Abstract
Gut microbiota represent an important bridge between environmental substances and host metabolism. Here we reported a comprehensive study of gut microbiota interaction with ochratoxin A (OTA), a major food-contaminating mycotoxin, using the combination of metagenomics and culture-based methods. Rats were given OTA (0, 70, or 210 μg/kg body weight) by gavage and fecal samples were collected at day 0 and day 28. Bacterial genomic DNA was extracted from the fecal samples and both 16S rRNA and shotgun sequencing (two main methods of metagenomics) were performed. The results indicated OTA treatment decreased the within-subject diversity of the gut microbiota, and the relative abundance of Lactobacillus increased considerably. Changes in functional genes of gut microbiota including signal transduction, carbohydrate transport, transposase, amino acid transport system, and mismatch repair were observed. To further understand the biological sense of increased Lactobacillus, Lactobacillus selective medium was used to isolate Lactobacillus species from fecal samples, and a strain with 99.8% 16S rRNA similarity with Lactobacillus plantarum strain PFK2 was obtained. Thin-layer chromatography showed that this strain could absorb but not degrade OTA, which was in agreement with the result in metagenomics that no genes related to OTA degradation increased. In conclusion, combination of metagenomics and culture-based methods can be a new strategy to study intestinal toxicity of toxins and find applicable bacterial strains for detoxification. When it comes to OTA, this kind of mycotoxin can cause compositional and functional changes of gut microbiota, and Lactobacillus are key genus to detoxify OTA in vivo.
Keywords: gut microbiota, ochratoxin A, metagenomics, Lactobacillus
Bacteria that inhabit the gastrointestinal tract play pivotal roles in host health. These bacteria link environmental factors (such as nutrients, medicinal drugs, and toxins) to host metabolism. With recent advances in next-generation sequencing technologies, metagenomics has enabled comprehensive analysis of the structure and function of gut microbial ecosystem, which is now referred to as the gut microbiota. These breakthroughs have provided increasing evidence that implicates gut microbiota in the etiology of host chronic metabolic diseases, including obesity (Devaraj et al., 2013), type 2 diabetes (Qin et al., 2012), and symptomatic atherosclerosis (Karlsson et al., 2012). Moreover, intakes of certain nutrients and pharmaceutical ingredients have been shown to trigger structural and functional changes in the gut microbiota, resulting in diverse metabolic impacts on the host (Cotillard et al., 2013; Looft et al., 2012; Zhang et al., 2012). However, how dietary intake of food mycotoxin modulates the structure and function of gut microbiota is largely unknown.
Mycotoxins are secondary metabolites of molds that have adverse effects on humans and animals, often resulting in illnesses and economic losses. Acute toxicity is generally characterized by rapid onset of toxic responses and death, whereas chronic toxicity is caused by low dose exposure over a long period of time, resulting in predisposition to cancers and other chronic diseases (Zain, 2011). Oral intake accounts for the majority of mycotoxin toxicity that is evoked by the consumption of contaminated grains or processed foods containing them. Most mycotoxins are absorbed in duodenum and jejunum with the bioavailability varying from 1% to 60% (Grenier and Applegate, 2013). As such, we propose that mycotoxins may interact with the gut microbiota before being absorbed in the gastrointestinal tract or throughout the entire intestine by non-absorbed toxins. On the one hand, the mycotoxins may alter the community structure and functional gene composition of the gut microbiota, causing indirect negative effects on the host. On the other hand, the mycotoxins can be metabolized or degraded by the microbiota so the toxicity may be decreased.
Traditional microbiological and molecular approaches have been employed to study the interaction between mycotoxins and gut microbiota. Wache et al. reported that deoxynivalenol, a mycotoxin produced by certain Fusarium species, had a moderate effect on cultivable bacteria and capillary electrophoresis-single strand conformation polymorphism patterns in the pig intestine (Wache et al., 2009). Boguhn et al. also found that Fusarium toxin-contaminated triticale altered the microbial community composition of the genus Clostridia in a simulated ruminant, which may cause a reduction in cellulolytic activity (Boguhn et al., 2010). Interestingly, a gut bacterium with 99.8% similarity to Eubacterium callanderi was isolated and found to be able to degrade Ochratoxin A (OTA) in culture (Schatzmayr et al., 2006). Nonetheless, these results displayed only a limited number of bacterial species and functional genes. Furthermore, whether bacteria can degrade mycotoxins in vivo remains to be confirmed. To address these issues, comprehensive interactions between the mycotoxins and the entire gut microbiota in vivo need to be studied in depth using metagenomics.
Metagenomics, mainly including 16S rRNA sequencing and shotgun sequencing, is defined as the application of modern genomics techniques to the study of communities of microbial organisms directly in their natural environments, bypassing the need for isolation and lab culture of individual species (Chen and Pachter, 2005). 16S rRNA sequencing is used for the detection of microbiota species composition, whereas shotgun sequencing is used when the functional genes of microbiota need to be researched. However, the results of metagenomics also need the function verification based on the culture. Although the metagenomics provides the target, the culture-based methods provide the proof.
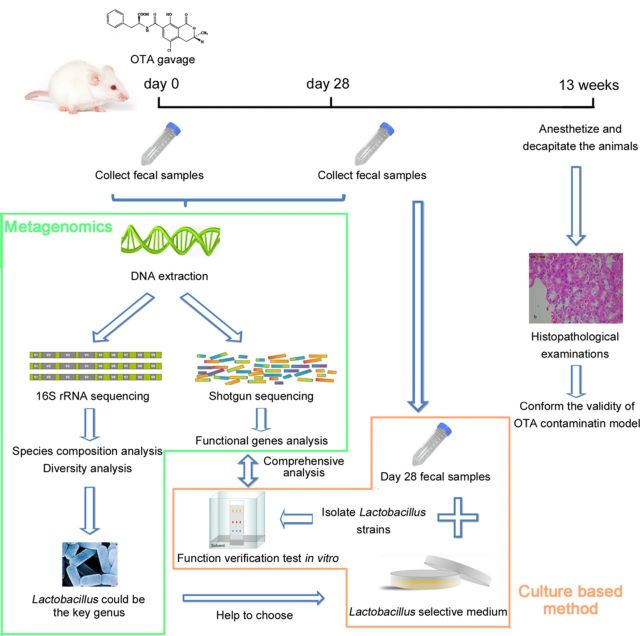
In this study, we researched the interactions between gut microbiota and OTA, a major food-contaminating mycotoxin, using the combination of metagenomics and culture-based methods. OTA was administered to male F344 rats by gavage. Fecal samples were collected before and 28 days after gavage. 16S rRNA and metagenomic approaches were used to comprehensively study changes in the taxonomic and genetic composition of the gut microbiota after exposure to OTA. The results of metagenomics indicated Lactobacillus could be the key genus in response to OTA, so the Lactobacillus strains were isolated and function verification test was carried out. The main experimental flow was shown in Figure 1.
