulation of transcription | DNA repair |
| Medtr3g117490 | FW | Zinc finger CCCH domain protein | NA | DNA/RNA binding |
| Medtr4g122000 | FW | Nucleic acid-binding-like | NA | DNA replication |
| Medtr1g007060 | FW | Exosome complex exonuclease RRP46 | RNA.processing.ribonucleases | RNA processing |
| Medtr2g101360 | FW | Proteinaceous RNase P | Pentatricopeptide (PPR) repeat | RNA processing |
| Medtr3g080260 | FW | Nodulin MtN21/EamA-like transporter | NA | Transporter (WAT1/auxin) |
| Medtr3g086440 | FW | Zinc induced facilitator-like | Transport.sugars | Transporter |
| Medtr4g127630 | FW | Metaxin-like protein, putative | Development.unspecified | Transporter (mitochondria) |
| Medtr4g130820 | C | Cyclic nucleotide-gated ion channel-like | Transport nucleotide/Ca regulated channel | Transporter |
| Medtr7g098800 | C | Drug resistance transporter-like ABC | Transport ABC.and multidrug resist systems | Transporter |
| Medtr7g111160 | C | Organic solute transporter ostalpha | NA | Transporter |
| Medtr3g055370 | C | Enhanced downy mildew (EDM2) | RNA.regulation of transcription | Transcription |
| Medtr5g078780 | C | CCAAT-binding factor | Development.unspecified | Transcription |
| Medtr4g127660 | FW | Actin filament bundling | Cell.organization.cytoskeleton.actin.binding | Cytoskeleton |
| Medtr4g006000 | FW | SDA1-like protein | NA | Cytoskeleton |
| Medtr1g102010 | FW | Phosphogluco-mutase | Glycolysis.plastid branch.(PGM) | Primary metabolism |
| Medtr3g071440 | FW | UDP-glucose pyrophosphorylase | NA | Primary metabolism |
| Medtr2g021720 | FW | Transmembrane protein, putative | Protein.synthesis.ribosomal.60 S subunit.L34 | Ribosome constituent |
| Medtr3g060660 | FW | Paramyosin | NA | Membrane protein |
| Medtr3g117420 | FW | 2OG-Fe(II) oxygenase family | Hormone metabolism.ethylene | Hormone synthesis |
| Medtr5g009110 | FW | Cytochrome P450 family 97 | Misc.cytochrome P450 | Carotenoid pathway |
| Medtr1g086070 | C | Cysteine desulfhydrase | NA | Cysteine degradation |
| Medtr3g114070 | C | Glutathione S-transferase-related | NA | Oxidative stress |
| Medtr7g099880 | C | Nuclear pore complex Nup205-like | NA | Nuclear trafficking |
| Medtr8g032340 | C | Signal recognition particle receptor FtsY | Protein.targeting.chloroplast | Protein targeting |
| Medtr3g086570 | FW | Growth regulator-like | Hormone metabolism.auxin.regulated | NA |
| Medtr5g096860 | FW | Armadillo/beta-catenin-like repeat | Armadillo/beta-catenin repeat family | NA |
| Medtr1te012110 | FW | Pol-like polyprotein%2Fretrotransposon | NA | Transposition |
| Medtr1te019330 | C | Reverse transcriptase | NA | Transposition |
| Medtr2te020490 | C | Transposable element | NA | Transposition |
| Medtr3te022330 | C | Gag-Pol polyprotein_2F_retrotransposon | NA | Transposition |
| Medtr3te056140 | FW | Transposable element | NA | Transposition |
| Medtr3te083220 | C | Copia-like polyprotein_2F_retrotransposon | NA | Transposition |
| Medtr7te061430 | FW | Transposable element | NA | Transposition |
| Medtr8te045990 | C | Transposable element | NA | Transposition |
| Medtr1g031590 | C | O-fucosyltransferase family | NA | NA |
| Medtr1g101990 | FW | GDP-fucose protein O-fucosyltransferase | NA | NA |
| Medtr3g080630 | FW | HAT_family_dimerization_domaina | NA | NA |
| Medtr7g078170 | FW | RIC1-like | NA | NA |
| Medtr8g020400 | FW | Animal MPPE1-like | NA | NA |
| Medtr5g091560 | FW | Hypothetical protein | NA | NA |
| Medtr6g005250 | C | Hypothetical protein | NA | NA |
| Medtr6g015240 | C | Hypothetical protein | NA | NA |
| Medtr6g057610 | FW | Hypothetical proteina | NA | NA |
| Medtr6g057790 | FW | Hypothetical protein | NA | NA |
| Medtr7g005960 | C | Hypothetical protein | NA | NA |
aGenes with Mt3.5 annotation (unavailable Mt4 annotation). More details on this gene list are provided in supplementary data S3, Supplementary Material online.
The identification of a sweep concerning a DNA topoisomerase I coding gene (Medtr3g114150) could well illustrate morphological adaptation at the species level. Indeed, two co-orthologs of this gene are present in Arabidopsis thaliana and a mutant analysis demonstrated that they play important roles in plant morphogenesis, notably in leaf and flower initiation (Takahashi et al. 2002). Therefore, it is likely that positive selection has acted on an ancient loss-of-expression (no evidence of expression from RNAseq and microarray data) or loss-of-function variant of DNA topoisomerase I gene, resulting in the strong morphological variation currently observable throughout M. truncatula distribution area (Bonnin et al. 2001; Julier et al. 2007; Espinoza et al. 2012). Adaptive loss-of-function mutations have already been identified in plant genes involved in flower color, flowering time, and disease-resistance (reviewed in Siol et al. 2010).
