Abstract
Exposure to the anticonvulsant drug valproic acid (VPA) is associated with an increased risk of congenital malformations. Although the mechanisms contributing to its teratogenicity are poorly understood, VPA has been shown to induce DNA double strand breaks (DSB) and to increase homologous recombination in vitro. The objective of the present study was to determine whether in utero exposure to VPA alters the frequency of intrachromosomal recombination and the expression of several genes involved in DSB repair in pKZ1 mouse embryos. Pregnant pKZ1 transgenic mice (GD 9.0) were administered VPA (500 mg/kg s.c.) and embryos were extracted and microdissected into the head, heart, and trunk regions 1, 3, 6, and 24 h after injection. Quantitative PCR was used to measure the tissue-specific expression of lacZ, a surrogate measure of recombination, Xrcc4, Rad51, Brca1, and Brca2, with Western blotting used to quantify Rad51, cleaved caspase-3 and cleaved-PARP protein. Increased recombination was only observed in the embryonic head following 6-h VPA exposure. VPA had no effect on Xrcc4 expression. Rad51, Brca1, and Brca2 expression rapidly decreased in head and trunk tissues after 1-h VPA exposure, followed by a subsequent increase in all tissues, although it was generally attenuated in the head and not due to differences in endogenous levels. Cleaved caspase-3 and cleaved-PARP expression was increased in all tissues 3 h following VPA exposure. This study indicates that the tissue-specific expression of several genes involved in DSB repair is altered following exposure to VPA and may be contributing to increased apoptosis.
Keywords: valproic acid, DNA double strand break repair, homologous recombination, nonhomologous end joining, transcriptional alteration
Mammalian embryonic development is a highly dynamic process involving complex spatial and temporal patterns of gene expression. Endogenous metabolites and exogenous drug or chemical exposures that perturb these cellular processes may induce developmental abnormalities (Wells et al., 2010). Transcriptional control of gene expression is mediated in part by epigenetic modifications to chromatin by histone acetyltransferases and histone deacetylases (HDACs) (Haberland et al., 2009). Inhibition of HDAC isoforms has been suggested to mediate teratogenesis, with global deletion of several HDACs in mice shown to result in embryolethality and developmental defects (Haberland et al., 2009). Although the exact mechanism is unknown, several HDAC inhibitors (HDACi) including valproic acid (VPA) are established teratogens in both animal models and humans (Kaneko et al., 1999).
VPA and several other HDACi are currently being explored as cancer therapeutics given their common ability to induce cell-cycle arrest, apoptosis and potentiate the efficacy of other chemotherapeutics and increase radiation sensitivity of several types of cancer cells in vitro and tumor xenografts in vivo (reviewed by Shankar and Srivastava, 2008). These effects are suggested to be mediated, in part, by transcriptional downregulation of DNA repair genes and consequently repair activity (Adimoolam et al., 2007; Zhang et al., 2007). HDACi-mediated alterations on DNA repair have also shown selectivity toward cancerous cells, with a minimal effect on normal cells (Lee et al., 2010). Although VPA has been investigated in several models of cancer, studies regarding its effect on DNA repair in the developing embryo are limited.
DNA repair processes are highly conserved among eukaryotes and are essential for normal development as they protect the genome from accumulating DNA damage, which can compromise genomic integrity (Taylor and Lehmann, 1998). With more than 15 human genetic disorders known to result from repair deficiencies, it is evident that unrepaired DNA damage can significantly alter normal cellular processes (Lehmann, 2003). Although many of these disorders are associated with an increased susceptibility to cancer, developmental defects are also observed. Additionally, polymorphisms in several DNA repair genes have been identified as risk factors for the development of structural birth defects including spina bifida and orofacial clefts (Olshan et al., 2005). DNA double strand breaks (DSBs) are among the most lethal types of DNA damage and can be repaired by both nonhomologous end joining (NHEJ) and homologous recombination (HR) (reviewed by Brandsma and Gent, 2012). The fundamental difference between these two processes is their dependence on a template and the fidelity of repair. NHEJ involves direct ligation of the DNA ends flanking the break site with no need for a homologous template, making this process inherently mutagenic. In contrast, HR is relatively error free as it requires a highly similar or identical undamaged template from which to reconstitute the original sequence. We have previously shown that VPA induces DSBs and increases the frequency of HR in vitro (Sha and Winn, 2010); however, the molecular basis and effect on recombination in vivo has yet to be determined.
During the period of organogenesis in the postimplantation embryo, DNA damage response genes have been shown to exhibit specific temporal and spatial patterns of expression (reviewed by Jaroudi and SenGupta 2007). However, many of these genes including several involved in HR are initially expressed only weakly during development (Jaroudi and SenGupta, 2007; Vinson and Hales, 2002). The importance of regulated DNA repair gene expression during embryonic development is evidenced by the targeted disruption of these genes in mice, which often results in embryolethality or altered growth and developmental malformations (Friedberg and Meira, 2006). This suggests that the ability of the postimplantation embryo to repair damage is highly dependent on the developmental stage and that factors that perturb normal expression patterns or damage DNA during a period of low repair expression could have significant detrimental effects including birth defects. Given that we have previously shown that VPA can increase the frequency of HR in vitro, the goal of this study was to determine the temporal and tissue-specific effects of VPA exposure on DNA recombination, the expression of DSB repair genes, and to examine markers of apoptosis in malformation-sensitive tissues of the postimplantation mouse embryo in vivo.