FIG. 1.
The main experimental flow. Rats were given OTA (0, 70, or 210 μg/kg body weight) by gavage and fecal samples were collected at day 0 and day 28. Bacterial genomic DNA was extracted from the fecal samples and both 16S rRNA and shotgun sequencing were performed. After finding the potential key genera by analysis of 16S rRNA sequencing data, the culture-based method was used to isolate the target genera. Then the reactions of these isolated strains to OTA were studied in vitro, and compared with their in vivo reaction indicated by the shotgun sequencing data.
MATERIALS AND METHODS
Animals
Male F344 rats (6–7 weeks old) were purchased from Vital River, Beijing, China. Animals were housed in stainless steel cages (three rats/cage) with ad libitum access to filtered tap water and chow diet (Keaoxieli, Beijing, China) in a specific pathogen-free animal room at The Supervision and Testing Center for GMO Food Safety, Ministry of Agriculture (Beijing, China) with the license number SYXK (Beijing) 2010–0036. We used male rats in this study, as it has been shown that they are more susceptible than female rats to OTA carcinogenicity (NationalToxicologyProgram, 1989). The animal room was maintained at a temperature of 22 ± 2°C and a relative humidity of 40–70%. The room was artificially illuminated (fluorescent light) with a 12 h light/dark cycle, and the rate of air exchange was 15 times/h. Body weight was recorded weekly. All animal protocols were approved by the Ethics Committee of China Agricultural University.
After a week of acclimatization, rats (six per group) were given OTA in doses of 0, 70, or 210 μg/kg body weight (referred to as the CK, LD, and HD group, respectively) in corn oil (Aladin, Shanghai, China) by gavage (5 days per week). Body weights were recorded every week. Fecal samples were collected before treatment (day 0) or 28 days after the OTA exposure (day 28) and stored at −80°C until further analyses. In total, we collected 36 samples from the three groups and two time points. Rats were anesthetized using chloral hydrate (6% wt/vol, 5 ml/kg) and decapitated at 13 weeks. The kidneys were removed and weighed, and histopathological examinations were performed.
Bacterial genomic DNA extraction and sequencing
Feces (0.5 g) were blended in 5 ml of sterile PBS. Bacterial components were separated by differential centrifugation according to Apajalahti's method (Fuchs et al., 2008). Next, genomic DNA was extracted using saturated phenol-chloroform from the bacterial components. For 16S rRNA sequencing, DNA from the 36 samples was conducted, respectively. The V4 region of the 16S rRNA was amplified by PCR and sequenced by using a MiSeq platform (Illumina, San Diego) according to the manufacturer's instructions. For shotgun sequencing, equal amount of DNA from the same group and time point was pooled to construct in total six sequencing libraries. The library construction and sequencing was carried out on the HiSeq 2000 platform (Illumina, San Diego) according to the manufacturer's protocol.
16S rRNA sequencing analysis
A total of 12,497,214 high-quality sequences were acquired from 36 samples with each of the samples containing 347,145 sequences on average. The number of sequences in this study was much larger than other studies with the same sequencing region (V4) and platform (MiSeq) (Lozupone et al., 2013). Rarefaction analysis was conducted using the software Mothur version 1.30.2 (Schloss et al., 2009) to assess the saturation of the datasets. The results show 10,000 sequences per sample were enough saturated for diversity analysis and a larger dataset was unnecessary. Thus, a sub-dataset with 10,000 unique sequences per sample was randomly generated for further analysis. Each sequence was annotated with taxonomic information using the classify.seqs command of the Mothur. For diversity analysis, operational taxonomic unit (OTU) based taxonomy cluster was performed using Mothur at 97% identity. Bray-Curtis similarity coefficients were calculated based on the OTU data and plotted on a nonmetric multidimensional scaling (NMDS) graph to show the similarity among samples using multivar/non-metric-MDS of the software PAST version 2.17 (Hammer et al., 2001). The NMDS showed the results visually but qualitatively. For quantitatively analysis, analysis of similarities (ANOSIM) was performed based on OTU data using multivar/one-way-ANOSIM of PAST with the distance measure set as Bray-Curtis. The α and β diversities of each group were calculated based on OTU data using diversity/diversity-indices and diversity/beta-diversity of PAST and compared in box plots.
Shotgun sequencing analysis
Sequences were dereplicated and analyzed by BLAST against the non-redundant database. The BLAST reports were parsed to extract clusters of orthologous groups (COG) information with reference to Looft's method (Looft et al., 2012). Rarefaction analysis was performed to assess the saturation of the COGs dataset using Mothur. COG frequencies were subsequently analyzed in ShotgunFunctionalizeR version 1.2–9 (Kristiansson et al., 2009), including the following three steps. First, the testGeneFamilies.dircomp function was used to perform gene-centric comparisons between the groups with (28d_LD, 28d_HD) and without OTA exposure (0d_CK, 0d_LD, 0d_HD, and 28d_CK). The strategy of comparisons between groups with and without treatment was referred to Looft's study (Looft et al., 2012). Second, the testGeneFamilies.regression function was used for regression analysis of the changes in COGs as a function of OTA dose. Samples from day 0 and day 28 were tested, respectively. Because 0 could cause calculation errors, the parameter “covariates” was set to 1, 70, and 210, instead of 0, 70, and 210. Third, the testGeneCategories.dircomp function was used to perform pathway-centric group comparisons between the groups with and without OTA exposure. All of the ShotgunFunctionalizeR analyses mentioned above were based on the Poisson distribution.