Several annotated gene functions predict a role in plant–microbe interactions and are particularly enriched in the list of swept genes identified in the Circum population (see table 1, and population-specific Gene Ontology (GO) terms in supplementary fig. S2, Supplementary Material online). In this population, 5 of 22 (23%) swept genes seem to be connected to immunity. First, a cyclic nucleotide-gated ion channel-like- CNGC -protein (Medtr4g130820) was assigned to the KEGG pathway “plant–pathogen interaction” and belongs to the functional class “transport cyclic nucleotide or calcium-regulated channels” which was significantly overrepresented in the swept gene list (table 2). Members of the CNGC family have indeed been reported to be involved in plant immunity, particularly AtCNGC11 and 12 (Yoshioka et al. 2006; Moeder et al. 2011). In addition, we identified an NBS-LRR type disease-resistance protein (Medtr7g071940) and a leucine-rich receptor-like kinase protein (Medtr3g041560) that are receptors involved in response to biotic stress (table 1). Interestingly, we also identified the “enhanced downy mildew protein”—EDM2 (Medtr3g055370, AT5G55390). In A. thaliana, EDM2 is required for disease-resistance against Hyaloperonospora parasitica, through direct or indirect regulation of the RPP7 (NBS-LRR) gene (Eulgem et al. 2007). Finally, we identified a drug-resistance transporter-like ABC domain protein (Medtr7g098800) also assigned to the GO term “death” (supplementary fig. S2, Supplementary Material online). In fact, one homolog of this gene in A. thaliana, PEN3/PDR8 (AT1G59870), was shown to contribute to nonhost resistance through exporting toxic materials to attempted invasion sites of pathogens that enter by direct penetration—that is, Blumeria graminis, Plectosphaerella cucumerina, Erysiphe pisi, and Phytophthora infestans (Stein et al. 2006). In another study, the loss of PDR8 was shown to cause hypersensitive response-like cell death (Kobae et al. 2006).
Table 2.
Functional Classes Significantly Affected by Selective Sweeps in Medicago truncatula Populations.
| Functional Class (Mercator) | Pop | Sample Freq. | Genome Freq. | P valuea | Gene Model | GO Terms (Gene Module) | KEGG Pathway | |
|---|---|---|---|---|---|---|---|---|
| Cell organization cytoskeleton actin binding | FW | 1 | 4 | 0.004 | Medtr4g127660 | Growth | (1) | — |
| Glycolysis plastid branch PGM | FW | 1 | 4 | 0.004 | Medtr1g102010 | — | (3) | Sugar and nucleotide metabolism |
| Hormone metabolism ethylene synthesis or degradation | FW | 1 | 22 | 0.019 | Medtr3g117420 | — | (4) | — |
| Protein synthesis ribosomal protein eukaryotic 60 S subunit L34 | FW | 1 | 11 | 0.006 | Medtr2g021720 | Structural molecule | (2) | — |
| Signaling phosphoinositides | FW | 1 | 17 | 0.025 | Medtr3g116250 | Symplast | (1) | Phosphatidylinositol signaling inositol phosph metabolism |
| Transport cyclic nucleotide or calcium regulated channels | C | 1 | 25 | 0.011 | Medtr4g130820 | — | Plant–pathogen interaction | |
| Protein targeting chloroplast | C | 1 | 43 | 0.023 | Medtr8g032340 | — | Protein export | |
| RNA regulation of transcription—DNA methyltransferases | C | 1 | 22 | 0.011 | Medtr4g123600 | — | — | |
aBin significance was corrected with an FDR threshold of 5%.
Expression of Swept Genes Reveals Potential Role in Interaction of Roots with Symbiotic Organisms
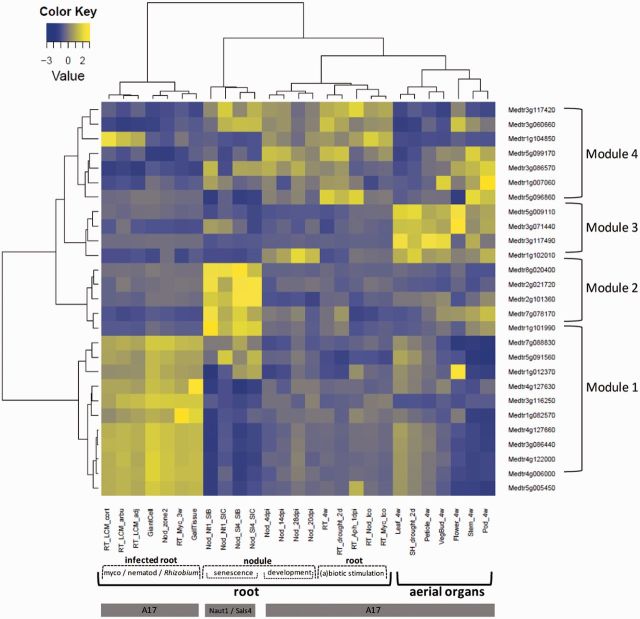
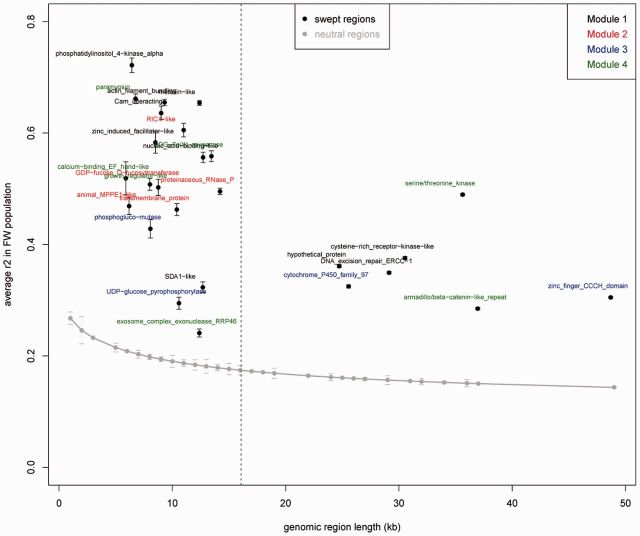
Medicago truncatula is a legume model for studying root–microbe mutualistic interactions with the nitrogen-fixing Rhizobia or with arbuscular mycorhizae (Jones et al. 2007; Parniske 2008). These interactions result from adaptation processes likely to have involved selective sweep events associated with genes expressed in the roots. To investigate this, expression of swept genes was examined by mining a large array-based transcriptomic data set available not only on root tissues colonized with either symbiotic or pathogenic microbes but also on aerial organs. This analysis was carried out on swept genes uncovered within the FW population since transcriptomic data mostly derived from the A17 line, which belongs to this population. Data were recovered from 27 of 34 (80%) swept genes and clustered into four distinct gene modules (fig. 4). In order to investigate selection strength, that is to pinpoint the putative main targets of selection in each module, we focused on genes associated with swept regions of size higher than the mean selective sweep size (i.e., 16 kb) in the FW population (fig. 5).