MATERIALS AND METHODS
pKZ1 transgenic mice
pKZ1 breeders were originally obtained from Dr Pamela Sykes (Department of Hematology and Genetic Pathology, Flinders University and Flinders Medical Centre; Bedford Park, SA, Australia). A colony of pKZ1 mice was maintained by outbreeding with C57Bl/6N mice (Taconic Farms) and was housed in a temperature-controlled room on a 12-h light/dark cycle. Standard rodent chow (Purina Rodent Chow, Ralston Purina International, Strathroy, Canada) and tap water was given ad libitum. Housing and breeding practices were conducted in accordance with guidelines set forth by the Canadian Council on Animal Care and experimental procedures approved by the Queen's University Animal Care Committee.
pKZ1 mouse chromosomal recombination assay
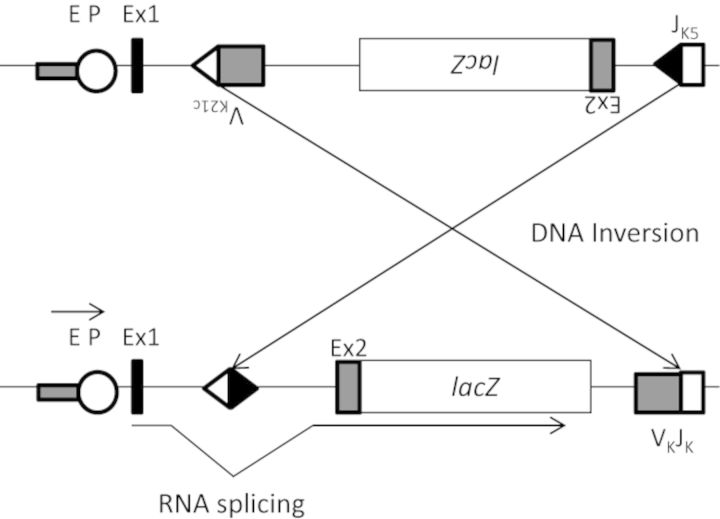
pKZ1 mice allow for the examination of somatic intrachromosomal recombination following exposure to DNA damaging agents by monitoring a stably expressed transgene (Sykes et al., 2006). The pKZ1 transgene is described in detail in Matsuoka et al. (1991). Briefly, the transgene is composed of an Escherichia coli lacZ gene situated in an inverse orientation with respect to its promoter that renders it nonfunctional (Fig. 1). The mouse immunoglobulin V(D)J recombination signals flank the inverted lacZ transgene and following induction of a DSB near the transgenic sequence, mediate recombination. This recombination inverts the lacZ gene into the correct orientation with respect to its promoter and allows for the gene to be expressed. Quantification of lacZ expression using quantitative real-time PCR (qRT-PCR) is indicative of the frequency of recombination events. This model is a surrogate measure of NHEJ activity as enzymes specific to this type of repair mediate recombination.
FIG. 1.
Intrachromosomal recombination reporter transgene stably integrated in pKZ1 mice. The reporter construct contains the E. coli lacZ gene situated in an inverse orientation with respect to its enhancer-promoter (EP) complex, which renders it nonfunctional. The JK5 and VK21C recombination signals that flank the lacZ gene invert it following a DNA DSB near the recombination reporter and enable lacZ transcription. The excess sequence 5′ to lacZ is removed by RNA splicing signals within exons 1 and 2 (Ex1 and Ex2). NHEJ proteins mediate the inversion of the lacZ gene into the correct orientation making this reporter a surrogate measure of DSB repair by NHEJ. Modified from Matsuoka et al. (1991).
pKZ1 genotyping
Transgenic mice were screened for the presence of the lacZ transgene at 21 days of age for breeding pKZ1 mice and at the time of sacrifice for experimental embryos using an E. coli lacZ-specific polymerase chain reaction. Tail clippings were taken from breeding pKZ1 mice and DNA extracted using the Qiagen DNeasy Blood and Tissue kit (Qiagen, Mississauga, ON). PCR reagents were purchased from Sigma-Aldrich (D4545/DNTP-10, St. Louis, MO) and primers were purchased from Invitrogen (Carlsbad, CA). Standard PCR genotyping was performed as described previously (Lau et al., 2009). Primer sequences were as follows: ZL1675: ATGAAAGCTGGCTACAGGAAGGCC and ZR2548: CACCATGCCGTGGGTTTCAATATT. PCR products were then separated by gel electrophoresis on a 2% agarose gel prepared using 1x tris-acetate ethylenediamine tetra-acetic acid buffer with 3% ethidium bromide (ICN Biomedicals, Aurora, OH) and visualized under an ultraviolet light. For embryonic genotyping, DNA was isolated from the yolk sac of each collected embryo and genotyped as above.
Breeding and animal treatment
pKZ1 mice were bred by housing three females with one male overnight, with breeding always between heterozygous (+/−) and wild-type (−/−) mice resulting in 50:50 (transgenic:nontransgenic) progeny. The presence of a vaginal plug the following morning was designated GD 1, and those females were separated from the colony and housed together. On the morning of GD9, dams were injected subcutaneously with a teratogenic dose of VPA (n = 3 for each time point, 500 mg/kg) (Sigma-Aldrich Canada Ltd., Oakville, ON) or the vehicle control (n = 3 for each time point, 0.9% saline). Dams were sacrificed 1, 3, 6, and 24 h after injection, and the embryos isolated. Embryos were dissected down to the yolk sac that was removed for genotyping and were further separated into the head, heart, and trunk tissue regions. Tissues from each litter were pooled according to genotype and RNA was extracted from lacZ positive tissues (approximately half the litter) for analysis, whereas protein was extracted from the remaining pooled tissues to assess the expression of Rad51 and the presence of apoptotic markers.