Lactobacillus species isolation and thin-layer chromatography
As Lactobacillus was identified to be the key genus in gut microbiota in response to OTA treatment in metagenomics study, and previous studies indicated that many Lactobacillus strains from fermented foods or wine could detoxify OTA (Del Prete et al., 2007; Piotrowska and Zakowska, 2005), we performed Lactobacillus species isolation and function exploration. 0.5 g fecal sample of day 28 HD group was thawed mildly and diluted with 10 ml 0.9% normal saline. Lactobacillus selective (LBS) agar (Land Bridge Technology, Beijing, China) was used to obtain Lactobacillus single colony at 37°C. MRS broth (Land Bridge Technology) was used for strain culture. The genomic DNA of the isolated strains was extracted using TIANamp bacterial DNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer's protocol. Whole 16S rRNA of the strains was amplified with the standard primer pair 8F/1492R and Sanger sequenced in Sangon Biotech (Shanghai, China). The 16S V4 regions of cultured strains were extracted to be aligned with the representative V4 region of Lactobacillus in metagenomics. The whole 16S rRNA of cultured strains were used to BLAST against the non-redundant database to identify taxonomy.
The effects of strains isolated in this study on OTA were detected by thin-layer chromatography (TLC) as Shi et al. described (Shi et al., 2013). Briefly, OTA was added to 7 ml (1) MRS broth, (2) overnight cultured broth, (3) supernate of overnight cultured broth, (4) supernate of ultrasonic broken overnight cultured broth, and (5) autoclaved supernate of ultrasonic broken overnight cultured broth. The final concentration of OTA in each solution was 6 μg/ml. After incubation overnight, 1.4 ml trichloromethane was added to each solution to extract the remaining OTA. Thirty microliter of each extraction was spotted on activated silica gel plate (Qingdao Haiyang Chemical, Qingdao, China). The plate was developed in developing solvent (methylbenzene: ethyl acetate: methanoic acid = 6:3:1, v:v:v) for 5 min, and photographed by ultraviolet imaging spectrograph at 360 nm wavelength. The brightness of bands of OTA was quantified using the software Quantity One version 4.62 (Bio-Rad Laboratories, Hercules).
RESULTS
Animal Pathology
The validity of the model was confirmed by the hallmark kidney pathology after OTA exposure. The detail data of the animal pathology have been described in a previous paper (Dai et al., 2014). Briefly, the mean body weights of rats in the high-dose group were slightly lower than those of rats in the CK group at 2, 3, 5, 7, and 10 (P < 0.05) weeks, but the loss of body weight was gradually recovered thereafter. The trend was similar to that described in Mantle's report (Mantle et al., 2011). As nephrotoxicity has been shown to be the primary form of OTA toxicity, we assessed the kidneys to confirm the reliability of our OTA-induced rat model. Kidney weights in the high-dose group were reduced by 30% (P < 0.05) 13 weeks after OTA administration. Histopathological examination demonstrated (1) cytoplasmic vacuolization in the outer stripe of the outer medulla in both the LD and HD groups, (2) karyomegaly in the tubular epithelium in an OTA dose dependent, and (3) no renal lesions in the CK group. These results were consistent with those of a previous study that demonstrated OTA-induced nephrotoxicity (NationalToxicologyProgram, 1989), validating our OTA-induced rat model.
Structural Changes by OTA Treatment in the Microbiota
Because gut microbiota in principle respond to OTA gavage prior to nephrotoxicity, we analyzed fecal samples 4 weeks after the treatment. A total of 12,497,214 high-quality sequences from the V4 region of the 16S rRNA were collected from a total of 36 fecal samples, each of which contained 347,145 ± 78,091 (means ± s.d.) sequences. In total 10,000 sequences per sample were used to create a sub-dataset for the convenience of annotation and calculation. The rarefaction curves showed that the sub-dataset was saturated (Supplementary fig. S1A), meaning that a larger dataset could bring no more information for diversity analysis and thus was unnecessary. Because each sample contained the same number of sequences in the sub-dataset, the values in taxonomy data and OTU data were relative ones.
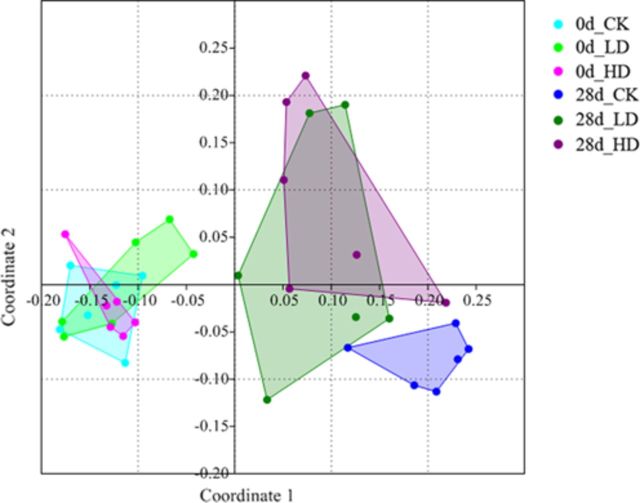
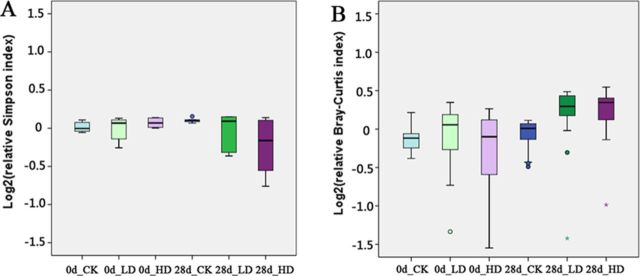
The Bray-Curtis indices were calculated for all samples, and a NMDS plot of these data revealed obvious changes in the taxonomic composition of gut microbiota over time and with treatment (Fig. 2). For significance testing of these changes, analysis of similarities (ANOSIM) was performed. On day 0, any pairwise comparisons were not different (Table 1), whereas on day 28, significant differences (P < 0.01) were found between CK and LD or HD, but not between LD and HD (P = 0.48; Table 1). Furthermore, the convex hulls around each group on the NMDS plot showed that the distributions of the 28d_LD group and the 28d_HD group were obviously more scattered than the others, indicating that OTA treatment might encourage the divergence of gut microbiota. To confirm these results, we further calculated α diversity (Simpson Index) and β diversity (Bray-Curtis Index) that represent within-subject and between-subject variations, respectively. The box plots of these indexes indicated a decrease in α-diversity in the HD groups (P < 0.1) and an increase in β-diversity in the LD and HD groups (P < 0.05 except the P value between 28d HD and 0d LD which was 0.064) on day 28 (Fig. 3).