Fig. 4.
Expression data clustering of genes under selective sweep in Medicago truncatula FW population. Rows represent the expression profile (from Affymetrix technology) of 27 gene models identified as being subjected to strong positive selection in the FW population. Columns represent the different selected conditions (RT, root; Nod, nodule; LCM, laser capture microdissection; cort, cortical cells; arbus, cells colonized by arbuscules; adj, adjacent cells; Aph, Aphanomyces euteiches; 4w, 4-week plant; lco, lipochitooligosaccharides). As indicated in the bottom of the figure most gene expression profiles were measured on the A17 reference genotype belonging to the FW population, with the exception of Naut1 (Nt1) and Sals4 (Sl4) wild genotype, inoculated with Sals B (SlB) and Sals C (SlC) Sinorhizobium meliloti genotypes. These two genotypes were not assigned to any of the two populations, but they carry the high-fitness variant at all genes (data not shown). Expression values were normalized among conditions and among genes. The resulting color code indicates highly expressed genes in yellow and weakly expressed genes in dark blue. Hierarchical clustering was performed with Euclidean distance and the Ward’s agglomeration criterium, which minimizes the total within-cluster variance.
Fig. 5.
Selection strength on genes of different regulatory modules, evaluated using sweep length and level of LD. Selection strength was evaluated using the length (x axis) and the average LD (r2 measure) of each sweep window in the FW population. The vertical dashed line indicates the average sweep length (∼16 kb). Gray dots and associated bars indicate the genome-wide average and standard error of the average r2 value for different lengths of genomic region (the same sizes as the swept regions). Black dots and associated bars indicate the average, and standard error of the average value of r2 in each swept region. The color code pinpoints genes of the four different modules of coregulated swept genes identified by hierarchical clustering of expression data (fig. 4).
The largest module, Module 1, corresponds to genes preferentially expressed in root tissues colonized with arbuscular fungus (Glomus intraradices—renamed Rhizophagus irregularis), in the infection zone of nodule (zone 2) and upon infection with the root-knot nematode Meloidogyne incognita, in giant cells and gall tissues (fig. 4). Expression of these genes is highly specific of these conditions, being weakly expressed in control roots or in aerial tissues, except for the AFG1-ATPase gene (Medtr1g012370), a swept gene at species level, which is also expressed in flowers. In Module 1, selection strength seems the highest for three genes: A cysteine-rich receptor kinase-like protein, the DNA excision repair protein ERCC-1, and a hypothetical protein (fig. 5). The cysteine-rich receptor kinase-like protein—CRK (Medtr5g005450)—is a member of the CRK receptor gene family whose functions have been related to abiotic and biotic stresses such as ozone levels and treatments with pathogen-associated molecular patterns (Chen et al. 2004; Wrzaczek et al. 2010). The DNA excision repair protein ERCC-1 (Medtr1g082570) is induced by DNA damage in plants (Vonarx et al. 1998; Kunz et al. 2005). The coregulation of ERCC-1 with the UV-B-induced AFG1-family ATPase coding gene (Brown et al. 2005) suggests that both genes might act in the same pathway of response to DNA-damages that can occur during plant–microbe interactions in root (Song and Bent 2014). However, occurrence of DNA damages during symbiotic interactions is currently unknown.
A nodule-specific module is Module 2, a cluster of five genes highly expressed in senescent (10 weeks) nodules (fig. 4). Although swept genes from Module 2 do not show signals of intense selection according to sweep lengths (fig. 5), the highest sweep length values concern the proteinaceous RNase P (Medtr2g101360) and a putative transmembrane protein (Medtr2g021720) belonging to the functional class “protein synthesis, ribosomal protein, eukaryotic 60 S subunit L34” which was significantly affected by positive selection (table 2). The homolog of the transmembrane protein coding gene in A. thaliana (AT3G06180), a predicted structural constituent of 60 S ribosomal proteins, was detected as a putative target of WRKY53 transcription factor which is involved in early events of leaf senescence (Miao et al. 2004). Proteinaceous RNase P homolog in A. thaliana (PRORP1, AT2G32230) is involved in the maturation of plant mitochondrial mRNAs but domain annotation also indicates antimicrobial properties typical from pentatricopeptides, suggesting in M. truncatula a putative role in the active rejection of Sinorhizobium meliloti in senescent nodule.