RNA extraction and qRT-PCR
RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen) and cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit according to the manufacturers' protocol including DNase treatment (Life Technologies, Burlington, ON). qRT-PCR reactions were carried out using primers designed against lacZ (reverse: 5′-ATGAACGGTCTGGTCTTTGC-3′ and forward: 5′-ACATCCAGAGGCACTTCACC-3′) and Hprt (forward: 5′-GTGCAACCATTGCCCTAAGT-3′ and reverse: 5′-CAGCCAGCATCTCAGGTGTA-3′), and specific RT2 qPCR primer assays (Qiagen) for mouse Rad51 (NM_011234), Xrcc4 (NM_028012), Brca1 (NM_009764), and Brca2 (NM_001081001). Although we and other groups have previously used X-gal staining to quantitate lacZ recombination, a qRT-PCR assay is more sensitive, allows for automated analysis of a larger number of samples, and removes any subjective bias in evaluating the tissues for the presence of staining. Primers were designed to span a 209-bp region within the E. coli lacZ gene (NCBI: NC_000913.2; 362455-365529). Reactions were performed with the RT2 SYBR® Green qPCR Mastermix (Qiagen) in triplicate using 2.0 μl of cDNA, corresponding to 20 μg of total RNA, and 0.6-μM lacZ, 0.3-μM Hprt, and 0.4-μM Qiagen primers (final concentration) in a 25-μl final volume. The PCR protocol consisted of one cycle for enzymatic activation (15 min at 95°C) followed by 40 amplification cycles (30 s at 94°C, 60 s at 63°C, 60 s at 72°C). Amplification specificity was verified using the dissociation curve and a no reverse transcription control. Five 10-fold serial dilutions of pooled whole embryo (GD 9) cDNA were used for calculation of the PCR efficiency, given by the equation E% = (101/slope − 1) × 100, where the slope was calculated from the linear regression of the log-transformed cDNA concentrations plotted against the Ct values in addition to correlation coefficient (R2) (Nolan et al., 2006). Target and reference efficiencies were determined to be between 90 and 110% with an R2 > 0.99, which is generally considered acceptable (Life Technologies). Given approximately equal efficiencies, the comparative CT method (2−ΔΔCt) (Livak & Schmittgen, 2001) was used to assess relative transcript levels (normalized to the Hprt internal control). For fold-changes less than one, the reciprocal value is reported.
Protein extraction and Western blotting
Pooled tissues from each litter were combined with an equal volume of 2X radioimmunoprecipitation assay (RIPA) buffer (100-mM tris-HCl pH 7.4, 300-mM NaCl, 2% Triton-X, 1% sodium deoxycholate, 2% sodium dodecyl sulfate) supplemented with protease and phosphatase inhibitors and agitated on ice for 30 min. Homogenates were centrifuged at 4°C for 10 min at 12,000 rpm and the supernatant collected. Protein was quantified using the Bradford protein assay according to the manufacturer's protocol (Bio-Rad Laboratories, Mississauga, ON) and an Ultrospec 3100 Pro scanning spectrophotometer (Biochrom Ltd, UK). Whole cell lysates (20 μg) were resolved by SDS-PAGE on an 8% polyacrylamide gel and transferred to a 0.45-μm PVDF membrane (Millipore Co., Bedford, MA) using overnight wet transfer. Membranes were blocked with 3% milk (w/v) in tris-buffered saline containing Tween (25-mM tris-HCl, 140-mM NaCl, 2-mM KCl, 0.05% (v/v) Tween 20) for 1 h at room temperature and probed overnight at 4°C with primary antibody. The same membrane was used to probe for each protein. Primary antibodies included: anti-Rad51 (ab88572, 1:1000; AbCam, Cambridge, MA), anti-cleaved caspase-3 (D175, 1:500; Cell Signaling Technology, Danvers, MA), anti-cleaved PARP (D214; Cell Signaling Technology), and alpha-tubulin (T5168, 1:2000; Sigma-Aldrich), which served as a loading control. Membranes were then washed and incubated with appropriate secondary antibodies, developed using Western Lightning Plus 40 ECL (Perkin Elmer, Waltham, MA), and quantified by densitometry (Taylor et al., 2013).
Statistical analyses
Statistical analysis for qRT-PCR and Western blotting was performed with a two-way analysis of variance, followed by the Bonferroni multiple comparison test for post hoc analysis (Prism 5.0, GraphPad Software Inc., San Diego, CA). P < 0.05 was designated as statistically significant and GraphPad Prism 5 was used to graph all results.
