Abstract
Eukaryotes were born of a chimeric union between two prokaryotes—the progenitors of the mitochondrial and nuclear genomes. Early in eukaryote evolution, most mitochondrial genes were lost or transferred to the nucleus, but a core set of genes that code exclusively for products associated with the electron transport system remained in the mitochondrion. The products of these mitochondrial genes work in intimate association with the products of nuclear genes to enable oxidative phosphorylation and core energy production. The need for coadaptation, the challenge of cotransmission, and the possibility of genomic conflict between mitochondrial and nuclear genes have profound consequences for the ecology and evolution of eukaryotic life. An emerging interdisciplinary field that I call “mitonuclear ecology” is reassessing core concepts in evolutionary ecology including sexual reproduction, two sexes, sexual selection, adaptation, and speciation in light of the interactions of mitochondrial and nuclear genomes.
Keywords: coadaptation, genomic conflict, sexual reproduction, sexual selection, adaptation, speciation
Introduction
About 2 billion years ago, a eubacterium joined with or was engulfed by an archaebacterium and so began the evolution of eukaryotes and complex life (Martin and Müller 1998; Williams et al. 2013). The years since this union have been one long negotiation between the symbiotic partners over how to partition cellular duties, which partner is responsible for maintaining which genes, and, above all, how to coordinate genomic products to enable system function (Lane 2011a). The archaebacterial genome gave rise to the nucleus and took command of information functions of the cell including regulatory pathways and the bulk of transcription and translation. The eubacterium became the mitochondrion and specialized in metabolic functions (Cotton and McInerney 2010). The relationship is built on a foundation of cooperation, but conflict is never far away.
To avoid redundancies, facilitate efficiencies, and perhaps escape the higher mutation rates and linkages in the mitochondrial (mt) genome, most genes originally located in the mitochondrion were either lost or transferred to the nuclear genome (Bar-Yaacov et al. 2012). This transfer and purging of genes was extensive—involving more than 1,000 genes and approximately 99% of the original eubacterial genome—but it was not quite complete. For reasons that have remained obscure until relatively recently, mitochondria retained a small set of unique genes (Allen 2003a). Every eukaryote that derives energy from oxidative phosphorylation (OXPHOS) has mt genes that code for OXPHOS function. This mt genome includes genes that code for proteins in the electron transport system as well as for components of the translational machinery needed to create OXPHOS proteins (Wallace 2007). Retention of some OXPHOS genes in the mt genome is essential because of the critical need for immediate transcriptional responsiveness to the local redox state of mitochondria (Allen 2003b; Lane and Martin 2010; Lane 2011a).
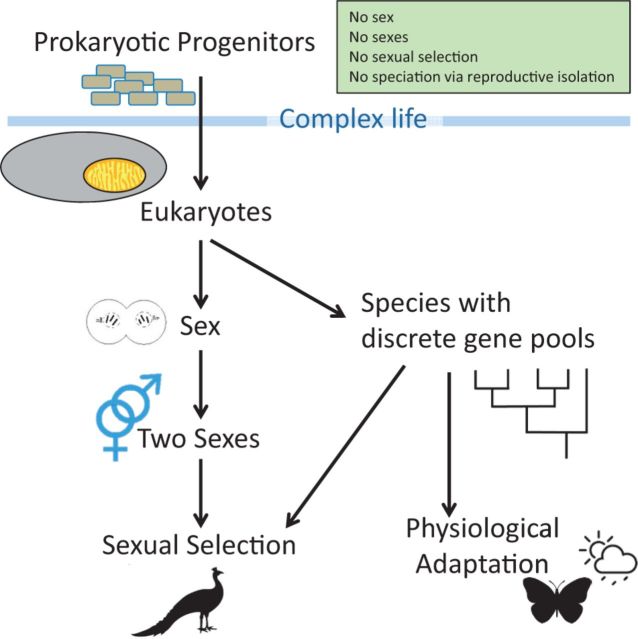
This seemingly trivial constraint necessitating the retention of a few dozen mt genes has enormous implications for the evolution of complex life (Lane 2005). The need for the mitochondrion to have its own genome has played a key role in the evolution of some of the most fundamental features of eukaryotes including sex, death, mate choice, pace of life, speciation through reproductive isolation, inherited disease, and adaptation to novel environments (fig. 1). There is a growing realization among evolutionary ecologists that the biological world simply cannot be understood without taking account of coevolution, coadaptation, and conflict between mt and nuclear genomes. The result is a new integrative discipline that I call mitonuclear ecology, the study of how the interactions between the mt and nuclear genomes shape the evolution and ecology of eukaryotes.
Fig. 1.
Mitonuclear ecology is the study of how the interactions of mt and nuclear genomes shaped the nature of complex life. Mitonuclear interactions are proposed to have been the driving force behind the evolution of such quintessential eukaryotic characteristics as sexual reproduction involving two mating types, sexual selection and ornamentation, reproductively isolated species, and physiological adaptation. Arrows show potential consequences of evolutionary milestones.
Two Genomes; One Soma
Eukaryotes have two genomes but only one soma. (Eukaryotes with chloroplasts have three genomes with the addition of chloroplast genes.) The vast majority of the structures and components of the mitochondrion is encoded by nuclear genes (N-mt genes) (Calvo and Mootha 2010). This means that every product of the mt genome functions in intimate association with products of the nuclear genome (Woodson and Chory 2008; Lane 2011b). Mitonuclear compatibility is a measure of the extent to which interacting mt and nuclear components reach their functional potential (Burton et al. 2013; Meiklejohn et al. 2013). Because mt gene products are closely tied to OXPHOS, the consequences of mitonuclear compatibility manifest as effects on cellular respiration, the core biochemical processes of eukaryotic life.
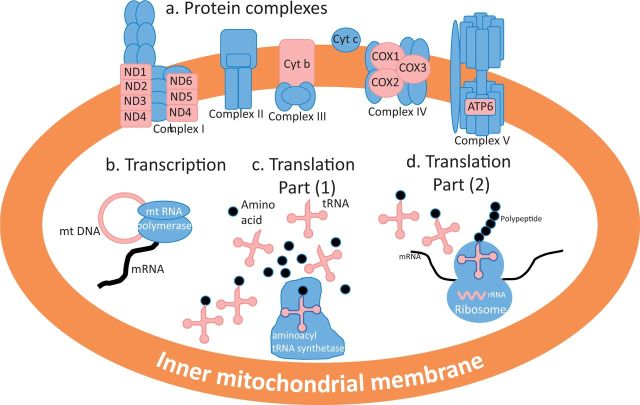
Mitonuclear compatibilities play out primarily in four distinct arenas through the interactions of 1) mt RNA polymerase/mt transcription factors (nuclear) and mt DNA (mt) in the transcription of mt genes, 2) aminoacyl tRNA synthetase (nuclear) and tRNAs (mt) in the loading of tRNAs with correct amino acids as a key step in the translation of mt genes, 3) ribosomal proteins (nuclear) and rRNA (mt) in the translation of mt genes into polypeptides, and 4) the nuclear and mt protein subunits of the complexes that make up the electron transport system and carry out OXPHOS (fig. 2; Rand et al. 2004; Burton and Barreto 2012). Recently discovered short open-reading frames within the mt genome code for signaling peptides that regulate such critical cellular functions as insulin sensitivity and apotosis (Lee et al. 2013, 2015). These mt-encoded peptides interact with nuclear encoded-receptors, presenting the potential for yet more mitonuclear interactions, although these receptor/peptide interactions have yet to be studied from the perspective of mitonuclear compatibility (Horan et al. 2013). It is through these varied mitonuclear interactions that OXPHOS function and hence mt activity is controlled, mitonuclear coevolution and coadaptation are necessitated, and organisms adapt to changing environments.