Module 4 shows less contrasted transcriptional profiles among conditions and organs but genes are mainly induced in root and during nodule development (fig. 4). Two genes of Module 4 show evidence of high selection strength: A serine/threonine kinase (Medtr1g104850) and an armadillo/beta-catenin-like repeat protein coding gene (Medtr5g096860) (fig. 5). Serine/threonine kinases are widely represented in plants. They are activated by receptors that sense various external factor and they act downstream to change metabolism, gene expression, and cell growth and division (Hardie 1999). The armadillo/beta-catenin-like repeat protein belongs to armadillo repeat proteins which are common in all eukaryotes but most of them have unknown functions (Coates 2003). However, in A. thaliana, two armadillo/beta-catenin homologs called ARABIDILLO-1 (AT2G44900) and -2 (AT3G60350) have been shown to promote lateral root formation (Coates et al. 2006) and can therefore lead to a better adaptation of the plant to its abiotic and biotic environment.
Only Module 3 is a cluster of genes preferentially expressed in aerial parts (fig. 4). Module 3 is involved in primary metabolism by including notably a phosphoglucomutase (PGM) (Medtr1g102010) and an UDP-glucose pyrophosphorylase (Medtr3g071440). In starch-storing plants, PGM provides the substrate phosphoglucose for starch synthesis or other pathways (Harrison et al. 2000). PGM is a central actor of the control of photosynthetic carbon flow (Streb et al. 2009). UDP-glucose pyrophosphorylase immediately acts upstream to PGM in the sucrose-to-starch conversion pathway, which could explain their coregulation. The control of photosynthetic carbon flow by PGM and other actors of the starch synthesis pathway is probably a highly conserved molecular feature in plants. A noteworthy feature of PGM is its increased expression with time (up to 4 weeks) in nodules infected with S. meliloti strain 1021, but not in roots of the M. truncatula A17 genotype (fig. 4). It has been shown that PGM plays a role in the availability of phosphoglucose for synthesis of required polysaccharides by rhizobia during nodulation (Lepek et al. 2002). A proteomic analysis also identified a PGM as a specific protein present in soybean root hairs after infection by Bradyrhizobium japonicum (Wan et al. 2005) suggesting that PGM may play a role in the Rhizobium-legume interaction by affecting the availability of host phosphoglucose to the Rhizobium. Considering the hypothesis that PGM is a phosphoglucose supplier to S. meliloti, a basic adaptive hypothesis is that selection has favored the expression of PGM in the nodule, which has increased the fitness of plants by improving symbionts nutrition thus their ability to fix nitrogen in optimal conditions. Remarkably, PGM was not expressed in 10-week nodules of M. truncatula Sals4 and Naut1 genotypes inoculated with either SalsB or SalsC S. meliloti genotypes (fig. 4, data from Heath et al. 2012), suggesting a temporal regulation of PGM expression in the nodule. In Module 3, PGM and UDP-glucose pyrophosphorylase show less intense selection strength, according to sweep length, than the cytochrome P450—Medtr5g009110—and the zinc finger CCCH domain protein—Medtr3g117490 (fig. 5). The two latter genes belong to large protein families. Cytochrome P450 proteins are used by plants in various biosynthetic and catabolic pathways (Schuler and Werck-Reichhart 2003). Zinc finger proteins have highly diverse functions, notably in DNA-binding and transcriptional activations (Laity et al. 2001), and Zinc finger CCCH domain proteins display considerable versatility in binding mode (DNA, RNA, or proteins). It is thus tempting to hypothesize that these proteins are potential transcriptional and/or posttranscriptional regulator of PGM expression in nodule, which could explain that they have undergone stronger selective sweep than PGM.
Overall, expression data indicate that approximately 70% of swept genes (19 over 27 genes analyzed) are preferentially expressed in root and nodule than in aerial organs (fig. 4). However, our selection of experimental conditions to analyze the expression of swept genes might appear biased toward root and nodule conditions, relative to experiments in aerial organs. This primarily reflects that the Medicago truncatula Gene Expression Atlas lacks available experiments focusing on aerial organs in different stress conditions, notably in biotic stress conditions. However, to investigate whether or not swept genes are mainly involved in root biology relative to all genes present in the microarray, we performed an enrichment test for root and nodule organs, separately. Table 3 shows that there is a clear enrichment for swept genes with “strong expression” in 50–75% of root conditions (P = 0.0077), relative to the microarray. We also found enrichment for the categories 0–25% and 75–100% of root conditions (P = 0.017 and 0.0358, respectively). For nodule organ, enrichment in highly expressed swept genes was found in 0–25% of nodule conditions (P = 0.0021) only. These results show that swept genes are significantly highly expressed in various root conditions, relative to the microarray, indicating a central role in root biology (developmental response to stress, response to infection by microorganisms). For nodule, we can hypothesize that some swept genes are significantly highly expressed in specific conditions (senescence or development; see fig. 4), relative to the microarray.
Table 3.