RESULTS
Effect of VPA on Recombination in Postimplantation Mouse Embryos
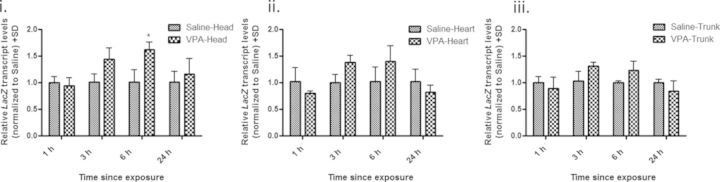
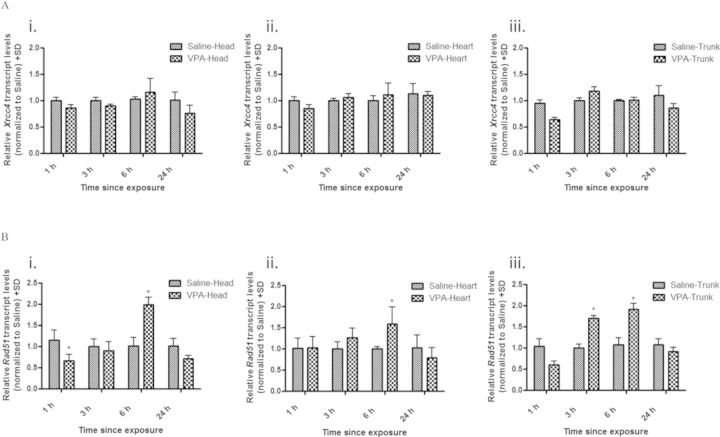
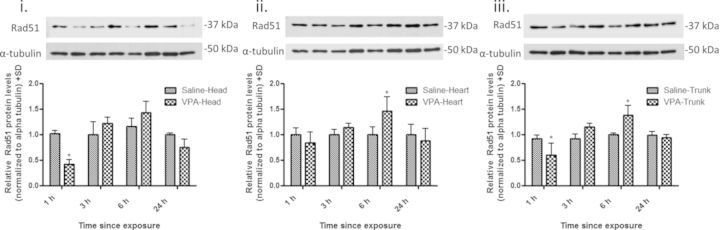
After a maternal injection of 500 mg/kg VPA, we assessed recombination in GD 9 postimplantation embryos after 1, 3, 6, and 24 h of exposure using lacZ transcript levels as a surrogate measure. A marginal increase in recombination was detected in all pKZ1 positive embryo tissues exposed to VPA compared with controls; however, a significant increase (1.62-fold) was only detected in the embryo head following a 6-h exposure (Fig. 2). With the pKZ1 recombination assay a surrogate for NHEJ activity, we examined the embryonic tissues for the mRNA expression of the NHEJ repair gene Xrcc4 and HR repair gene Rad51. Consistent with a minimal effect on NHEJ-mediated recombination, Xrcc4 transcript levels did not differ from controls in any of the tissues or at any time points examined following VPA exposure (Fig. 3A). Given the lack of change in mRNA, we did not extend our analysis to Xrcc4 protein levels. In contrast, Rad51 exhibited a biphasic and tissue-specific pattern of expression (Fig. 3B). Decreased transcript levels were observed in the embryo head (1.52-fold decrease) and trunk (1.67-fold decrease) following 1-h exposure to VPA with no effect observed in the heart tissue. This decrease in transcript levels, however, was only statistically significant in the head. By 6 h, Rad51 was significantly increased in the head (2.0-fold increase), heart (1.60-fold increase), and trunk (1.92-fold increase) tissues with expression returning to control levels by 24 h. Rad51 protein similarly followed this biphasic trend in expression (Fig. 4).
FIG. 2.
VPA increases intrachromosomal recombination in embryonic head tissue. qRT-PCR analysis of lacZ expression in embryonic head (i), heart (ii), and trunk (iii) tissues following maternal administration of VPA (500 mg/kg) for 1, 3, 6, or 24 h. All data were quantified using the delta-delta Ct method utilizing hypoxanthine-guanine phosphoribosyltransferase as the housekeeping gene control. * indicates a significant difference from saline-treated controls (p < 0.05).
FIG. 3.
VPA alters Rad51 but not Xrcc4 transcript levels in embryonic tissues. qRT-PCR analysis of Xrcc4 (A) and Rad51 (B) expression in embryonic head (i), heart (ii), and trunk (iii) tissues following maternal administration of VPA (500 mg/kg) for 1, 3, 6, and 24 h. All data were quantified using the delta-delta Ct method utilizing hypoxanthine-guanine phosphoribosyltransferase as the housekeeping gene control. * indicates a significant difference from saline-treated controls (p < 0.05).
FIG. 4.
VPA alters Rad51 protein expression in embryonic tissues. Western blot analysis for Rad51 protein in embryonic head (i), heart (ii), and trunk (iii) tissues following maternal administration of VPA (500 mg/kg) for 1, 3, 6, and 24 h. Expression was normalized to the β-actin loading control. * indicates a significant difference from saline-treated controls (p < 0.05).
VPA Exposure Alters the Expression of Other HR Genes
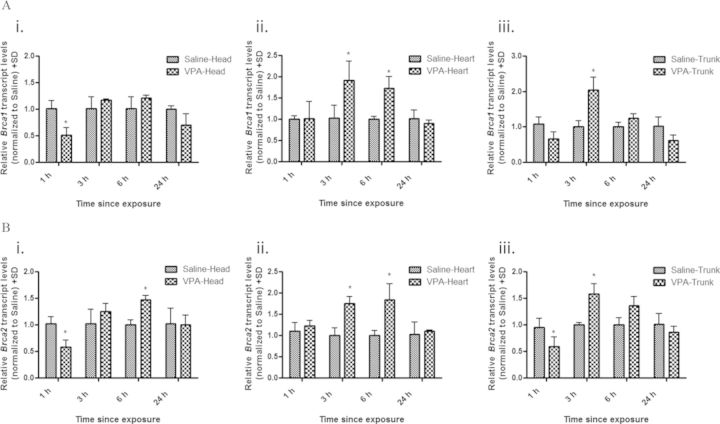
To determine if VPA alters the expression of other genes involved in HR, we further examined the embryonic head, heart, and trunk tissues for the transcript levels of Brca1 and Brca2. As with Rad51 expression, Brca1 and Brca2 exhibited a similar biphasic response to VPA exposure (Fig. 5). Brca1 and Brca2 expression was decreased in the head (1.96- and 1.72-fold decrease, respectively) and trunk (1.45- and 1.69-fold decrease, respectively) tissues following 1-h VPA exposure, with no effect on the heart. This was followed by a significant increase in both Brca1 and Brca2 expression in the heart (1.90- and 1.75-fold increase, respectively) and trunk (2.04- and 1.58-fold increase, respectively) tissues by 3-h exposure which returned to control levels by 24 h. In the head tissue, Brca2 expression was significantly increased after 6-h VPA exposure (1.47-fold increase), whereas Brca1 expression was not significantly increased compared with controls at any time point examined.