FIG. 2.
NMDS analysis of Bray-Curtis similarity coefficients calculated from 16S rRNA sequencing data from individual animals revealed changes in the taxonomic composition of the gut microbiota over time and with treatment. CK, control; LD, low-dose, 70 μg OTA/Kg body weight; HD, high-dose, 210 μg OTA/Kg body weight.
TABLE 1. Pairwise Comparisons P Values by ANOSIM Analysis Based on Bray-Curtis Index.
| 0d_CK | 0d_LD | 0d_HD | 28d_CK | 28d_LD | 28d_HD | |
|---|---|---|---|---|---|---|
| 0d_CK | 0 | 0.7516 | 0.3359 | 0.0030 | 0.0018 | 0.0030 |
| 0d_LD | 0.7516 | 0 | 0.2749 | 0.0031 | 0.0023 | 0.0022 |
| 0d_HD | 0.3359 | 0.2749 | 0 | 0.0015 | 0.0023 | 0.0019 |
| 28d_CK | 0.0030 | 0.0031 | 0.0015 | 0 | 0.0053 | 0.0072 |
| 28d_LD | 0.0018 | 0.0023 | 0.0023 | 0.0053 | 0 | 0.4813 |
| 28d_HD | 0.0030 | 0.0022 | 0.0019 | 0.0072 | 0.4813 | 0 |
FIG. 3.
The box plots of α-diversity (Simpson index) and β-diversity (Bray-Curtis index) in gut microbiota of rats with or without OTA treatment. The circles represent the outliers and the asterisks represent the extremes.
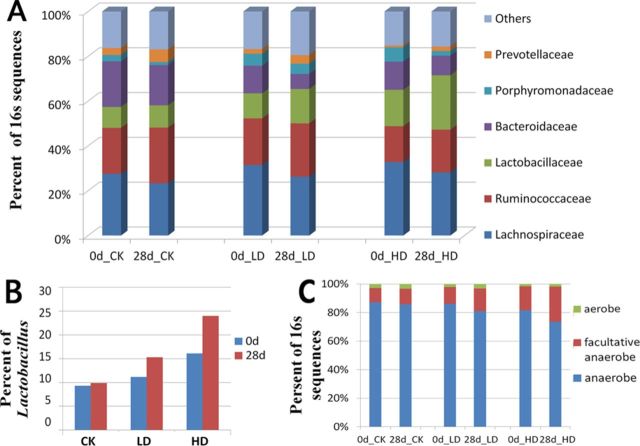
We identified Lachnospiraceae, Ruminococcaceae, Lactobacillaceae, and Bacteroidaceae as the most abundant families in gut microbiota of the rat (Supplementary table S1). Specific changes in the microbial community associated with OTA treatment included an increase of Lactobacillaceae and a decrease of Bacteroidaceae in relative abundance. Comparing day 0 and day 28, the populations of Bacteroidaceae were decreased and Lactobacillaceae were increased 28 days after gavage in OTA treatment rats but not or with a much less extent in control rats (Fig. 4A). At the genus level, Bacteroides, Dorea, Escherichia, Oribacterium, Ruminococcus, and Syntrophococcus were the most significantly decreased genera after OTA gavage (P < 0.01), whereas the increase in relative abundance of Lactobacillus was particularly striking (P <0.01, Fig. 4B, Supplementary table S1). Moreover, previous studies based on culture-dependent methods have reported that some mycotoxins, such as deoxynivalenol, could alter the ratio of aerobic and anaerobic bacteria, so we detected whether OTA had similar effect in vivo. The genera in our 16S rRNA sequencing data were annotated as aerobic, facultative anaerobic, or anaerobic according to the Bergey's Manual of Systematic Bacteriology. The aggregate percentage showed that facultative anaerobes increased whereas anaerobes decreased after gavage (Fig. 4C). Therefore, we have specifically identified facultative anaerobe, in particular, Lactobacillus, as the key gut microbiota enriched after OTA treatment.
FIG. 4.
Taxonomic composition changes in the gut microbiota as evidenced by 16S rRNA gene sequencing. (A) Changes in microbiota composition at the family level. (B) Changes in Lactobacillus abundance. (C) Changes in the proportions of aerobic, facultative anaerobic, and anaerobic bacteria.
Functional Changes by OTA Treatment in the Microbiota
16S rRNA sequencing was performed on each individual animal, whereas shotgun sequencing was performed on each group. The microbiota genomic DNA from the same group and time point was pooled for shotgun sequencing as Looft et al. did (Looft et al., 2012). A total of 234,682,342 shotgun sequences were obtained from six samples (with 39,113,724 ± 6,607,468 sequences per sample). Then, COG annotation and subsequent analyses were performed by using 200,000 randomly generated sequences, which were sufficient to meet the sample size requirements based on the rarefaction curves (Supplementary fig. S1B). In contrast to the taxonomic composition of the gut microbiota, the functional structure seemed relatively stable among groups and over time, as evidenced by the proportion of each COG category (Supplementary fig. S2). For a more detailed exploration of this observation, ShotgunFunctionalizeR gene-centric analyses were performed. The results revealed stability among groups, as there were no significantly different COGs among the three samples collected at day 0. Additionally, the results revealed relative stability over time, as there were only 36 COGs exhibiting significant differences between day 0 and 28d_CK samples (Supplementary table S2).