Analysis of Swept Gene Expression Enrichment in Root and Nodule, Relative to Medicago truncatula Expression Microarray.
| Organ | % of Experimentsa | % of Highly Expressed Swept Genes | % of Highly Expressed Microarray Genes | P value |
|---|---|---|---|---|
| Root | 0–25 | 0.32 | 0.16878 | 0.0170* |
| Root | 25–50 | 0.32 | 0.66986 | 0.9996ns |
| Root | 50–75 | 0.32 | 0.14939 | 0.0077** |
| Root | 75–100 | 0.04 | 0.01196 | 0.0358* |
| Nodule | 0–25 | 0.40 | 0.17780 | 0.0021** |
| Nodule | 25–50 | 0.24 | 0.17316 | 0.1276ns |
| Nodule | 50–75 | 0.36 | 0.59998 | 0.9868ns |
| Nodule | 75–100 | 0.00 | 0.04906 | 0.7157ns |
aThe analysis deals with 11 root conditions and 9 nodule conditions.
*P ≤ 0.05; **P ≤ 0.01; ns, not significant.
Comparison with Other Genome Scans in M. truncatula
Comparison of the list of 190 gene models covering all candidate regions for a selective sweep uncovered in this study with the list of candidate genes under positive selection identified in the first M. truncatula population genomics survey of selection (Paape et al. 2013) pinpoints eight common regions among which four were detected in the same populations (supplementary data S4, Supplementary Material online).
First, both analyses identified in both populations the two gene models Medtr3g114130 and Medtr3g114140, which flank the DNA topoisomerase gene (Medtr3g114150), therefore increasing the evidence that this region has undergone strong positive selection at M. truncatula species scale. Second, the cysteine-rich receptor-like protein kinase 25 (Medtr1g104890) identified by Paape et al. (2013) in the FW population lies in a candidate region of our analysis where the best candidate is the serine/threonine kinase—Medtr1g104850 (Module 4, fig. 4). The zinc-induced facilitator-like protein coding gene (Medtr3g086440, Module 1) was also found in both analyses in the FW population. Finally, only one gene was found by the two analyses in the C population: The O-fucosyltransferase family protein (Medtr1g031590).
Two candidate genes of our study were also identified in the first genome-wide association study performed in M. truncatula (Stanton-Geddes et al. 2013). They include a high mobility group B-like protein (Medtr7g005970) and a magnesium transporter CorA family protein (Medtr5g091570), respectively, associated with trichome density and S. meliloti nodule occupancy in upper root. Interestingly, the magnesium transporter CorA family protein gene model is located next to a ubiquitin carboxyl-terminal hydrolase (Medtr5g091550) identified in Paape et al. (2013) and to the hypothetical protein gene model Medtr5g091560 which belongs to our gene Module 1.
It was shown in M. truncatula that purifying (negative) selection acts more significantly on broadly expressed genes compared with nonexpressed genes or genes with tissue-specific expression, as the former are enriched with deleterious mutations (Paape et al. 2013). In agreement with this, in our study the expression of genes under strong positive selection looks more tissue-specific or heterogeneous among tissues and experimental conditions (fig. 4). This suggests that molecular adaptation may result either 1) from advantageous de novo gene expression in a specific tissue in a given condition or 2) from the co-opting of genes with tissue-specific expression to other tissues. The PGM gene (Medtr1g102010) seems to fit the last hypothesis as it is mainly expressed in aerial organs together with other genes of Module 1 but it is also uniquely expressed during nodule development. The serine/threonine kinase (Medtr1g104850), by contrast, is more specifically expressed in root, suggesting putative de novo recruitment.
Conclusions
Understanding how contemporary species adapt to changing environments (biotic and abiotic) requires a thorough identification of the genes that respond to positive selective pressures. Consecutively, exploring candidate genes functional annotation and expression data enables the prediction of which molecular functions and putative regulatory network are involved in adaptation. This study reports the first genome-wide selective sweep analysis with high marker density (>15 million SNPs) performed on a representative sample in a plant species, M. truncatula. We demonstrate that selective sweeps identified in the Circum population are more population-specific than those identified in the Far West population, suggesting that the latter represent more ancient adaptations that are more likely to be shared between M. truncatula populations. We also detected two species-wide selective sweeps clearly indicating ancient selective events.
Through the combined analysis of transcriptomic data and genomic features of swept regions (swept length and LD), we could identify four putative gene regulatory networks (called “Module”) involved in adaptation of (at least) the Far West M. truncatula population to its root-associated biotic environment. Interestingly, these Modules mainly concern not only the adaptive control of root symbiotic interaction with Rhizobia (carbon flow in nodule, nodule senescence) but also the adaptation of root cells upon infection with soil microorganisms (DNA damage repair and cell organization processes) and root development upon stress perception. This suggests that root-associated microorganisms may have triggered high selective pressures on the root system of M. truncatula.
Finally, a more theoretical issue that our study highlights is that the number of selective sweep events in a genome does not probably represent an equivalent number of selective pressures. Indeed, a particular phenotypic response (cellular, physiological, and morphological) seldom results from the action of a single gene, but is rather polygenic. Hence, any identified module of coregulated swept genes might represent a group of coadapted genes; that is a group of genes co-responding to the same selective pressure, because involved in the same phenotypic expression. Although polygenic selection with additive effects on the phenotype may tend to generate groups of coadapted genes, it seems rather challenging to detect them using population genomics alone (see, e.g., Berg and Coop 2014). Another hypothesis explaining the existence of coregulated genes identified by selective sweep signatures is the action of epistatic selection; that is the favoring of individuals bearing a specific combination of mutations at different genes (see, e.g., Takahasi and Tajima 2005). As a perspective of this work, assessing epistatic selection signatures either in identified modules of coregulated genes or in groups of detected swept genes could allow identifying networks of coadapted genes in M. truncatula and other species.