FIG. 5.
VPA alters transcript levels of Brca1 and Brca2 in embryonic tissues. qRT-PCR analysis of Brca1 (A) and Brca2 (B) expression in embryonic head (i), heart (ii), and trunk (iii) tissues following maternal administration of VPA (500 mg/kg) for 1, 3, 6, and 24 h. All data were quantified using the delta-delta Ct method utilizing hypoxanthine-guanine phosphoribosyltransferase as the housekeeping gene control. * indicates a significant difference from saline-treated controls (p < 0.05).
DNA Repair Genes are Differentially Expressed in Postimplantation Embryos
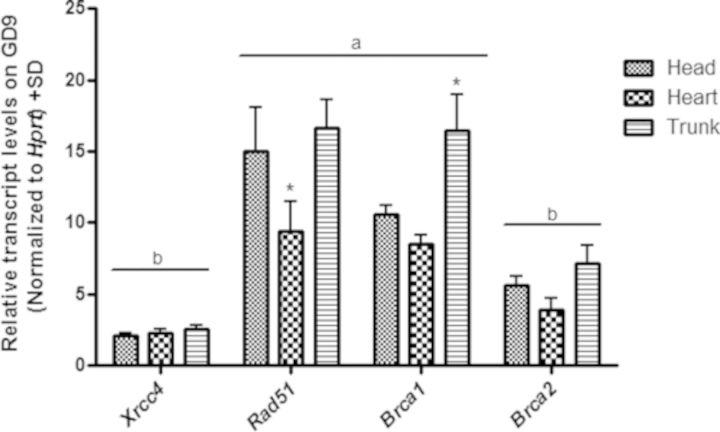
As differences in gene expression were observed following VPA exposure in the embryonic tissues examined, we compared the normal tissue-specific differences in expression for Xrcc4, Rad51, Brca1, and Brca2 on days 9 and 10 of gestation in control embryos (Fig. 6). No significant differences in transcript levels were observed in any of the tissues between GD9 and GD10; however, there was a general trend toward increased expression on GD10 (data not shown). On GD9, gene expression was generally different between the tissues with the exception of Xrcc4 and Brca2. Compared with the head and trunk, expression was significantly lower in the heart (1.61- and 1.77-fold decrease, respectively) for Rad51, whereas expression of Brca1 was significantly increased in the trunk compared with the head and heart tissues (1.56- and 1.94-fold increase, respectively). Differences in normal transcript levels were also observed between genes with Xrcc4 and Brca2 expression significantly lower in all tissues compared with Rad51 and Brca1.
FIG. 6.
DNA repair genes are differentially expressed in postimplantation embryonic tissues. qRT-PCR analysis of Xrcc4, Rad51, Brca1, and Brca2 expression in embryonic head, heart, and trunk tissues. All data were expressed relative to the hypoxanthine-guanine phosphoribosyltransferase housekeeping gene control. * indicates a significant difference from the other tissues examined (p < 0.05). Genes in (a) are significantly higher compared with (b), p < 0.05.
VPA Differentially Induces Apoptosis in Embryo Tissues
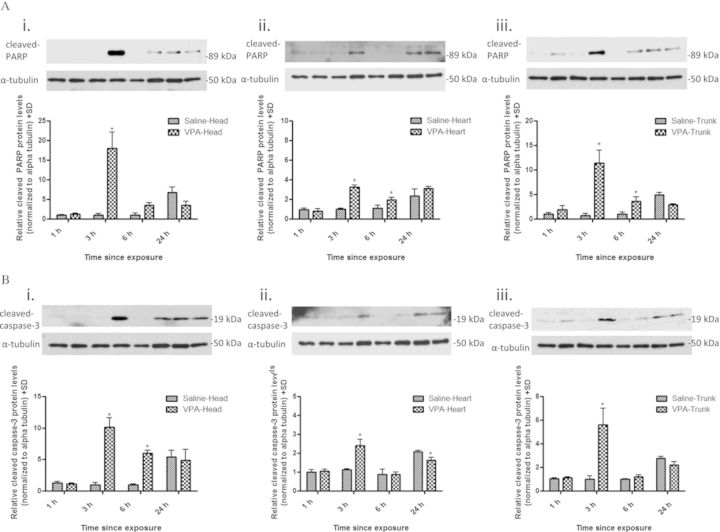
As a downstream consequence of unrepaired DNA damage is apoptosis, we examined activated caspase-3 and PARP protein expression in different embryonic tissues following maternal VPA exposure as they become cleaved during apoptosis (Fig. 7). Expressions of cleaved caspase-3 and PARP were significantly increased in the embryonic head (10.13- and 18.06-fold increase), heart (2.40- and 3.24-fold increase), and trunk (5.59- and 11.45-fold increase) by 3 h following maternal exposure to VPA. Although there was a significant increase in expression for cleaved caspase-3 and cleaved PARP in all embryonic tissues examined, the magnitude of this increase differed significantly between the tissues.
FIG. 7.