Because of the stability discussed above, we were able to combine the three samples from day 0 and the 28d_CK sample together to form an OTA-unexposed group (n = 4). When the toxic effects of OTA were studied, the OTA-unexposed group was compared with the OTA-exposed group (28d_LD, 28d_HD). This strategy, which enlarges the sample size per group, had been employed in a previous metagenomic study (Looft et al., 2012). The gene-centric group comparison between the OTA-exposed and OTA-unexposed groups revealed 155 significantly different COGs (adjusted P-value <0.05). After excluding the COGs that changed with time, 138 COGs that were affected by OTA remained (Table 2 and Supplementary table S3). To identify themes among these 138 differentially represented COGs, an enrichment analysis was performed based on COG categories and COG pathways; however, no significantly enriched category or pathway was found.
TABLE 2. Partial COGs Differentially Represented (P < 0.05) in the OTA-Exposed and OTA-Unexposed Groups Detected by ShotgunFunctionalized R Gene-Centric Analysis of Metagenomic Sequencing Data. (Only Different COGs Belong to the Four COG Pathways That We Focus on Are List the Here. The Full List of 138 Different COGs Is Given in Supplementary table S5.).
| GeneFamily | Annotation | Adj P-value |
|---|---|---|
| *Amino acid transport and metabolism* | ||
| COG1113 | Gamma-aminobutyrate permease and related permeases | 6.05E-10 |
| COG4166 | ABC-type oligopeptide transport system, periplasmic component | 2.57E-08 |
| COG0531 | Amino acid transporters | 8.05E-08 |
| COG1114 | Branched-chain amino acid permeases | 3.18E-03 |
| COG1003 | Glycine cleavage system protein P (pyridoxal-binding), C-terminal domain | 4.28E-03 |
| COG0477 | Permeases of the major facilitator superfamily | 6.17E-03 |
| COG2195 | Di- and tripeptidases | 1.26E-02 |
| COG0404 | Glycine cleavage system T protein (aminomethyltransferase) | 1.67E-02 |
| COG0339 | Zn-dependent oligopeptidases | 1.85E-02 |
| COG1703 | Putative periplasmic protein kinase ArgK and related GTPases of G3E family | 2.12E-02 |
| COG1166 | Arginine decarboxylase (spermidine biosynthesis) | 2.68E-02 |
| COG1932 | Phosphoserine aminotransferase | 3.29E-02 |
| COG2866 | Predicted carboxypeptidase | 3.75E-02 |
| COG4690 | Dipeptidase | 3.94E-02 |
| *Carbohydrate transport and metabolism* | ||
| COG0395 | ABC-type sugar transport system, permease component | 5.92E-10 |
| COG4209 | ABC-type polysaccharide transport system, permease component | 1.16E-08 |
| COG3525 | N-acetyl-beta-hexosaminidase | 1.19E-08 |
| COG2814 | Arabinose efflux permease | 2.58E-04 |
| COG2407 | L-fucose isomerase and related proteins | 4.09E-04 |
| COG3957 | Phosphoketolase | 5.31E-04 |
| COG4975 | Putative glucose uptake permease | 3.21E-03 |
| COG3405 | Endoglucanase Y | 4.43E-03 |
| COG0176 | Transaldolase | 6.94E-03 |
| COG1175 | ABC-type sugar transport systems, permease components | 8.24E-03 |
| COG0580 | Glycerol uptake facilitator and related permeases (Major Intrinsic Protein Family) | 1.41E-02 |
| COG1554 | Trehalose and maltose hydrolases (possible phosphorylases) | 1.41E-02 |
| COG2376 | Dihydroxyacetone kinase | 1.59E-02 |
| COG0366 | Glycosidases | 2.28E-02 |
| COG1449 | Alpha-amylase/alpha-mannosidase | 3.66E-02 |
| COG2190 | Phosphotransferase system IIA components | 4.28E-02 |
| *Replication, recombination and repair* | ||
| COG3666 | Transposase and inactivated derivatives | 1.12E-07 |
| COG3436 | Transposase and inactivated derivatives | 2.34E-06 |
| COG0675 | Transposase and inactivated derivatives | 1.63E-05 |
| COG3547 | Transposase and inactivated derivatives | 1.84E-05 |
| COG0582 | Integrase | 3.58E-04 |
| COG0514 | Superfamily II DNA helicase | 1.11E-03 |
| COG3328 | Transposase and inactivated derivatives | 1.11E-02 |
| COG0249 | Mismatch repair ATPase (MutS family) | 1.48E-02 |
| *Signal transduction mechanisms* | ||
| COG3292 | Predicted periplasmic ligand-binding sensor domain | 5.65E-05 |
| COG4753 | Response regulator containing CheY-like receiver domain and AraC-type DNA-binding domain | 8.41E-04 |
| COG0784 | FOG: CheY-like receiver | 1.04E-02 |
| COG2205 | Osmosensitive K+ channel histidine kinase | 1.67E-02 |
| COG1716 | FOG: FHA domain | 1.94E-02 |
| COG0394 | Protein-tyrosine-phosphatase | 1.99E-02 |
| COG2310 | Uncharacterized proteins involved in stress response, homologs of TerZ and putative cAMP-binding protein CABP1 | 2.95E-02 |
| COG3434 | Predicted signal transduction protein containing EAL and modified HD-GYP domains | 3.45E-02 |
| COG2337 | Growth inhibitor | 3.94E-02 |
Regression analysis was then performed to search for the COGs that changed with the concentration gradient. Eight COGs were found to be significant in the regression analysis among the three samples from day 28 (adjusted P < 0.05). Most of these COGs were among the 138 COGs with the highest levels of significance in the group comparative analysis, and none were significant in the regression analysis of samples from 0 day (Supplementary table S3). Of the eight COGs that changed with the concentration of OTA, 3 COGs (COG1113, COG4166, COG0531) belonged to the COG pathway Amino acid transport and metabolism, three COGs (COG3666, COG3436, COG0675) belonged to the COG pathway Replication, recombination and repair, and two COGs (COG0395, COG2190) belonged to the COG pathway Carbohydrate transport and metabolism.