Fig. 2.
A summary of key points of physical interaction of products of the mt genome (red) and the nuclear genome (blue), all of which occur on or within the inner mt membrane. Interactions of animal mt and N-mt genomes concern protein subunits of the electron transport system in complexes I, III, IV, and V (a) as well as interactions in the transcriptional (b) and translational (c, d) mechanisms needed to produce electron transport system proteins. There are two distinct mitonuclear interactions involved in translation.
The concept of coadapted gene complexes began within the context of the interactions of nuclear genes (Dobzhansky 1946; Orr 2005a) as biologist realized that the interdependencies of genes made it essential that sets of genes be inherited together. A growing body of research, however, suggests that the most significant coadapted gene complexes for eukaryotes are N-mt and mt genes (Lane 2011b; Burton et al. 2013; Foley et al. 2013). The interactions between N-mt and mt gene products are intimate, and their functional match must be exact (Lane 2011b; Pierron et al. 2012). The consequences of poor compatibility between mt and N-mt genes are reduced coupling of the electron transport system (Brand and Nicholls 2011) that results in lower ATP output and increased free radical production (Lane 2011b; Barreto and Burton 2013a) both of which result in significant loss of fitness (Ellison and Burton 2006). Different life histories may select for tighter or looser coupling of the electron transport system (Lane 2011c, 2014), but the need for mitonuclear coordination is universal. Poor compatibility of mt and N-mt genes can also adversely affect transcription of mt genes and translation of mt genes into proteins, which in turn affects cellular respiration (Ellison and Burton 2008; Burton and Barreto 2012). Because populations can evolve idiosyncratic changes to interacting mt and nuclear elements, coadaptation of mt and N-mt genes can form significant barriers to gene flow between populations (Gershoni et al. 2009; Chou and Leu 2010; Burton and Barreto 2012).
Sex Linkage, Coevolution, and Genomic Conflict
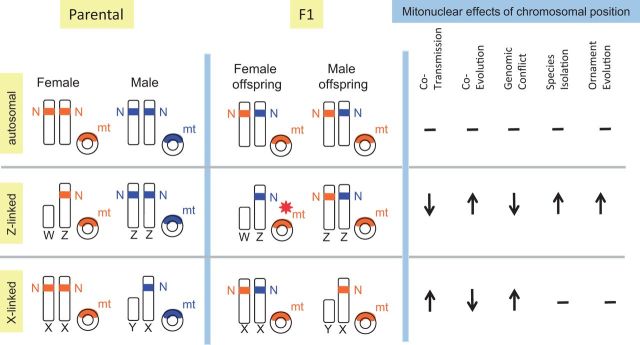
To maintain coadapted mitonuclear complexes, the genes comprising such complexes should be cotransmitted across generations, at least so far as is possible when two genomes are in play (Rand et al. 2004; Rogell et al. 2014). mt genes are, with few exceptions, nonrecombining and maternally transmitted, so cotransmission of mt and N-mt genes occurs through maternal lineages. Emerging evidence suggests that the chromosomal position of the N-mt genes has important consequences for cotransmission of coadapted mt and N-mt genes (Rand et al. 2004; Drown et al. 2012), for the capacity for coevolution of mt and N-mt genes (Rand et al. 2004; Hill 2014a), for genomic conflict between mt and N-mt genes (Drown et al. 2012; Crespi and Nosil 2013), for the efficacy of mate choice (Hill and Johnson 2013), and for the effects of hybridization on the viability of offspring (Burton et al. 2006; Foley et al. 2013; Hill and Johnson 2013; fig. 3).
Fig. 3.
The effects of sex linkage of N-mt genes on cotransmission, coevolution, genomic conflict, speciation, and ornamentation. Compatibility of nuclear and mt genes is color-coded: Blue is compatible with blue and orange with orange. The effects on F1 crosses are shown. The red star indicates offspring with total mt/N-mt incompatibility. Assuming that function is retained so long as one set of compatible genes is retained, then negative F1 effects occur only with Z linkage. For autosomal position and X linkage, incompatibility and function loss do not occur until the F2 hybrid generation.
By convention, chromosomal sex determination systems in which the female is the homogametic sex are designated XY, whereas sex determination systems in which the male is the homogametic sex are designated as ZW. Sex linkage of N-mt genes can affect the probability of cotransmission of mt and N-mt genes because unequal distribution of sex chromosomes between males and females leads to enhanced or diminished association between sex-linked genes and maternally transmitted mt genes (fig. 3). Cotransmission of N-mt and mt genes is facilitated if N-mt genes are positioned on the X chromosome because mt genes are maternally transmitted and the X chromosome is 67% maternally transmitted; consequently, mt genes and X-linked N-mt genes tend to pass together between mothers and daughters (Drown et al. 2012; fig. 3). In contrast, positioning N-mt genes on any particular autosome neither facilitates nor hinders cotransmission with mt genes because autosomes and mt genes are transmitted independently. Thus, strictly from the standpoint of promoting the cotransmission of coadapted N-mt and mt genes, N-mt genes should be positioned on the X chromosome (Rogell et al. 2014). However, recent studies have found that N-mt genes occur on X chromosomes less frequently than expected by chance in mammals and Caenorhabditis elegans but not different than expected by chance in other XY taxa that were examined (Drown et al. 2012; Dean et al. 2014). In mammals, this underrepresentation of N-mt genes on the X chromosome could simply be a consequence of underrepresentation of N-mt genes on the autosomes that evolved into the mammalian sex chromosomes (Dean et al. 2015). To date, N-mt genes have not been found to be overrepresented on the X in any taxa.
Given the advantages of the cotransmission of mt and N-mt genes that result from positioning N-mt genes on X chromosomes, why are few N-mt genes on the X chromosome? The answer seems to be that the advantage of cotransmission of coadapted mt and N-mt genes is not the only factor affecting chromosomal architecture. The cotransmission of N-mt and mt genes resulting from X-linkage of N-mt genes necessarily involves linkage of mt and N-mt genes and hence restriction of independent evolution (Rand et al. 2004; Hill 2014a; fig. 3). Coevolution of genes is facilitated when interacting genes have the freedom to evolve independently (Barton and Charlesworth 1998). X-linkage and cotransmission of mt and N-mt genes therefore inhibit coevolution of interacting mt and N-mt genes (Hill 2014a). Coevolution of mt and N-mt genes seems essential because, in many eukaryotes, mt genes are subject to mutation rates that are much higher than those of nuclear genes (Lynch 1997) and the mt genome is haploid and transmitted clonally. With high mutation rate and no recombination, the mt genome is subjected to perpetual mutational erosion (Wallace 2010a). It has been proposed that N-mt genes must constantly coevolve with mt genes to compensate for deleterious mt mutations (Osada and Akashi 2012; Burton and Barreto 2012; Levin et al. 2014). Coevolution of mt and N-mt genes may also play an important role in physiological adaptation to novel environments (Pierron et al. 2012). Thus, the benefits of cotransmission that results from X-linkage of N-mt genes may be countered by the costs of restricted coevolution of mt and N-mt genes (Drown et al. 2012; Hill 2014a).