VPA increases the expression of cleaved caspase-3 and cleaved PARP in embryo tissues. Western blot analysis for cleaved PARP (A) and cleaved caspase-3 (B) protein in embryonic head (i), heart (ii), and trunk (iii) tissues following maternal administration of VPA (500 mg/kg) for 1, 3, 6, and 24 h. Expression was normalized to the β-actin loading control. * indicates a significant difference from saline-treated controls (p < 0.05).
DISCUSSION
The present study evaluated the tissue-specific effects of VPA exposure on recombination and DSB repair mediators in postimplantation mouse embryos in vivo. Although error-prone NHEJ is the predominant mechanism of DSB repair in mature somatic cells of higher eukaryotes, HR often serves as a backup in situations of NHEJ deficiencies (Allen et al., 2003). This hierarchy of DSB repair activation appears to be different during embryonic development with evidence suggesting a predominance of HR during early embryogenesis and the switch to NHEJ occurring later (Chiruvella et al., 2012). With early embryogenesis a key period of susceptibility to structural malformations, the efficient repair of DNA damage is crucial for normal embryonic development. Therefore, the reliance on HR during this early embryonic period would maximize the high-fidelity repair of DNA and prevent malformations. Our findings that NHEJ-mediated recombination is marginally activated following VPA exposure supports a predominance of HR-mediated repair of DSBs, with NHEJ serving as a backup. Additionally, the significant increase in NHEJ-mediated recombination activity exclusively in the embryo head following maternal VPA exposure could reflect a higher level of error-prone repair in this tissue and could provide a mechanistic basis for the 1–2% incidence of neural tube defects associated with VPA exposure (Nau et al., 1991).
The Rad51-mediated strand invasion of a homologous template represents a critical step in HR, and is mediated in part by Brca1 and Brca2 that function to recruit Rad51 and facilitate the formation of the invasive nucleoprotein filament (O'Donovan and Livingston, 2010). Although changes in the expression of DNA repair genes are not necessarily indicative of altered repair capacity, this is one mechanism by which it could be regulated. This is supported by studies demonstrating that cells deficient in Rad51, Brca1, or Brca2 have reduced HR activity (Magwood et al., 2013; Snouwaert et al., 1999; Xia et al., 2001). Although we did not examine HR activity directly, our finding that 1-h maternal exposure to VPA generally decreases the expression of Rad51, Brca1, and Brca2 in the embryonic head and trunk suggests that HR activity and consequently the ability of the embryo to respond to DNA damage may be similarly limited in these tissues. The lack of change in gene expression in the embryonic heart at this time point may be the result of an earlier peak period of tissue sensitivity to VPA for heart malformations in mice which has been demonstrated to occur on GD7 (Sonoda et al., 1993). HDAC inhibition is frequently accompanied by downregulation of DNA repair genes including Rad51 (Adimoolam et al., 2007) and Brca1 (Zhang et al., 2007). It is uncertain whether transcription mediates this downregulation of DNA repair genes, and nontranscriptional targets of HDACi have been proposed (Taddei et al., 2005). Although the exact mechanism is not clear, our observed decreases in Rad51, Brca1, and Brca2 gene expression are likely a specific effect of the HDACi action of VPA. We also observed a differential increase in Rad51, Brca1, and Brca2 mRNA and Rad51 protein levels in embryonic tissues following 3-h maternal exposure to VPA. The increased expression of Brca1 and Brca2 in the embryonic heart and trunk tissues likely reflects a cellular response to maintain genomic integrity and repair damage, as previous studies have found transcriptional induction of these genes following exposure to genotoxicants and other stresses (De Siervi et al., 2010; Volcic et al., 2012). Notably, we did not observe the same increase in expression in the embryonic head tissue at this time point, suggesting that normal safeguards to maintain genome integrity may not be in place. This may be due to the differential uptake and elimination of VPA from mouse embryo tissues, with the neuroepithelium shown to have a higher uptake and slower elimination as compared with other tissues (Dencker and D'Argy, 1990). Increased Rad51 gene transcript or protein levels are not frequently observed following DSB induction, with HR activation in higher eukaryotes involving increased nuclear localization of mediator proteins including Rad51, Brca1, and Brca2 (Chen et al., 1997). Several studies using in vitro models of DSB repair have demonstrated that transient increases in Rad51 protein levels (2–4-fold) can increase the frequency of HR considerably (Vispe et al., 1998) and promote the repair of DSBs using alternative repair pathways that can induce genomic instability (Richardson et al., 2004). We observed a statistically significant VPA-induced increase in Rad51 protein levels in the heart and trunk (∼1.5-fold), and a nonsignificant increase in the head (∼1.42-fold), which likely reflects the slightly increased endogenous levels quantified in the saline controls at this time point. Although these increases are less than what has been shown in vitro, it remains possible that this increase is also associated with increased HR, especially because we have previously demonstrated that VPA increases the frequency of HR in vitro. Given that the threshold for Rad51 protein levels required to switch between normal strand invasion in HR and altered repair pathways is still unclear, further studies are needed using in vivo models of HR during development.
DNA repair and genotoxic stress response genes have been previously shown to exhibit differing basal levels of expression in the whole rat embryo and yolk sac, with expression often increasing throughout organogenesis (Vinson and Hales, 2002). We examined several genes essential to HR and NHEJ in the mouse embryo head, heart, and trunk tissues on GD9 and 10 to assess whether differences in basal gene expression could account for the observed tissue-specific transcriptional responses following VPA exposure. Consistent with the previous study by Vinson and Hales, we observed differences in basal levels of expression between the genes. The NHEJ gene Xrcc4 was significantly lower compared with both HR genes Rad51 and Brca1, and is in accordance with a predominance of HR over NHEJ at this stage of development (Chiruvella et al., 2012). Although increased gene expression for Brca1 and Brca2 was observed at an earlier time point in the heart and trunk tissue as compared with the head following VPA exposure, this was not due to higher basal levels of gene expression in these tissues. Therefore, following VPA exposure, embryonic head tissue displays a delayed activation of Brca1 and Brca2 that is not due to low endogenous gene transcript levels and suggests increased susceptibility to damage in this tissue.