Finally, a pathway-centric analysis was performed to further analyze which COG pathways were represented differently between the OTA-exposed and unexposed groups, based on the entire COG dataset using the testGeneCategories.dircomp function of ShotgunFunctionalizeR. The results showed that Signal transduction mechanisms, Amino acid transport and metabolism and Cell motility were three of the most significant COG pathways, with Carbohydrate transport and metabolism and Replication, recombination and repair also on the list of significantly different COG pathways (Supplementary table S3).
Changes in Genera or COGs Related to OTA Degradation
Many gut bacterial species with OTA degradation or adsorption abilities have been identified in vitro using culture-dependent methods in previous studies; however, whether these species play a role in vivo has yet to be confirmed. Here, we focused on the five genera that are known to be involved in OTA detoxification, Lactobacillus (Del Prete et al., 2007; Fuchs et al., 2008; Mechoud et al., 2012; Piotrowska and Zakowska, 2005), Bifidobacterium (Skrinjar et al., 1996), Acinetobacter (Hwang and Draughon, 1994), Bacillus (Petchkongkaew et al., 2008), and Eubacterium (Schatzmayr et al., 2006). The relative abundance of Lactobacillus increased, as described in the previous section. By contrast, the other genera of bacteria appeared not to play a role in OTA detoxification, as their relative abundances were quite low and did not increase when the gut microbiota were exposed to OTA.
Four particular groups of enzymes were examined in this study, including proteases and peptidases, esterases, lipases, and dioxygenases, because of their potential OTA degradation activities (Abrunhosa et al., 2006; Hwang and Draughon, 1994; Rodriguez et al., 2011; Stander et al., 2000; Wegst and Lingens, 1983). Proteases, peptidases, esterases, and lipases can hydrolyze the peptide bond of OTA, forming phenylalanine and the much less toxic OTα, whereas the dioxygenases can catalyze the attack of the phenyl moiety of OTA to form a dihydrodiol derivative, which is also transformed into OTα via a series of further metabolic reactions. According to our gene-centric analyses, only four COGs related to the protease and peptidase enzyme groups increased following exposure to OTA; however, none were able to hydrolyze OTA due to their substrate specificity annotated in KEGG and the structure of OTA.
Lactobacillus Species Isolation and Function Exploration
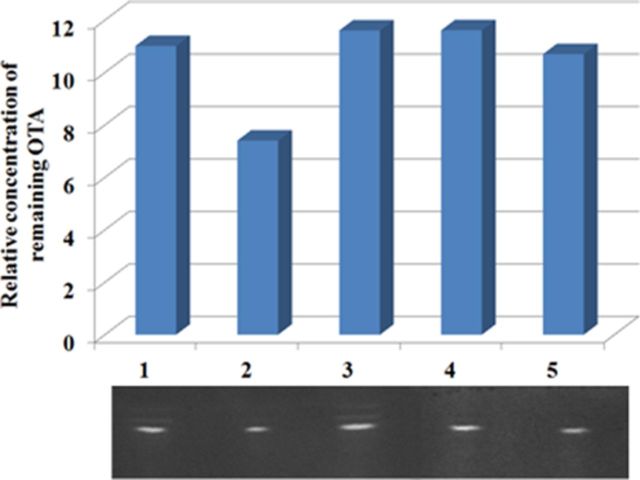
Whole length 16S rRNAs of 10 strains isolated by LBS agar were Sanger sequenced. Their sequences were identical, and the V4 region had a 99.0% similarity to the representative sequence of Lactobacillus obtained in metagenomics (Fig. 5). Blast analysis showed the 16S rRNA sequence of isolated strain had a 99.8% similarity to Lactobacillus plantarum strain PFK2 16S rRNA (Sequence ID: gb|DQ295035.1). TLC showed that overnight cultured broth could decrease the concentration of OTA, whereas if the bacterial body was removed by centrifugation, no matter ultrasonication and autoclaving or not, the concentration of OTA remained unchanged (Fig. 6). The results of TLC implicated that the isolated strain had the ability to detoxify OTA, and the detoxification mechanism could be absorption, instead of degradation. These results were in agreement with the results of metagenomics that though the relative abundance of Lactobacillus increased, no increase of OTA degradation related genes was observed.
FIG. 5.
Alignment of isolated strain 16S rRNA V4 region and representative sequence of Lactobacillus in metagenomics.
FIG. 6.
The effects of isolated strains on OTA. OTA was added to (1) MRS broth, (2) overnight cultured broth, (3) supernate of overnight cultured broth, (4) supernate of ultrasonic broken overnight cultured broth, and (5) autoclaved supernate of ultrasonic broken overnight cultured broth. After incubation overnight, the remaining OTA was detected by TLC. The brightness of the bands was analysis by the software Quantity One and plotted in the bar diagram.
DISCUSSION
Overall, we developed a new strategy to study intestinal toxicity of toxins and find applicable bacterial strains for detoxification. Metagenomics, including 16S rRNA sequencing and shotgun sequencing, can detect the structural and functional changes of gut microbiota accurately and sensitively. The results of metagenomics can be informative screenings or indications for further exploration of the treasure of gut microbiota. When the target genus or species is acquired, the isolation and culture of them will be much more feasible, compared with untargeted large-scale culture. Then, the function verification tests of isolated strains can be performed, and the results of these tests may lead to deeper understandings of functional changes of gut microbiota.
The 16S rRNA sequencing of each individual sample made the diversity analysis realized, but in the shotgun sequencing, pool of samples from the same group and time point was the usual practice (Looft et al., 2012) due to the high cost of this kind of sequencing. The metagenomic results of this study indicate that the gut microbiota had a highly variable species composition and a relatively stable functional composition among groups and across time, which is similar to the results of metagenomic studies on pigs (Looft et al., 2012) and human beings (Consortium, 2012). This phenomenon may be due to the selective effect of host. The functions of gut microbiota, compared with species composition, are more important to the host in normal status, so the host may control the functions of gut microbiota more tightly, in unknown ways.