Materials and Methods
Inference of Selective Sweeps and SNP Data
Evidence for positive selection was determined using an HMM as described by Boitard et al. (2009). This method exploits the fact that allele frequencies are distorted around a locus under selection, compared with neutrality. We note that the method used here is designed to identify selective sweeps but not balancing selection, another form of positive selection. Contrary to classical approaches like Watterson’s theta or Tajima’s D, the HMM method uses the full allele frequency spectrum rather than a summary statistic computed from this spectrum, resulting in increased detection power (Kim and Stephan 2002). Briefly, in this model, the number of copies of the derived allele at SNP i, denoted Y(i), is taken as the observed state at this SNP. Each SNP i is also assumed to have a hidden state X(i), which can take three different values: “Selection,” for SNPs very close to a swept site; “neutral,” for SNPs far away from any swept site; and “intermediate,” for SNPs in between. These three values are associated with different allele frequency distributions. The “neutral” allele frequency distribution is estimated using all SNPs in the genome. Allele frequency distributions in states “intermediate” and “selection” are deduced from this “neutral” distribution using the derivations in Nielsen et al. (2005) and are typically more skewed toward low- and high-allele frequencies. The hidden states X(i) form a Markov chain along the genome with a per base pair probability, p, of switching state. Under this HMM, the most likely sequence of hidden states can be predicted from the sequence of observed states using the Viterbi algorithm. Each set of consecutive SNPs with the predicted state “selection” is called a sweep window. Besides, applying the backward–forward algorithm to the same HMM provides, for each SNP i, the posterior probability q(i) of hidden state “selection”.
A detailed description of the bioinformatic pipeline used for SNP calling in the Medicago truncatula HapMap project can be found in Stanton-Geddes et al. (2013) and Bonhomme et al. (2014). The raw data set contains 16,515,723 SNPs. For selective sweep analysis, we used the same SNP data set in populations FW and C, thereby reducing the data set to a total of 15,746,610 SNPs due to allele fixation in either of the two populations. The ancestral alleles of 1,283,721 SNPs (8%) could be specified using outgroup species (M. littoralis, M. turbinata, and M. murex), allowing the use of the unfolded allele frequency spectrum. Using even a small proportion of derived alleles is important because it avoids detecting regions under negative selection. Indeed, high frequency-derived alleles cannot be caused by negative selection, contrary to low frequency-derived alleles. For the remaining 14,462,889 SNPs, we used the folded allele count as the observed state, which means that Y(i) and [n(i) − Y(i)] (with n(i) equal to the haplotypes sample size) were considered as the same observation.
The type I error of the above method, that is, the probability that it detects a sweep window in a population that has evolved under neutrality, depends on parameter p (see Boitard et al. 2009 for more details). To control the genome-wide number of false positives, we simulated 1,000 samples of length 100 kb under neutral evolution using the ms software (Hudson 2002) and adjusted p so that sweeps are detected in only 1% of these samples. We performed this calibration for each population, with a sample size equal to the median number of nonmissing alleles per SNP in the observed data (135 in C, 70 in FW). We used the same proportion of unfolded sites as in the observed data (7.8%). Population-scaled mutation and recombination rates also had to be specified for these ms simulations. We estimated the mutation rate using Watterson’s theta (Watterson 1975) and obtained, respectively, 0.00498 and 0.00674 mutations per generation per bp in C and FW. We used a scaled recombination rate of 0.0018 per generation per bp, as reported in (Branca et al. 2011). The ms commands used to generate neutral C-like and FW-like samples were, respectively, “ms 135 1000 -t 498 –r 180 100000” and “ms 70 1000 -t 674 –r 180 100000.” The transition probabilities p obtained from this calibration were 5.96046 × 10−9 in C and 4.0745 × 10−11 in FW.
LD and Genetic Differentiation Analyses
We used the r2 and D′ measures to quantify levels of LD in swept regions and in genome-wide regions of the same size. For each population (FW and C) and swept region, we computed these measures for all SNP pairs involving SNP with MAF ≥0.05 located in the region. For this purpose we used the software plink (Purcell et al. 2007)—version 1.9, available at https://www.cog-genomics.org/plink2/ (last accessed April 23, 2015), with the options “–chr my.chr –from-bp my.start –to-bp my.end –r2 dprime –maf 0.05 –ld-window 50 000 –ld-window-kb 50 –ld-window-r2 0,” where my.chr, my.start and my.end indicate the location of the sweep window. Then, we did the same for all genome-wide SNP pairs involving SNP with MAF ≥0.05 and whose physical distance was below L, the size of the sweep in kilobase. plink options for these computations were “–r2 dprime –maf 0.05 –ld-window 50 000 –ld-window-kb L –ld-window-r2 0.” The properties of LD measures in a given sweep window or in genome-wide regions of the same length were obtained by computing averages and standard deviations among these pairs.
Genetic differentiation was estimated using the FLK statistics which is related to Fst but accounts for the phylogenetic structure of populations and the heterogeneity of genetic drift through the estimation of a population kinship matrix (Bonhomme et al. 2010). FLK values and P values in each swept region were obtained from the software hapflk, available at https://forge-dga.jouy.inra.fr/projects/hapflk (last accessed April 23, 2015). The average genome-wide FLK value was obtained from the same program using option, “—thin 0.01,” which samples at random 1% of the SNP genome-wide. Again, only SNP with MAF ≥0.05 was used for this analysis.