VPA exposure in pregnant C57Bl/6N mice is known to induce a higher incidence of skeletal malformations than neural tube defects (Beck, 1999; Downing et al., 2010). However, knockout mice for several of the HR repair genes we examined in this study show increased incidences of exencephaly, spina bifida and microcephaly that are all associated with defects in neurogenesis (reviewed in Friedberg and Meira, 2006), with skeletal defects, not reported. This suggests that although HR repair plays a large role in the formation of the neural tube, defects in this process may not entirely account for skeletal abnormalities observed following VPA exposure. Although it is interesting that VPA alters the expression of HR repair genes in the embryo trunk, skeletal defects that are observed following exposure are likely a result of the multiple cellular pathways affected by VPA including DNA repair, and ongoing studies are working to clarify this complex interaction.
The DNA damage response comprises DNA repair, cell-cycle checkpoint control, and apoptosis that serve to prevent genomic instability. Failure to repair damaged DNA often results in apoptosis and can be quantified using markers of apoptosis including activated caspase-3 and PARP which become cleaved during the initiation of the apoptotic cascade (Mirkes, 2002). Although programmed cell death is an essential component of normal embryonic development, excessive apoptosis of embryonic cells is recognized as contributing to structural defects (Mirkes, 2002). The expression of cleaved caspase-3 and cleaved PARP was significantly increased in all tissues following 3-h maternal exposure to VPA, with expression consistently highest in the head and lowest in the heart. This is in agreement with our previous study that observed increased cleaved caspase-3 following 3-h maternal exposure to VPA in whole embryo homogenates, with expression localized primarily to the neuroepithelium using immunohistochemistry (Tung and Winn, 2011). These observed differences in relative expression of apoptotic markers between the tissues may also inversely reflect their capacity to repair DNA. It should be noted that increased apoptotic cell death could potentially result in a decreased ability to detect increased NHEJ, however, given the lack of any changes in mRNA expression of Xrcc4 following VPA exposure in any of the tissues at any of the examined time points, this was likely not an issue in the current study.
In summary, our results demonstrate a tissue-specific biphasic response of HR repair genes in the embryo following VPA exposure which may reflect a transition from low HR activity and limited repair potential to a state of hyper-recombination and genomic instability which may be an early initiator of aberrant apoptosis. Ongoing studies are investigating the tissue-specific HR repair capacity and the persistence of DSBs in the developing embryo following VPA exposure to further understand the involvement of DNA repair in VPA-mediated teratogenicity.
FUNDING
Canadian Institutes for Health Research (MOP115188 to L.W.); Ontario Graduate Scholarship (to C.L.).
Acknowledgments
The authors would like to thank Christine Belanger for technical assistance with mouse treatments and sample collection.
REFERENCES
- Adimoolam S., Sirisawad M., Chen J., Thiemann P., Ford J. M., Buggy J. J. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19482–19487. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C., Halbrook J., Nickoloff J. A. Interactive competition between homologous recombination and non-homologous end joining. Mol. Cancer Res. 2003;1:913–920. [PubMed] [Google Scholar]
- Beck S. L. Contributions of dam and conceptus to differences in sensitivity to valproic acid among C57 black and SWV mice. Reprod. Toxicol. 1999;13:353–360. doi: 10.1016/s0890-6238(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Brandsma I., Gent D. C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integrity. 2012;3:9. doi: 10.1186/2041-9414-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Nastasi A., Shen Z., Brenneman M., Crissman H., Chen D. J. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat. Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- Chiruvella K. K., Sebastian R., Sharma S., Karande A. A., Choudhary B., Raghavan S. C. Time-dependent predominance of nonhomologous DNA end-joining pathways during embryonic development in mice. J. Mol. Biol. 2012;417:197–211. doi: 10.1016/j.jmb.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Dencker L. N. H., D'Argy R. Marked accumulation of valproic acid in embryonic neuroepithelium of the mouse during early organogenesis. Teratology. 1990;41:699–706. doi: 10.1002/tera.1420410606. [DOI] [PubMed] [Google Scholar]
- De Siervi A., De Luca P., Byun J. S., Di L. J., Fufa T., Haggerty C. M., Vazquez E., Moiola C., Longo D. L., Gardner K. Transcriptional autoregulation by BRCA1. Cancer Res. 2010;70:532–542. doi: 10.1158/0008-5472.CAN-09-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C., Biers J., Larson C., Kimball A., Wright H., Ishii T., Gilliam D., Johnson T. Genetic and maternal effects on valproic acid teratogenesis in C57BL/6J and DBA/2J mice. Toxicol. Sci. 2010;116:632–639. doi: 10.1093/toxsci/kfq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Meira L. B. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair. 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Haberland M., Montgomery R. L., Olson E. N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroudi S., SenGupta S. DNA repair in mammalian embryos. Mutat. Res. 2007;635:53–77. doi: 10.1016/j.mrrev.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Battino D., Andermann E., Wada K., Kan R., Takeda A., Nakane Y., Ogawa Y., Avanzini G., Fumarola C., et al. Congenital malformations due to antiepileptic drugs. Epilepsy Res. 1999;33:145–158. doi: 10.1016/s0920-1211(98)00084-9. [DOI] [PubMed] [Google Scholar]