Another negative consequence of cotransmission of mt and N-mt genes when N-mt genes are positioned on the X chromosome is genomic conflict (Drown et al. 2012; Rogell et al. 2014). Genomic conflict gives rise to the “mother’s curse” because, when transmission of genetic elements is primarily through female lines, selection can favor mutations that are beneficial to females regardless of whether they are detrimental to males (Frank 1996; Gemmell et al. 2004). The result can be an accumulation of alleles that are detrimental to males in mt and X-linked N-mt genes. Selection for avoidance of mother’s curse might select for the positioning of N-mt genes on chromosomes other than the X (Drown et al. 2012). Alternatively, it has been proposed that cotransmission of male and female mitochondria could stem the evolution of the mother’s curse (Kuijper et al. 2015). Paternal transmission of mitochondria varies from essentially zero in some animals to abundant transmission in some plants (Gyllensten et al. 1991; McCauley 2013). More paternal mt contribution across generations subjects the mt genome to selection against male-damaging alleles, and models show that transmission of both male and female mitochondria could restrict the evolution of alleles that are detrimental to males in the mt genome (Kuijper et al. 2015). The role of genomic conflict in mitonuclear coevolution is a topic of growing interest among evolutionary biologists (Wolff et al. 2014).
Relative to putting N-mt genes on the X chromosome, positioning these genes on the Z chromosome presents entirely different selection pressures. Males carry two Z chromosomes whereas female carry only one. Thus, the Z chromosome is 67% paternally transmitted and genes on the Z chromosome are cotransmitted with mitochondria only 33% of the time—less than autosomes (Hill 2014a; fig. 3). There are important consequences of these cotransmission rates if N-mt genes are positioned on the Z. First, there can be no mothers curse involving N-mt genes on the Z because male transmission of the Z subjects male-detrimental genes to negative selection. Second, coevolution is facilitated because, with reduced cotransmission, mt and N-mt genes have greater capacity to evolve independently (Drown et al. 2012; Hill 2014a). Third, the benefits gained from enhanced coevolution and no sexual conflict may be countered by a loss of fitness lost from reduced cotransmission and diminished coadaptation. The limited data available on sex linkage of N-mt genes indicate that N-mt genes are present on the Z at the rate expected by chance (Drown et al. 2012; Dean et al. 2014). It seems likely that a need to balance the conflicting costs and benefits of sex linkage of N-mt genes has guided the evolution of chromosomal positioning of those genes, but at present there are too few data on the positions of N-mt genes that interact with mt genes—and particularly that subset of N-mt genes that engage in close functional interactions with mt genes—to fully evaluate the evolutionary pressures that determine chromosomal positions of the genes that determine respiratory function.
Whether N-mt genes are positioned on autosomes or sex chromosomes can also affect gene flow in the face of mitonuclear incompatibilities between populations. Crossing Tigriopus californicus copepods from geographically isolated populations results in respiratory dysfunction and fitness loss in hybrids because of incompatibilities in mt and N-mt genes between different populations (Ellison and Burton 2006). Importantly, these fitness effects are not revealed until the F2 generation (Burton et al. 2006) because there is no sex linkage of N-mt genes in copepods (Foley et al. 2013). With N-mt genes located on autosomes, all individuals in an F1 hybrid generation carry both maternal and paternal N-mt genes (Burton et al. 2013; fig. 3), and the single copy of a compatible N-mt gene can mask effects of paternally contributed incompatible N-mt genes in the first generation. Thus, in taxa in which N-mt genes are located on autosomes or on X chromosomes, postzygotic selection against hybrid pairings that yield mitonuclear incompatibilities is delayed for a generation. Such a generational delay in negative fitness effects will permit nuclear genes to recombine and diffuse over population boundaries (Toews and Brelsford 2012). In contrast, if N-mt genes are positioned on Z chromosomes, the negative effects of hybrid dysfunction are experienced in the F1 generation because females receive their only N-mt genes from their father (fig. 3). Chromosomal position of N-mt genes could thus play a key role in speciation (Hill and Johnson 2013), especially in organisms with ZW sex-determination systems.
The evolutionary forces that shape genomic distribution of N-mt genes as well as the effects of chromosomal position of N-mt genes is an area of active research, and the perspective of mitonuclear genomic interactions is changing the interpretation of genomic architecture.
Sex and Sexual Selection
Two profound consequences of the interactions of mt and nuclear genomes are sexual reproduction and sexual selection. Understanding the evolution of these core characteristics of eukaryotes has presented among the most enduring challenges to evolutionary biologists. The great evolutionary thinkers of the past 150 years from Darwin to Fisher to Maynard Smith spent careers pondering these topics, and yet sexual reproduction and sexual selection remain incompletely understood. New theories for the evolution of sexual reproduction and female mate choice propose that a key missing piece to these evolutionary puzzles is a consideration of the need to facilitate the cotransmission of coadapted mt and N-mt genes.
The fundamental paradox of sex is that it invokes a 2-fold cost through the 50% reduction in genetic representation of a parent in its offspring (Maynard Smith 1978). Theoretically, a sexually reproducing individual could double its reproductive output by reproducing asexually, and yet multigenerational asexual reproduction is rare in eukaryotes (Maynard Smith 1978). The key advantages of sex are that it allows for recombination, which enhances selection on alleles at one locus independent of alleles at other loci thereby promoting genetic diversity (Otto and Michalakis 1998; Otto and Barton 2001; McGaugh et al. 2012).
Sexual reproduction with recombination may have evolved during the initial stages of eukaryotic evolution when the “host” nuclear genome was bombarded by genetic elements from the “endosymbiont” mt genome (Lane 2014). The resulting insertions of numerous genetic elements into the nuclear genome may have led to the evolution of both eukaryotic introns and spliceosomes (Martin and Koonin 2006; Roy and Gilbert 2006) with recombination playing a key role by allowing deleterious insertions to be selected against independent of noncorrupted portions of the genome. Because it is hard to test such hypotheses for the origin of sexual reproduction, evolutionary biologists have focused on the question of why sexual systems persist when asexual alternatives seem to evolve readily, even in the most complex eukaryotes. In recent decades, the consensus explanation has been that sexual reproduction is necessary to generate sufficient genetic diversity to keep pace with environmental uncertainty, particularly related to pathogens (Crespi and Schwander 2012). This is known as the Red Queen Hypothesis (Lively et al. 1990).