The OTA treatment caused remarkable changes in the microbial community structure. The decrease in α-diversity indicated that OTA could inhibit the growth of gut microbiota in vivo; however, the ultimate status of the gut microbiota following OTA exposure was not identical among individuals, as indicated by the β-diversity, which was possibly related to differences in the initial structures of the microbial communities. This decrease in the diversity of the gut microbiota is dangerous to the host's health. Many studies on intestinal and immune diseases, such as Crohn's disease (Manichanh et al., 2006), ulcerative colitis (Nemoto et al., 2012), viral diarrhea (Ma et al., 2011), eczema (Ismail et al., 2012; Wang et al., 2008), and allergic disease (Bisgaard et al., 2011), have revealed reductions in the diversity of the gut microbiota.
The changes of the species composition indicated that different bacterial strains had different sensitivity to OTA. Bacteroidaceae may be more sensitive than Lactobacillaceae to the inhibitory effects of OTA, and anaerobic strains may be more sensitive than facultative anaerobic strains. Lactobacillus exhibited the strongest resistance to OTA. A previous culture-dependent in vitro study tested the antibacterial effects of different kinds of mycotoxin on 10 different strains (Ali-Vehmas et al., 1998). Different degrees of inhibition by mycotoxin were observed in different strains; however, L. plantarum and L. casei were not affected, indicated Lactobacillus was more resistant to mycotoxin. The increase in facultative anaerobes is one of the key changes in gut microbiota that has been observed in elderly populations and that may be related to aging (Woodmansey, 2007). Therefore, we can predict that the changes in the community structure of the gut microbiota caused by OTA may have some negative effects on the host health.
The functional genes of the gut microbiota were also affected by the OTA treatment. Signal transduction mechanism was the most significant COG pathway affected by OTA, according to the pathway-centric analyses. Changes in signal transduction, such as changes in the expression of MAPKs and in calcium homeostasis, have been shown to induce the carcinogenicity of OTA in eucaryon (Marin-Kuan et al., 2008). So it is appropriate to predict that the changes of signal transduction caused by OTA in prokaryote may be one of the potential ways through which the growth of some bacteria strains was inhibited. However, owing to differences in the signal transduction systems between bacteria and eucaryon, much more studies need to be conducted to confirm this prediction.
COGs related to amino acid and oligopeptide transporters increased significantly, which may be a protection mechanism generated by the bacteria. As OTA is a competitive inhibitor with respect to phenylalanine, the increased intake of phenylalanine due to the increased expression of amino acid transporters may counteract the effects of OTA. Supplementation with phenylalanine has been proven to reverse the toxicity of OTA in animals and cell lines (Creppy et al., 1979; Stoev et al., 2002; Zanic-Grubisic et al., 2000). Additionally, the relative abundance of COGs related to oligopeptidases, which also belonged to the COG pathway Amino acid transport and metabolism, increased as well. The synergistic increase in oligopeptide transporters and oligopeptidases may cause an increase in the intracellular phenylalanine concentration of bacteria to resist the effects of OTA. Transporter systems appeared to be the most sensitive to OTA. In addition to the amino acid transporters, carbohydrate transporters were also affected. COGs related to the ABC-type carbohydrate transport system decreased, whereas other types of carbohydrate transport systems, such as COG0580 (Glycerol uptake facilitator and related permeases) and COG4975 (Putative glucose uptake permease), increased. The effects of OTA on the carbohydrate transport system may explain how OTA inhibits the growth of some bacteria. The effects of OTA on the transport systems for amino acids and carbohydrates have also been observed in human epithelial intestinal cell lines; however, the pathway through which OTA regulates the abundance of genes involved in these transport systems remains unclear.
As genotoxicity is one of the major toxic effects of OTA, it is understandable that the COG pathway Replication, recombination and repair is affected when the gut microbiota is exposed to OTA. Particularly, there were five COGs named Transposase and inactivated derivatives that changed significantly, indicating that transposases may be neglected factors in the carcinogenicity network of OTA. The changes in the transposases varied, with COG3666 and COG0675 increasing and COG3436, COG3547, and COG3328 decreasing. Thus, OTA may modulate the proportions of different types of transposases, leading to changes in genome stability. Additionally, the mismatch repair ATPase (MutS family) increased, perhaps as a response to the oxidative DNA damage caused by OTA.
Though many types of gut bacteria have been shown to hydrolyze OTA to OTα in vitro, only bacteria belonging to the genus Lactobacillus played a role in vivo in this OTA-induced rat model. There has been a debate on the OTA detoxification mechanism of Lactobacillus (Del Prete et al., 2007; Fuchs et al., 2008). Our results suggested that absorption, rather than hydrolysis, was more likely to be the main way by which Lactobacillus detoxifies OTA. Though the Lactobacillus strain isolated in this study was less efficient in OTA detoxification compared with a Bacillus strain isolated in our previous study (Shi et al., 2013), the Lactobacillus strain could be more compatible to the host.
CONCLUSIONS
Combination of metagenomics and culture-based methods can be a new strategy to study intestinal toxicity of toxins and find applicable bacterial strains for detoxification. When it comes to OTA, this kind of mycotoxin can cause compositional and functional changes of gut microbiota, and Lactobacillus are key genus to detoxify OTA in vivo.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
Acknowledgments
We thank all members of Kunlun Huang's laboratory, who contributed to the Program for New Century Excellent Talents in University (2014FG046).