- Lau A., Belanger C. L., Winn L. M. In utero and acute exposure to benzene: Investigation of DNA double-strand breaks and DNA recombination in mice. Mutat. Res. 2009;676:74–82. doi: 10.1016/j.mrgentox.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Choy M. L., Ngo L., Foster S. S., Marks P. A. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Magwood A. C., Malysewich M. J., Cealic I., Mundia M. M., Knapp J., Baker M. D. Endogenous levels of Rad51 and Brca2 are required for homologous recombination and regulated by homeostatic re-balancing. DNA Repair. 2013;12:1122–1133. doi: 10.1016/j.dnarep.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Nagawa F., Okazaki K., Kingsbury L., Yoshida K., Muller U., Larue D. T., Winer J. A., Sakano H. Detection of somatic DNA recombination in the transgenic mouse brain. Science. 1991;254:81–86. doi: 10.1126/science.1925563. [DOI] [PubMed] [Google Scholar]
- Mirkes P. E. 2001 Warkany lecture: To die or not to die, the role of apoptosis in normal and abnormal mammalian development. Teratology. 2002;65:228–239. doi: 10.1002/tera.10049. [DOI] [PubMed] [Google Scholar]
- Nau H., Hauck R. S., Ehlers K. Valproic acid-induced neural tube defects in mouse and human: aspects of chirality, alternative drug development, pharmacokinetics and possible mechanisms. Pharmacol. Toxicol. 1991;69:310–321. doi: 10.1111/j.1600-0773.1991.tb01303.x. [DOI] [PubMed] [Google Scholar]
- Nolan T., Hands R. E., Bustin S. A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- O'Donovan P. J., Livingston D. M. BRCA1 and BRCA2: Breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 2010;31:961–967. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]
- Olshan A. F., Shaw G. M., Millikan R. C., Laurent C., Finnell R. H. Polymorphisms in DNA repair genes as risk factors for spina bifida and Orofacial clefts. Am. J. Med. Genet. A. 2005;135:268–273. doi: 10.1002/ajmg.a.30713. [DOI] [PubMed] [Google Scholar]
- Richardson C., Stark J. M., Ommundsen M., Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- Schmittgen L. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Sha K., Winn L. M. Characterization of valproic acid-initiated homologous recombination. Birth Defects Res. B Dev. Reprod. Toxicol. 2010;89:124–132. doi: 10.1002/bdrb.20236. [DOI] [PubMed] [Google Scholar]
- Shankar S., Srivastava R. K. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv. Exp. Med. Biol. 2008;615:261–298. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- Snouwaert J. N., Gowen L. C., Latour A. M., Mohn A. R., Xiao A., DiBiase L., Koller B. H. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- Sonoda T., Ohdo S., Ohba K., Okishima T., Hayakawa K. Sodium valproate-induced cardiovascular abnormalities in the Jcl:ICR mouse fetus: peak sensitivity of gestational day and dose-dependent effect. Teratology. 1993;48:127–132. doi: 10.1002/tera.1420480206. [DOI] [PubMed] [Google Scholar]
- Sykes P. J., Morley A. A., Hooker A. M. The PKZ1 recombination mutation assay: A sensitive assay for low dose studies. Dose Response. 2006;4:91–105. doi: 10.2203/dose-response.05-035.Sykes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Roche D., Bickmore W. A., Almouzni G. The effects of histone deacetylase inhibitors on heterochromatin: Implications for anticancer therapy. EMBO Rep. 2005;6:520–524. doi: 10.1038/sj.embor.7400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. M., Lehmann A. R. Conservation of eukaryotic DNA repair mechanisms. Int. J. Radiat. Biol. 1998;74:277–286. doi: 10.1080/095530098141429. [DOI] [PubMed] [Google Scholar]
- Taylor S. C., Berkelman T., Yadav G., Hammond M. A defined methodology for reliable quantification of Western blot data. Mol. Biotechnol. 2013;55:217–226. doi: 10.1007/s12033-013-9672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung E. W., Winn L. M. Valproic acid-induced DNA damage increases embryonic p27(KIP1) and caspase-3 expression: A mechanism for valproic-acid induced neural tube defects. Reprod. Toxicol. 2011;32:255–260. doi: 10.1016/j.reprotox.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Vinson R. K., Hales B. F. DNA repair during organogenesis. Mutat. Res. 2002;509:79–91. doi: 10.1016/s0027-5107(02)00223-3. [DOI] [PubMed] [Google Scholar]
- Vispe S., Cazaux C., Lesca C., Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volcic M., Karl S., Baumann B., Salles D., Daniel P., Fulda S., Wiesmuller L. NF-kappaB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic Acids Res. 2012;40:181–195. doi: 10.1093/nar/gkr687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells P. G., Lee C. J., McCallum G. P., Perstin J., Harper P. A. Receptor- and reactive intermediate-mediated mechanisms of teratogenesis. Handb. Exp. Pharmacol. 2010;196:131–162. doi: 10.1007/978-3-642-00663-0_6. [DOI] [PubMed] [Google Scholar]
- Xia F., Taghian D. G., DeFrank J. S., Zeng Z. C., Willers H., Iliakis G., Powell S. N. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Carr T., Dimtchev A., Zaer N., Dritschilo A., Jung M. Attenuated DNA damage repair by trichostatin A through BRCA1 suppression. Radiat. Res. 2007;168:115–124. doi: 10.1667/RR0811.1. [DOI] [PubMed] [Google Scholar]