A new idea for the benefits of sexual reproduction that is being developed by Justin Havird, Matthew Hall, and Damian Dowling focuses on mitonuclear coevolution and particularly on the imperative for nuclear genes to compensate for deleterious mt mutations. Because the mt genome is subject to much higher rates of mutation than the nuclear genome (Lynch 1997) and is typically transmitted without the potential for recombination, a mutational load builds in mitochondria, resulting in inefficiencies in respiration and decreased fitness (Denver et al. 2000; James and Ballard 2003). Complementary changes in N-mt genes can compensate for deleterious changes in mt genes (Frank 1996; Barreto and Burton 2013b), thus increasing fitness. A new theory proposes that the key advantages of sexual reproduction lie in the increased capacity of N-mt genes to keep pace with evolutionary changes in mt genes through rapid recombination of nuclear genes to find compensatory combinations for mitonuclear complexes (Havird et al. 2015). Evolution of nuclear genes is most responsive to coadaptation with mt genomes if there is minimal selective interference between loci (Keightley and Otto 2006). In other words, sexual reproduction with recombination enables the evolution of individual genes that promote mitonuclear coadaptation, avoiding selective sweeps of entire chromosomes (Lane 2014). This is the Red Queen Hypothesis with a mitonuclear twist—sex with recombination gives rise to the genetic diversity in N-mt genes that enables perpetual coevolution between mt and N-mt genes. However, this mitonuclear coadaptation model for the evolution of sexual reproduction is not mutually exclusive of the Red Queen model focused on the need for genomic diversity to keep evolutionary pace with pathogens and other environmental changes. The Red Queen likely runs for both reasons. Future research can determine whether mitonuclear coadaptation or diversity of nuclear genes is the bigger driver in the evolution of sex, but a focus on mitonuclear interactions is already stimulating new thinking with regard why sexual reproduction is the dominant strategy of eukaryotes.
It is not only the evolution of sex that has puzzled evolutionary biologists but also the evolution of two mating types (Hurst and Hamilton 1992; Greiner et al. 2014), and one of the most exciting insights from consideration of mitonuclear interactions is a new explanation for two sexes. Sexual reproduction requires two partners to contribute genetic material but that does not mean that there need be only two mating types. Because we live in a two-sex system and all of the organisms with which we are most familiar also exist in two-sex systems, any other mating system is nearly unimaginable. But there are clear drawbacks to having only two sexes. With two sexes, half of the individuals in a population are eliminated as mates (Lane 2005). Starting from a two-mating-type system, if a mutant mating type arises that is compatible with both original mating types, it will spread through the population unless there is some benefit to only two mating types (Fisher 1958). By the same logic, a fourth, fifth, and so on mating type would also evolve, and we should expect there to be many mating types. So why is two mating types by far the common pattern in eukaryotes?
The answer seems to be that two mating types is the best system to ensure that there is uniparental inheritance of mt genes (Hoekstra 2000). The potential for conflict is already substantial when one nuclear genome must coexist with one mt genome. When more than one mt type with functional differences in protein-coding regions is present in an organism, the resulting genomic conflicts among mitochondria can completely derail the function of the individual (Innocenti et al. 2011; Lane 2012). Therefore, there are substantial benefits to having only one mt type transmitted to a zygote each generation and this is facilitated by having exactly two mating types (Hurst and Hamilton 1992). But perhaps even more vital than avoiding genomic conflict among mitochondria is ensuring coadaptation between mt and nuclear genes. Lane (2005, 2014) proposed that the best way to ensure mitonuclear compatibility in offspring that result from two parental nuclear genotypes (through sexual reproduction) is to test the novel, recombined nuclear genotype against but a single mt genotype. Selection among zygotes that each have a single mt genome matched to a single nuclear genome facilitates selection for the most fit mt genome and the best mitonuclear coadaptation. In contrast, if multiple mt genomes are transmitted each generation, then selection against deleterious or less fit mt genes is ineffective, mt mutations accumulate, and mitonuclear coadaptation declines. This hypothesized imperative for transmission of only one mt type was recently modeled and, as predicted, models showed that two mating types best enabled the maintenance of mitonuclear coadaptation and respiratory function across generations for most taxa (Hadjivasiliou et al. 2012, 2013). Models also predicted the situations in which biparental inheritance of mitochondria would be favored (Hadjivasiliou et al. 2013). Mitonuclear genetic interactions may thus create the impetus for two sexes and for all of the complexities that come with coordinating the union of two individuals.
The evolution of two mating types engaging in sexual reproduction seems to have been an essential step in the evolution of eukaryotes, enabling the maintenance of coadapted mitonuclear gene complexes. But coadaptation also relies critically on matching gametes from appropriate gene pools that share compatible mt and N-mt genes (Gershoni et al. 2009; Hill 2014a). The evolution of two mating types inevitably gives rise to sexual selection—competition for access to individuals of the opposite mating types (Andersson 1994; Parker 2014). Because females tend to invest more resources in offspring than males, females typically serve as the choosy sex, and female mate choice is the driving force in sexual selection (Andersson and Simmons 2006). Evolutionary biologists have long recognized that females benefit by choosing conspecific rather than heterospecific mates, and that such female choice gives rise to selection for male traits such as sounds, colors, shapes, chemical signals, and electrical discharges that unambiguously signal species identity (Wallace 1889; Ryan and Rand 1993; Ritchie 2007; Mendelson and Shaw 2012). Until recently, the benefits of mating with conspecifics was explained by invoking compatibility among nuclear genotypes (e.g., Servedio 2001), but the imperative to choose a mate that will provide N-mt genes that are compatible with mt genes, and hence that will sire fully functional offspring, has recently been proposed to be a driving force in the evolution of female choice for ornamental traits in animals (Hill and Johnson 2013). In essence, this new hypothesis proposes that female mate choice for ornaments arises from the need for prezygotic sorting of compatible mt and N-mt gene pools and that ornaments should be distinctive and readily recognizable signals of the boundaries between pools of compatible mt and N-mt genes. Consistent with this hypothesis, ornamentation in animals tends to have modest variation within species and large and quantitative differences between species (Dalrymple et al. 2015). Moreover, if mate choice evolves in response to postzygotic selection to ensure mitonuclear compatibility, then sexual selection will be a follower rather than an initiator of speciation. This prediction is in opposition to the growing literature proposing that mating preferences can promote speciation by creating prezygotic barriers to gene flow (Ritchie 2007; Safran et al. 2013). These are clearly testable alternative ideas for the interaction of sexual selection and speciation and should be the focus of future research.
A mitonuclear-focused interpretation of female mate choice and male ornamentation holds the promise to explain why sex determination systems of taxa affect the potential for sexual selection. Across taxa as disparate as insects, amphibians, mammals and birds, ZW taxa are more ornamented than XY taxa (Reeve and Pfennig 2003). This fascinating and largely unexplained pattern makes sense in light of the need for mitonuclear coadaptation. When N-mt genes are on autosomes or X-linked, the negative fitness consequences for choosing a mate from outside of a species boundary can be completely masked in the F1 generation; they are not revealed until F2 and subsequent generations (Burton et al. 2006; fig. 3). This generational delay in fitness consequences of hybrid pairing will weaken selection for female choice for signals of species identity. Hence, female choice for signals of species identity evolves most effectively if N-mt genes are Z linked, and the serious fitness costs of mismatching mitonuclear genes are revealed in the F1 generation (Hill and Johnson 2013; fig. 3). The presumption at present is that the sex determination system of a taxon can affect the potential for sexual selection; however, it is also plausible that selective pressure for better sorting of prospective mates for mitonuclear compatibility could influence the evolution of sex determination systems.