Functional Gene Annotation and Transcriptome Data Analyses
Biological functions associated with the genes under adaptive evolution were investigated using automated annotation tools. First, a global approach using GO terms was carried out, based on M. truncatula protein sequence similarity with A. thaliana, using The Arabidopsis Information Resource –TAIR- (http://www.arabidopsis.org/, last accessed April 23, 2015). Second, a multidatabase-automated sequence annotation approach was used through the Mercator annotation tool (http://mapman.gabipd.org/web/guest/mercator, last accessed April 23, 2015). By searching the TAIR 10 and SwissProt/UniProt Plant databases, Mercator allowed the assignment of a functional class (bin) to each of the 50,894 gene models identified in Mt4.0 M. truncatula genome version (http://www.jcvi.org/medicago/display.php?pageName= General& section=Download, last accessed April 23, 2015). The distribution of functional classes in the candidate gene list and in the genome allowed us to test whether the swept genes list was statistically enriched in some functional classes using the hypergeometric distribution. Finally, to gain additional insight into gene functions, we investigated correlated transcriptional profiles and putative tissue-specific expression among adaptive genes, through hierarchical clustering of Affymetrix gene expression data from 27 selected experimental conditions available at the Medicago truncatula Gene Expression Atlas (http://mtgea.noble.org/v3/, last accessed April 23, 2015; Benedito et al. 2008). To test for putative swept genes enrichment in root or nodule conditions relative to the microarray, we implemented a test based on the hypergeometric distribution, on root and nodule separately, because of the specific nature of nodules (only formed during symbiosis with Rhizobia). The idea was to compare the proportion of swept genes with high expression in root (or nodule) experimental conditions, relative to the proportion of genes in the Medicago Affymetrix microarray which were also highly expressed in the same experimental conditions. To do this, we first normalized microarray gene expression data in each experimental condition to obtain a mean expression level of 0 and a standard deviation of 1. Then, for root and nodule experimental conditions separately (11 and 9, respectively), we calculated for each gene in the microarray (50,894 genes, including the 27 swept genes) the proportion of experimental conditions in which the expression of the gene was higher than the mean expression level of the gene across all 27 conditions (i.e., “strong expression”). We then calculated in our sample (27 swept genes) and in the overall sample (50,894 genes) the proportion of genes which had strong expression in a given range of conditions (e.g., between 0 and 50% of root conditions, or between 50% and 100% of root conditions). We then compared the proportion in the sample with the one in the microarray using a hypergeometric distribution in order to see whether the differences were statistically significant.
Supplementary Material
Supplementary data S1–S4 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Université Paul Sabatier, and the French laboratory of Excellence project (LABEX) “TULIP” (ANR-10-LABX-41). The authors thank the bioinformatics platform Toulouse Midi-Pyrenees (Genotoul).
References
- Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5–21. doi: 10.1104/pp.107.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben C, Toueni M, Montanari S, Tardin MC, Fervel M, Negahi A, Saint-Pierre L, Mathieu G, Gras MC, Noël D, et al. Natural diversity in the model legume Medicago truncatula allows identifying distinct genetic mechanisms conferring partial resistance to Verticillium wilt. J Exp Bot. 2013;64:317–332. doi: 10.1093/jxb/ers337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitard S, Schlötterer C, Futschik A. Detecting selective sweeps: a new approach based on hidden Markov models. Genetics. 2009;181:1567–1578. doi: 10.1534/genetics.108.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme M, André O, Badis Y, Ronfort J, Burgarella C, Chantret N, Prosperi JM, Briskine R, Mudge J, Debéllé F, et al. High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 2014;201:1328–1342. doi: 10.1111/nph.12611. [DOI] [PubMed] [Google Scholar]
- Bonhomme M, Chevalet C, Servin B, Boitard S, Abdallah J, Blott S, Sancristobal M. Detecting selection in population trees: the Lewontin and Krakauer test extended. Genetics. 2010;186:241–262. doi: 10.1534/genetics.110.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin I, Ronfort J, Wozniak F, Olivieri I. Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae) Mol Ecol. 2001;10:1371–1383. doi: 10.1046/j.1365-294x.2001.01278.x. [DOI] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L, et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci U S A. 2011;108:E864–E870. doi: 10.1073/pnas.1104032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Fan B, Du L, Chen Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol. 2004;56:271–283. doi: 10.1007/s11103-004-3381-2. [DOI] [PubMed] [Google Scholar]
- Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 2003;13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- Coates JC, Laplaze L, Haseloff J. Armadillo-related proteins promote lateral root development in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:1621–1626. doi: 10.1073/pnas.0507575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Chantret N, Loridon K, Ronfort J, Bataillon T. Molecular adaptation in flowering and symbiotic recognition pathways: insights from patterns of polymorphism in the legume Medicago truncatula. BMC Evol Biol. 2011;11:229. doi: 10.1186/1471-2148-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djébali N, Jauneau A, Ameline-Torregrosa C, Chardon F, Jaulneau V, Mathé C, Bottin A, Cazaux M, Pilet-Nayel ML, Baranger A, et al. Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Mol Plant Microbe Interact. 2009;22:1043–1055. doi: 10.1094/MPMI-22-9-1043. [DOI] [PubMed] [Google Scholar]