Sexual selection has the potential to enable mitonuclear coadaptation far beyond avoiding the blunder of mixing incompatible mitonuclear genes between species. In sexually reproducing organisms, nuclear genotypes are created anew each generation with new mutations introduced and new recombinations of N-mt genes generated (Lane 2011b, 2014). Some of these new genotypes will be more compatible with mt genomes than others (Dowling et al. 2007; Arnqvist et al. 2010). Females should benefit by recognizing and avoiding those prospective mates with poorly functioning mitonuclear genes (arising from mutation, recombination, or gene flow). The benefits to females for choosing males with fully functional cellular respiration favor mate choice for indicator traits in males that signal OXPHOS efficiency (Hill 2011; Hill and Johnson 2013). Recent work suggests that signaling cellular respiration may be a common feature of male ornaments (Johnson and Hill 2013; Hill 2014b), but more research is needed. In species in which males are monogamous and invest heavily in reproduction, males as well as females should assess mitonuclear function in prospective mates, and mate choice by males for mt function in females could explain gaudy ornamental plumage in many monogamous species of birds (Ligon 1999). These new ideas that consider mate choice in light of the need for mitonuclear coadaptation can potentially integrate well with conventional models of sexual selection (Kuijper et al. 2012) perhaps clarifying long-standing puzzles like the lek paradox (Hill and Johnson 2013). Already indicator models of sexual selection propose that females choose males for good genes (Hamilton and Zuk 1982). The mitonuclear compatibility model of sexual selection simply emphasizes the assessment of genes needed for mitonuclear coadaptation. Moreover, once selection for signals of mitonuclear type has been fixed in populations, a runaway process (Arnold 1983) can give rise to the fantastic ornamentation of many animal taxa (Hill and Johnson 2013).
Adaptation and Speciation
According to these new hypotheses, two mating types and female mate choice both evolved, at least in part, to ensure mitonuclear coadaptation across generations. In ever-changing environments, however, stasis in energy production systems over evolutionary time is untenable. Populations must adapt to changing environments or suffer serious fitness consequences (Orr 2005b). Until recently, any variation in mt genes was proposed to be neutral, with negligible contributions of the mt genome to adaptive changes in response to the environment (Ballard and Kreitman 1995; Galtier et al. 2009). A rapidly expanding literature, however, indicates that mt genes are under strong selection, both stabilizing and directional (Hahn 2008; Stoeckle and Thaler 2014; Garvin et al. 2015), and that the evolution of mt genes is fundamental to environmental adaptation (Dowling et al. 2008; Wallace 2013; Levin et al. 2014; Morales et al. 2015). For instance, the capacity to fly over the Himalayan Mountains and achieve a much shorter migration route in Bar-headed Geese (Anser indicus) arose through a single amino acid substitution in the mt gene COX3 (cytochrome c oxidase subunit 3) which is a core subunit of Complex IV of the electron transport system (Scott et al. 2011, 2015). The new mt genotype allows the Bar-headed Goose, unlike other taxa in their clade, to function in the low-oxygen environment of very high altitudes. This Bar-headed Goose study is an example of a growing literature linking functional changes in mt genes within populations, including humans, to better attunement to environmental conditions (Wallace 2010b, 2013; Cheviron and Brumfield 2011; Wilson et al. 2013). Moreover, standing variation in mt haplotypes within populations is now being linked to respiratory function and fitness (Kurbalija Novičić et al. 2015).
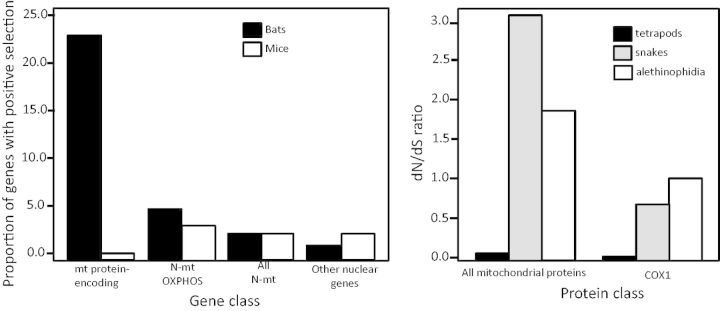
Presumably, unpredictable and variable environments select for haplotype diversity in some populations. mt evolution underlies not only small physiological adjustments to thermal environment or altitude but also major adaptive radiations. Recent studies link functional changes in key mt genes through natural selection to radiations associated with flight in bats (Shen et al. 2010) and large prey consumption in snakes (Castoe et al. 2008; fig. 4).
Fig. 4.
The role of genetic changes to respiratory chain function in major adaptive radiations. In the transition from terrestrial locomotion to flight that accompanied the evolution of bats, there was strong selection for changes to the functional capacity of the respiratory chain to allow for increased ATP production to power flight muscles (Shen et al. 2010). In bats, 23% of the mt genes and about 5% of N-mt genes that code for OXPHOS proteins show evidence of positive selection, a proportion that is much higher than for non-OXPHOS N-mt genes or non-mt nuclear genes (left). Moreover, these rates of positive selection on OXPHOS genes are significantly higher in bats than in mice (left). Similarly in the evolution of snakes, nonsynonymous changes in mt OXPHOS genes, and particularly in cytochrome c oxidase subunit 1, suggest that a major shift in respiratory function accompanied the major shifts in body plan and prey consumption (Castoe et al. 2008) (right). The Alethinophidia lineage is the clade within all snakes with the greatest adaptive shifts. Deducing whether respiratory chain innovation leads or follows adaptive radiation will be a fascinating focus of future studies. Left panel redrawn from Shen et al. (2010) and right panel drawn from data in Castoe et al. (2008).
Adaptation to environmental conditions is a topic intimately connected to speciation. In eukaryotes, the maintenance of discrete pools of coadapted genes is the cornerstone of species concepts (Coyne and Orr 2004; Price 2007). By conventional models of speciation, when populations are isolated, divergence of genomes leads to reproductive isolation because there is disruption of coadapted gene complexes when the diverged genotypes are blended in hybrid offspring (Coyne and Orr 2004; Turelli and Moyle 2007). Until recently, the genomic interactions that gave rise to incompatibilities and reduced fitness of hybrids were proposed to involve nuclear genes. mt genotypes were viewed as neutral markers of divergence (Gerber et al. 2001; Dowling et al. 2008).
Consideration of the coadaptation of mt and N-mt genes and the speed with which mitonuclear incompatibilities can evolve is now stimulating novel hypotheses for the process of speciation. This new take on speciation proposes that once gene flow is disrupted, the high mutational rate of mt genes will lead to rapid and unpredictable divergences between populations in coadapted mt and N-mt gene complexes (Gershoni et al. 2009; Chou and Leu 2010). These diverged mt and N-mt genotypes are then incompatible with the mt/N-mt combinations from other populations creating postzygotic barriers to gene flow and the opportunity for speciation (Burton and Barreto 2012). Observations from model systems such as copepods (Burton et al. 2013), yeast (Chou and Leu 2010), Drosophila (Sackton et al. 2003; Meiklejohn et al. 2013), parasitoid wasps (Ellison et al. 2008), seed beetles (Arnqvist et al. 2010), and fish (Bolnick et al. 2008) provide empirical support that loss of viability resulting from mitonuclear incompatibilities in crosses between diverged populations can serve as a postzygotic isolating mechanism.
Studies by Burton and colleagues (Ellison and Burton 2006; Burton et al. 2013a) on T. californicus copepods provide unique insight into the potential role of mitonuclear interactions in population structure and speciation. These researchers demonstrated that there are incompatibilities between N-mt and mt genes among populations of copepods along the Pacific coast of California that reduce the viability of hybrid offspring from interpopulation crosses. These mitonuclear incompatibilities likely evolved in large part due to drift, but also in response to selection for different thermal environments. Tigriopus californicus copepods from warm water habitats are more heat tolerant than copepods from cooler water habitats (Pereira et al. 2014). Insight into how such divergent populations can remain a single species with a shared evolutionary history came from experiments crossing individuals from copepod populations with different thermal tolerances. Although most hybrid offspring from F2 and later generations showed significant loss of fitness due to mitonuclear incompatibilities, occasional hybrid mitonuclear combinations from diverged, heat-tolerant populations had greater tolerance to heat stress than any parental population (Pereira et al. 2014). It seems that even though most hybrid crosses result in lost fitness, rare advantageous mitonuclear interactions that significantly increased fitness can maintain sufficient gene flow to resist speciation. More model systems, including especially model systems with the potential for sex linkage and hence F1 incompatibilities (copepods have no sex chromosomes), are needed to more fully test the dynamics of mitonuclear interactions in population structure and speciation. Toward this end, it is noteworthy that recent studies have found that some bird taxa are the result of hybridization events, with novel mitonuclear combinations playing a key role in both in isolation of the new hybrid gene pool and generation of a fitness advantages in some environments for the new hybrid taxa (Toews et al. 2014; Trier et al. 2014).
In light of the role of mitonuclear compatibility in the divergence of populations, the strong association between COX1 genotype (the most widely used DNA barcode gene for animals) and species boundaries (Hebert and Gregory 2005; Bucklin et al. 2011) is especially intriguing. COX1 is one of three mt genes that form the catalytic center of complex IV in the electron transport system—the complex that controls the flow of electrons and the reduction of oxygen (Arnold 2012; Pierron et al. 2012). Cytochrome c oxidase subunits produced by mt genes are intimately associated with N-mt genes such that mitonuclear coadaptation is essential for proper respiratory function (Schmidt et al. 2001; Pierron et al. 2012). An emerging idea is that specific mt genes that serve as DNA barcodes, including especially COX1, are not simply neutral markers of allopatric speciation. Rather, COX1 may be such an effective marker of species boundaries because it plays a role in speciation (Lane 2009). By this idea, divergence in interacting mt and nuclear genes between populations creates postzygotic barriers to gene flow (Gershoni et al. 2009; Lane 2009; Chou and Leu 2010). Such incompatibilities in mt/N-mt genes might evolve much more rapidly than incompatibilities among nuclear genes because mt genes mutate at a higher rate and the coadaptations of mt/N-mt genes accommodate less divergence (Burton and Barreto 2012). DNA barcoding with mt genes has been successful in identifying species boundaries for most metazoan groups (Hebert and Gregory 2005), including among closely related and recently diverged species (Hebert et al 2003; Baker et al. 2009). It is intriguing to speculate that mt genes are the best markers of species boundaries because speciation in metazoans is a consequence of the disruption of gene flow resulting from mitonuclear interactions. This mitonuclear speciation hypothesis predicts that gene flow between species should greater for nuclear genes than for mt genes or N-mt genes as was observed among populations of the Eastern Yellow Robin (Eopsaltria australis) (Morales et al. 2015). The role of mitonuclear interactions in speciation is an exciting area of current research.
Conclusions
The need for local regulation of OXPHOS necessitated the retention of an mt genome, and the imperative for coadaptation of mt and nuclear genomes changed the fundamental nature of the organism. Sexual reproduction with two mating types became essential to maintain mitonuclear coadaptation across generations. Under pressure for maintaining mitonuclear coadaptation, female mate choice evolved for male ornaments that signaled mitonuclear type as well as OXPHOS efficiency. Divergence in key OXPHOS genes under selection for respiratory adaptation created barriers to gene flow leading to speciation, with species boundaries defined by ornamentation. The chromosomal position of N-mt genes affected the cotransmission of coadapted mt/N-mt complexes as well as the potential for coevolution of mt and N-mt genes, and these sex-linkage effects hold large implications for how chromosomal sex determination might affect patterns of evolution. Such an integration of the interactions of mt and nuclear genomes into theories of sex, sexual selection, adaptation, and speciation is largely speculative at this point, but mitonuclear ecology holds exciting potential to transform our understanding of the most basic characteristics of eukaryotes.
Acknowledgments
The author thanks Justin Havird, Steve Dobson, Ryan Weaver, Becca Koch, Wendy Hood, two anonymous reviewers, and the participants of the 2014 graduate seminar “Mitonuclear Ecology” at Auburn University for stimulating discussions. His interest in the role of mitonuclear interactions in eukaryotic evolution was ignited by books and essays by Nick Lane.
References
- Allen JF. 2003a. Why chloroplasts and mitochondria contain genomes. Comp Funct Genomics. 4:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. 2003b. The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci. 358:19–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. 1994. Sexual selection. Princeton (NJ): Princeton University Press. [Google Scholar]
- Andersson M, Simmons LW. 2006. Sexual selection and mate choice. Trends Ecol Evol. 21:296–302. [DOI] [PubMed] [Google Scholar]
- Arnold S. 2012. The power of life—cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 12:46–56. [DOI] [PubMed] [Google Scholar]
- Arnold SJ. 1983. Sexual selection: the interface of theory and empiricism. In: Bateson PPG, editor. Mate choice. Cambridge: Cambridge University Press; p. 67–107. [Google Scholar]
- Arnqvist G, Dowling DK, Eady P, Gay L, Tregenza T, Tuda M, Hosken DJ. 2010. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution 64:3354–3363. [DOI] [PubMed] [Google Scholar]
- Baker AJ, Tavares ES, Elbourne RF. 2009. Countering criticisms of single mitochondrial DNA gene barcoding in birds. Mol Ecol Resour. 9:257–268. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Kreitman M. 1995. Is mitochondrial DNA a strictly neutral marker? Trends Ecol Evol. 10:485–488. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D, Blumberg A, Mishmar D. 2012. Mitochondrial-nuclear co-evolution and its effects on OXPHOS activity and regulation. Biochim Biophys Acta-Gene Reg Mech. 1819:1107–1111. [DOI] [PubMed] [Google Scholar]
- Barreto FS, Burton RS. 2013a. Elevated oxidative damage is correlated with reduced fitness in interpopulation hybrids of a marine copepod. Proc R Soc Lond B Biol Sci. 280:20131521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto FS, Burton RS. 2013b. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 30:310–314. [DOI] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. 1998. Why sex and recombination? Science 281:1986–1990.. [PubMed] [Google Scholar]
- Bolnick DI, Turelli M, Lopez-Fernández H, Wainwright PC, Near TJ. 2008. Accelerated mitochondrial evolution and “Darwin's corollary”: asymmetric viability of reciprocal F1 hybrids in Centrarchid fishes. Genetics 178:1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Nicholls D. 2011. Assessing mitochondrial dysfunction in cells. Biochem J. 435:297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucklin A, Steinke D, Blanco-Bercial L. 2011. DNA barcoding of marine metazoa. Ann Rev Mar Sci. 3:471–508. [DOI] [PubMed] [Google Scholar]