- Espinoza LeC, Huguet T, Julier B. Multi-population QTL detection for aerial morphogenetic traits in the model legume Medicago truncatula. Theor Appl Genet. 2012;124:739–754. doi: 10.1007/s00122-011-1743-0. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Tsuchiya T, Wang XJ, Beasley B, Cuzick A, Tör M, Zhu T, McDowell JM, Holub E, Dangl JL. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49:829–839. doi: 10.1111/j.1365-313X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:97–131. doi: 10.1146/annurev.arplant.50.1.97. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Mould RM, Leech MJ, Johnson SA, Turner L, Schreck SL, Baird KM, Jack PL, Rawsthorne S, Hedley CL, et al. The rug3 locus of pea encodes plastidial phosphoglucomutase. Plant Physiol. 2000;122:1187–1192. doi: 10.1104/pp.122.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath KD, Burke PV, Stinchcombe JR. Coevolutionary genetic variation in the legume-rhizobium transcriptome. Mol Ecol. 2012;21:4735–4747. doi: 10.1111/j.1365-294X.2012.05629.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics. 2005;170:1401–1410. doi: 10.1534/genetics.104.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier B, Huguet T, Chardon F, Ayadi R, Pierre JB, Prosperi JM, Barre P, Huyghe C. Identification of quantitative trait loci influencing aerial morphogenesis in the model legume Medicago truncatula. Theor Appl Genet. 2007;114:1391–1406. doi: 10.1007/s00122-007-0525-1. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y, Sekino T, Yoshioka H, Nakagawa T, Martinoia E, Maeshima M. Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol. 2006;47:309–318. doi: 10.1093/pcp/pcj001. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Anderson HJ, Osmond MJ, Vonarx EJ. Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ Mol Mutagen. 2005;45:115–127. doi: 10.1002/em.20094. [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Lepek VC, D’Antuono AL, Tomatis PE, Ugalde JE, Giambiagi S, Ugalde RA. Analysis of Mesorhizobium loti glycogen operon: effect of phosphoglucomutase (pgm) and glycogen synthase (g/gA) null mutants on nodulation of Lotus tenuis. Mol Plant Microbe Interact. 2002;15:368–375. doi: 10.1094/MPMI.2002.15.4.368. [DOI] [PubMed] [Google Scholar]
- McVean G. The structure of linkage disequilibrium around a selective sweep. Genetics. 2007;175:1395–1406. doi: 10.1534/genetics.106.062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55:853–867. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- Moeder W, Urquhart W, Ung H, Yoshioka K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol Plant. 2011;4:442–452. doi: 10.1093/mp/ssr018. [DOI] [PubMed] [Google Scholar]
- Nallu S, Silverstein KA, Zhou P, Young ND, Vandenbosch KA. Patterns of divergence of a large family of nodule cysteine-rich peptides in accessions of Medicago truncatula. Plant J. 2014;78:697–705. doi: 10.1111/tpj.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape T, Bataillon T, Zhou P, J Y Kono T, Briskine R, Young ND, Tiffin P. Selection, genome-wide fitness effects and evolutionary rates in the model legume Medicago truncatula. Mol Ecol. 2013;22:3525–3538. doi: 10.1111/mec.12329. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfort J, Bataillon T, Santoni S, Delalande M, David JL, Prosperi JM. Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol. 2006;6:28. doi: 10.1186/1471-2229-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Samac DA, PeÑUela S, Schnurr JA, Hunt EN, Foster-Hartnett D, Vandenbosch KA, Gantt JS. Expression of coordinately regulated defence response genes and analysis of their role in disease resistance in Medicago truncatula. Mol Plant Pathol. 2011;12(8):786–798. doi: 10.1111/j.1364-3703.2011.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Annu Rev Plant Biol. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- Siol M, Wright SI, Barrett SC. The population genomics of plant adaptation. New Phytol. 2010;188:313–332. doi: 10.1111/j.1469-8137.2010.03401.x. [DOI] [PubMed] [Google Scholar]
- Song J, Bent AF. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog. 2014;10:e1004030. doi: 10.1371/journal.ppat.1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton-Geddes J, Paape T, Epstein B, Briskine R, Yoder J, Mudge J, Bharti AK, Farmer AD, Zhou P, Denny R, et al. Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS One. 2013;8:e65688. doi: 10.1371/journal.pone.0065688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sanchez-Rodriguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Egli B, Eicke S, Zeeman SC. The debate on the pathway of starch synthesis: a closer look at low-starch mutants lacking plastidial phosphoglucomutase supports the chloroplast-localized pathway. Plant Physiol. 2009;151:1769–1772. doi: 10.1104/pp.109.144931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Matsuhara S, Abe M, Komeda Y. Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell. 2002;14:2085–2093. doi: 10.1105/tpc.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi KR, Tajima F. Evolution of coadaptation in a two-locus epistatic system. Evolution. 2005;59:2324–2332. [PubMed] [Google Scholar]
- Thompson J. Plant evolution in the Mediteranean. Oxford: Oxford University Press; 2005. [Google Scholar]
- Trujillo DI, Silverstein KA, Young ND. Genomic characterization of the LEED.PEEDs, a gene family unique to the medicago lineage. G3 (Bethesda) 2014;4:2003–2012. doi: 10.1534/g3.114.011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Sartorel E, Jardinaud MF, Chardon F, Genin S, Huguet T, Gentzbittel L, Petitprez M. Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula. Mol Plant Microbe Interact. 2007;20:159–167. doi: 10.1094/MPMI-20-2-0159. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarx EJ, Mitchell HL, Karthikeyan R, Chatterjee I, Kunz BA. DNA repair in higher plants. Mutat Res. 1998;400:187–200. doi: 10.1016/s0027-5107(98)00043-8. [DOI] [PubMed] [Google Scholar]
- Wan J, Torres M, Ganapathy A, Thelen J, DaGue BB, Mooney B, Xu D, Stacey G. Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol Plant Microbe Interact. 2005;18:458–467. doi: 10.1094/MPMI-18-0458. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010;10:95. doi: 10.1186/1471-2229-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. The chimeric Arabidopsis cyclic nucleotide-gated ion channel11/12 activates multiple pathogen resistance responses. Plant Cell. 2006;18:747–763. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.