- Burton RS, Barreto FS. 2012. A disproportionate role for mtDNA in Dobzhansky–Muller incompatibilities? Mol Ecol. 21:4942–4957. [DOI] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. 2006. The sorry state of F-2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am Nat. 168:S14–S24. [DOI] [PubMed] [Google Scholar]
- Burton RS, Pereira RJ, Barreto FS. 2013. Cytonuclear genomic interactions and hybrid breakdown. Ann Rev Ecol Evol Syst. 44:281–302. [Google Scholar]
- Calvo SE, Mootha VK. 2010. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 11:25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD. 2008. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS One 3:e2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron Z, Brumfield R. 2011. Genomic insights into adaptation to high-altitude environments. Heredity 108:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JY, Leu JY. 2010. Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. Bioessays 32:401–411. [DOI] [PubMed] [Google Scholar]
- Cotton JA, McInerney JO. 2010. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc Natl Acad Sci U S A. 107:17252–17255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. New York: Sinauer Associates, Inc. [Google Scholar]
- Crespi B, Nosil P. 2013. Conflictual speciation: species formation via genomic conflict. Trends Ecol Evol. 28:48–57. [DOI] [PubMed] [Google Scholar]
- Crespi B, Schwander T. 2012. Asexual evolution: do intragenomic parasites maintain sex? Mol Ecol. 21:3893–3895. [DOI] [PubMed] [Google Scholar]
- Dalrymple RL, Hui FK, Flores-Moreno H, Kemp DJ, Moles AT. 2015. Roses are red, violets are blue–so how much replication should you do? An assessment of variation in the colour of flowers and birds. Biol J Linn Soc Lond. 114:69–81. [Google Scholar]
- Dean R, Zimmer F, Mank JE. 2014. The potential role of sexual conflict and sexual selection in shaping the genomic distribution of mito-nuclear genes. Genome Biol Evol. 6:1096–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Zimmer F, Mank JE. 2015. Deficit of mito-nuclear genes on the human X chromosome predates sex chromosome formation. Genome Biol Evol. 7:636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. 2000. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science 289:2342–2344. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1946. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics 31:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Hailer F, Arnqvist G. 2007. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster . Genetics 175:235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol. 23:546–554. [DOI] [PubMed] [Google Scholar]
- Drown DM, Preuss KM, Wade MJ. 2012. Evidence of a paucity of genes that interact with the mitochondrion on the x in mammals. Genome Biol Evol. 4:875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C, Niehuis O, Gadau J. 2008. Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J Evol Biol. 21:1844–1851. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Burton RS. 2006. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution 60:1382–1391. [PubMed] [Google Scholar]
- Ellison CK, Burton RS. 2008. Genotype-dependent variation of mitochondrial transcriptional profiles in interpopulation hybrids. Proc Natl Acad Sci U S A. 105:15831–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1958. The genetical theory of natural selection. New York: Dover. [Google Scholar]
- Foley B, Rose C, Rundle D, Leong W, Edmands S. 2013. Postzygotic isolation involves strong mitochondrial and sex-specific effects in Tigriopus californicus, a species lacking heteromorphic sex chromosomes. Heredity 111:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S. 1996. Mitochondria and male disease. Nature 383:224. [DOI] [PubMed] [Google Scholar]
- Galtier N, Nabholz B, Glémin S, Hurst G. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4550. [DOI] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Sazanov LA, Gharrett AJ. 2015. Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J Zool Syst Evol Res. 53:1–17. [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. 2004. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 19:238–244. [DOI] [PubMed] [Google Scholar]
- Gerber AS, Loggins R, Kumar S, Dowling TE. 2001. Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Annu Rev Genet. 35:539–566. [DOI] [PubMed] [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D. 2009. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays 31:642–650. [DOI] [PubMed] [Google Scholar]
- Greiner S, Sobanski J, Bock R. 2014. Why are most organelle genomes transmitted maternally? Bioessays 37:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U, Wharton D, Josefsson A, Wilson AC. 1991. Paternal inheritance of mitochondrial DNA in mice. Nature 352:255–257. [DOI] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Lane N, Seymour RM, Pomiankowski A. 2013. Dynamics of mitochondrial inheritance in the evolution of binary mating types and two sexes. Proc R Soc Lond B Biol Sci. 280:20131920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. 2012. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc R Soc Lond B Biol Sci. 279:1865–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2008. Toward a selection theory of molecular evolution. Evolution 62:255–265. [DOI] [PubMed] [Google Scholar]
- Hamilton WD, Zuk M. 1982. Heritable true fitness and bright birds: a role for parasites? Science 218:384–386.. [DOI] [PubMed] [Google Scholar]
- Havird JC, Hall MD, Dowling DK. 2015. Mitochondria, mutations and sex: a new hypothesis for the evolution of sex based on mitochondrial mutational erosion. bioRxiv doi: http://dx.doi.org/10.1101/019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Gregory TR. 2005. The promise of DNA barcoding for taxonomy. Syst Biol. 54:852–859. [DOI] [PubMed] [Google Scholar]
- Hebert PD, Ratnasingham S, de Waard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 270:S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. 2011. Condition-dependent traits as signals of the functionality of vital cellular processes. Ecol Lett. 14:625–634. [DOI] [PubMed] [Google Scholar]
- Hill GE. 2014a. Sex linkage of nuclear-encoded mitochondrial genes. Heredity 112:469–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. 2014b. Cellular respiration: the nexus of stress, condition, and ornamentation. Integr Comp Biol. 54:645–657. [DOI] [PubMed] [Google Scholar]
- Hill GE, Johnson JD. 2013. The mitonuclear compatibility hypothesis of sexual selection. Proc R Soc Lond B Biol Sci. 280:20131314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RF. 2000. Evolutionary origin and consequences of uniparental mitochondrial inheritance. Hum Reprod. 15:102–111. [DOI] [PubMed] [Google Scholar]
- Horan MP, Gemmell NJ, Wolff JN. 2013. From evolutionary bystander to master manipulator: the emerging roles for the mitochondrial genome as a modulator of nuclear gene expression. Eur J Hum Genet. 21:1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Hamilton WD. 1992. Cytoplasmic fusion and the nature of sexes. Proc R Soc Lond B Biol Sci. 247:189–194. [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK. 2011. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332:845–848. [DOI] [PubMed] [Google Scholar]
- James AC, Ballard JWO. 2003. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JD, Hill GE. 2013. Is carotenoid ornamentation linked to the inner mitochondria membrane potential? A hypothesis for the maintenance of signal honesty. Biochimie 95:436–444. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Otto SP. 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443:89–92. [DOI] [PubMed] [Google Scholar]
- Kuijper B, Lane N, Pomiankowski A. 2015. Can paternal leakage maintain sexually antagonistic polymorphism in the cytoplasm? J Evol Biol. 28:468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper B, Pen I, Weissing FJ. 2012. A guide to sexual selection theory. Annu Rev Ecol Evol Syst. 43:287–311. [Google Scholar]
- Kurbalija Novičić Z, Immonen E, Jelić M, AnĐelković M, Stamenković-Radak M, Arnqvist G. 2015. Within-population genetic effects of mtDNA on metabolic rate in Drosophila subobscura. J Evol Biol. 28:338–346. [DOI] [PubMed] [Google Scholar]
- Lane N. 2005. Power, sex, suicide: mitochondria and the meaning of life. Oxford: Oxford University Press. [Google Scholar]
- Lane N. 2009. On the origin of bar codes. Nature 462:272–274. [DOI] [PubMed] [Google Scholar]
- Lane N. 2011a. Energetics and genetics across the prokaryote-eukaryote divide. Biol Direct. 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. 2011b. Mitonuclear match: optimizing fitness and fertility over generations drives ageing within generations. Bioessays 33:860–869. [DOI] [PubMed] [Google Scholar]
- Lane N. 2011c. The costs of breathing. Science 334:184–185. [DOI] [PubMed] [Google Scholar]
- Lane N. 2012. The problem with mixing mitochondria. Cell 151:246–248. [DOI] [PubMed] [Google Scholar]
- Lane N. 2014. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 6:a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467:929–934. [DOI] [PubMed] [Google Scholar]
- Lee C, Yen K, Cohen P. 2013. Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol Metab. 24:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim S-J, Mehta H, Hevener AL, de Cabo R. 2015. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin L, Blumberg A, Barshad G, Mishmar D. 2014. Mito-nuclear co-evolution: the positive and negative sides of functional ancient mutations. Front Genet. 5:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon JD. 1999. The evolution of avian breeding systems. Oxford: Oxford University Press. [Google Scholar]
- Lively CM, Craddock C, Vrijenhoek RC. 1990. Red Queen hypothesis supported by parasitism in sexual and clonal fish. Nature 344:864–866. [Google Scholar]
- Lynch M. 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 14:914–925. [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV. 2006. Introns and the origin of nucleus–cytosol compartmentalization. Nature 440:41–45. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392:37–41. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. Cambridge: Cambridge University Press. [Google Scholar]
- McCauley DE. 2013. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 200:966–977. [DOI] [PubMed] [Google Scholar]
- McGaugh SE, Heil CS, Manzano-Winkler B, Loewe L, Goldstein S, Himmel TL, Noor MA. 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10:e1001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. 2013. An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9:e1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson TC, Shaw KL. 2012. The (mis)concept of species recognition. Trends Ecol Evol. 27:421–427. [DOI] [PubMed] [Google Scholar]
- Morales HE, Pavlova A, Joseph L, Sunnucks P. 2015. Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol. [DOI] [PubMed] [Google Scholar]
- Orr HA. 2005a. The genetic basis of reproductive isolation: insights from Drosophila. Proc Natl Acad Sci U S A. 102:6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 2005b. The genetic theory of adaptation: a brief history. Nat Rev Genet. 6:119–127. [DOI] [PubMed] [Google Scholar]
- Osada N, Akashi H. 2012. Mitochondrial–nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome c oxidase complex. Mol Biol Evol. 29:337–346. [DOI] [PubMed] [Google Scholar]
- Otto SP, Barton NH. 2001. Selection for recombination in small populations. Evolution 55:1921–1931. [DOI] [PubMed] [Google Scholar]
- Otto SP, Michalakis Y. 1998. The evolution of recombination in changing environments. Trends Ecol Evol. 13:145–151. [DOI] [PubMed] [Google Scholar]
- Parker GA. 2014. The sexual cascade and the rise of pre-ejaculatory (Darwinian) sexual selection, sex roles, and sexual conflict. Cold Spring Harb Perspect Biol. 6:a017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RJ, Barreto FS, Burton RS. 2014. Ecological novelty by hybridization: experimental evidence for increased thermal tolerance by transgressive segregation in Tigriopus californicus. Evolution 68:204–215. [DOI] [PubMed] [Google Scholar]
- Pierron D, Wildman DE, Huttemann M, Markondapatnaikuni GC, Aras S, Grossman LI. 2012. Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochim Biophys Acta Bioenerg. 1817:590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. 2007. Speciation in birds. London: Roberts and Company Publishers. [Google Scholar]
- Rand DM, Haney RA, Fry AJ. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19:645–653. [DOI] [PubMed] [Google Scholar]
- Reeve HK, Pfennig DW. 2003. Genetic biases for showy males: are some genetic systems especially conducive to sexual selection? Proc Natl Acad Sci U S A.. 100:1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MG. 2007. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 38:79–102. [Google Scholar]
- Rogell B, Dean R, Lemos B, Dowling DK. 2014. Mito-nuclear interactions as drivers of gene movement on and off the X-chromosome. BMC Genomics 15:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. 2006. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 7:211–221. [DOI] [PubMed] [Google Scholar]
- Ryan MJ, Rand AS. 1993. Species recognition and sexual selection as a unitary problem in animal communication. Evolution 47:647–657. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Haney RA, Rand DM. 2003. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution 57:2315–2325. [DOI] [PubMed] [Google Scholar]
- Safran RJ, Scordato ES, Symes LB, Rodriguez RL, Mendelson TC. 2013. Contributions of natural and sexual selection to the evolution of premating reproductive isolation: a research agenda. Trends Ecol Evol. 28:643–650. [DOI] [PubMed] [Google Scholar]
- Schmidt TR, Wu W, Goodman M, Grossman LI. 2001. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol Biol Evol. 18:563–569. [DOI] [PubMed] [Google Scholar]
- Scott GR, Hawkes LA, Frappell PB, Butler PJ, Bishop CM, Milsom WK. 2015. How bar-headed geese fly over the Himalayas. Physiology 30:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GR, Schulte PM, Egginton S, Scott AL, Richards JG, Milsom WK. 2011. Molecular evolution of cytochrome c oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 28:351–363. [DOI] [PubMed] [Google Scholar]
- Servedio MR. 2001. Beyond reinforcement: the evolution of premating isolation by direct selection on preferences and postmating, prezygotic incompatibilities. Evolution 55:1909–1920. [DOI] [PubMed] [Google Scholar]
- Shen Y-Y, Liang L, Zhu Z-H, Zhou W-P, Irwin DM, Zhang Y-P. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci U S A. 107:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle MY, Thaler DS. 2014. DNA barcoding works in practice but not in (neutral) theory. PLoS One 9:e100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews DP, Brelsford A. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 21:3907–3930. [DOI] [PubMed] [Google Scholar]
- Toews DP, Mandic M, Richards JG, Irwin DE. 2014. Migration, mitochondria, and the yellow-rumped warbler. Evolution 68:241–255. [DOI] [PubMed] [Google Scholar]
- Trier CN, Hermansen JS, Sætre G-P, Bailey RI. 2014. Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid Italian sparrow and its parent species. PLoS Genet. 10:e1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. 2007. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176:1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AR. 1889. Darwinism. London: Macmillan. [Google Scholar]
- Wallace DC. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 76:781–821. [DOI] [PubMed] [Google Scholar]
- Wallace DC. 2010a. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 51:440–450. [DOI] [PubMed] [Google Scholar]
- Wallace DC. 2010b. Bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci U S A. 107:8947–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. 2013. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos Trans R Soc Lond B Biol Sci. 368:20120267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236. [DOI] [PubMed] [Google Scholar]
- Wilson RE, Peters JL, McCracken KG. 2013. Genetic and phenotypic divergence between low—and high—altitude populations of two recently diverged cinnamon teal subspecies. Evolution 67:170–184. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Lond B Biol Sci. 369:20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J. 2008. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 9:383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]