REFERENCES
- Abrunhosa L., Santos L., Venâncio A. Degradation of ochratoxin A by proteases and by a crude enzyme of Aspergillus niger. Food Biotechnol. 2006;20:231–242. [Google Scholar]
- Ali-Vehmas T., Rizzo A., Westermarck T., Atroshi F. Measurement of antibacterial activities of T-2 toxin, deoxynivalenol, ochratoxin A, aflatoxin B1 and fumonisin B1 using microtitration tray-based turbidimetric techniques. Zentralbl. Veterinarmed. A. 1998;45:453–458. doi: 10.1111/j.1439-0442.1998.tb00848.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard H., Li N., Bonnelykke K., Chawes B. L., Skov T., Paludan-Muller G., Stokholm J., Smith B., Krogfelt K. A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011;128:646–652. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- Boguhn J., Neumann D., Helm A., Strobel E., Tebbe C. C., Danicke S., Rodehutscorda M. Effects of concentrate proportion in the diet with or without Fusarium toxin-contaminated triticale on ruminal fermentation and the structural diversity of rumen microbial communities in vitro. Arch Anim. Nutr. 2010;64:467–483. doi: 10.1080/1745039X.2010.511515. [DOI] [PubMed] [Google Scholar]
- Chen K., Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S. P., Kong L. C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Creppy E. E., Lugnier A. A., Beck G., Roschenthaler R., Dirheimer G. Action of ochratoxin A on cultured hepatoma cells–reversion of inhibition by phenylalanine. FEBS Lett. 1979;104:287–290. doi: 10.1016/0014-5793(79)80834-0. [DOI] [PubMed] [Google Scholar]
- Dai Q., Jue Z., Qi X., Xu W., He X., Guo M., Dweep H., Cheng W., Luo Y., Xia K., et al. MicroRNA profiling of rats with ochratoxin A nephrotoxicity. BMC Genomics. 2014;15:333. doi: 10.1186/1471-2164-15-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete V., Rodriguez H., Carrascosa A. V., de las Rivas B., Garcia-Moruno E., Munoz R. In vitro removal of ochratoxin A by wine lactic acid bacteria. J. Food Prot. 2007;70:2155–2160. doi: 10.4315/0362-028x-70.9.2155. [DOI] [PubMed] [Google Scholar]
- Devaraj S., Hemarajata P., Versalovic J. The human gut microbiome and body metabolism: Implications for obesity and diabetes. Clin. Chem. 2013;59:617–628. doi: 10.1373/clinchem.2012.187617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Sontag G., Stidl R., Ehrlich V., Kundi M., Knasmuller S. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008;46:1398–1407. doi: 10.1016/j.fct.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Grenier B., Applegate T. J. Modulation of intestinal functions following mycotoxin ingestion: Meta-analysis of published experiments in animals. Toxins (Basel) 2013;5:396–430. doi: 10.3390/toxins5020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D., Ryan P. Past: Paleontological Statistics Software Package for education and data analysis. Paleontología Electrónica. 2001;4:1–9. [Google Scholar]
- Hwang C., Draughon F. A. Degradation of ochratoxin A by Acinetobacter calcoaceticus. J. Food Prot. 1994;57:410–414. doi: 10.4315/0362-028X-57.5.410. [DOI] [PubMed] [Google Scholar]
- Ismail I. H., Oppedisano F., Joseph S. J., Boyle R. J., Licciardi P. V., Robins-Browne R. M., Tang M. L. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr. Allergy Immunol. 2012;23:674–681. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- Karlsson F. H., Fak F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D., Backhed F., Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012;3:1245–1252. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E., Hugenholtz P., Dalevi D. ShotgunFunctionalizeR: An R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- Looft T., Johnson T. A., Allen H. K., Bayles D. O., Alt D. P., Stedtfeld R. D., Sul W. J., Stedtfeld T. M., Chai B., Cole J. R., et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl Acad. Sci. U.S.A. 2012;09:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A., Li M., Campbell T. B., Flores S. C., Linderman D., Gebert M. J., Knight R., Fontenot A. P., Palmer B. E. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantle P. G., Nicholls A. W., Shockcor J. P. H NMR spectroscopy-based metabolomic assessment of uremic toxicity, with toxicological outcomes, in male rats following an acute, mid-life insult from ochratoxin a. Toxins (Basel) 2011;3:504–519. doi: 10.3390/toxins3060504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Kuan M., Cavin C., Delatour T., Schilter B. Ochratoxin A carcinogenicity involves a complex network of epigenetic mechanisms. Toxicon. 2008;52:195–202. doi: 10.1016/j.toxicon.2008.04.166. [DOI] [PubMed] [Google Scholar]
- Ma C., Wu X., Nawaz M., Li J., Yu P., Moore J. E., Xu J. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr. Microbiol. 2011;63:259–266. doi: 10.1007/s00284-011-9972-7. [DOI] [PubMed] [Google Scholar]
- Mechoud M. A., Juarez G. E., de Valdez G. F., Rodriguez A. V. Lactobacillus reuteri CRL 1098 and Lactobacillus acidophilus CRL 1014 differently reduce in vitro immunotoxic effect induced by Ochratoxin A. Food Chem. Toxicol. 2012;50:4310–4315. doi: 10.1016/j.fct.2012.07.070. [DOI] [PubMed] [Google Scholar]
- NationalToxicologyProgram. Toxicology and carcinogenesis Studies of ochratoxin A (CAS no. 303–47–9) in F344/N rats (gavage studies) Natl Toxicol. Program Tech. Rep. Ser. 1989;358:1–142. [PubMed] [Google Scholar]
- Nemoto H., Kataoka K., Ishikawa H., Ikata K., Arimochi H., Iwasaki T., Ohnishi Y., Kuwahara T., Yasutomo K. Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig. Dis. Sci. 2012;57:2955–2964. doi: 10.1007/s10620-012-2236-y. [DOI] [PubMed] [Google Scholar]
- Petchkongkaew A., Taillandier P., Gasaluck P., Lebrihi A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 2008;104:1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x. [DOI] [PubMed] [Google Scholar]
- Piotrowska M., Zakowska Z. The elimination of ochratoxin A by lactic acid bacteria strains. Pol. J. Microbiol. 2005;54:279–286. [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rodriguez H., Reveron I., Doria F., Costantini A., De Las Rivas B., Munoz R., Garcia-Moruno E. Degradation of ochratoxin a by Brevibacterium species. J. Agric. Food Chem. 2011;59:10755–10760. doi: 10.1021/jf203061p. [DOI] [PubMed] [Google Scholar]
- Schatzmayr G., Zehner F., Taubel M., Schatzmayr D., Klimitsch A., Loibner A. P., Binder E. M. Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 2006;50:543–551. doi: 10.1002/mnfr.200500181. [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Liang Z., Li J., Hao J., Xu Y., Huang K., Tian J., He X., Xu W. Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14. J. Sci. Food Agric. 2013 doi: 10.1002/jsfa.6507. [DOI] [PubMed] [Google Scholar]
- Skrinjar M., Rasic J. L., Stojicic V. Lowering of ochratoxin A level in milk by yoghurt bacteria and bifidobacteria. Folia Microbiol. (Praha) 1996;41:26–28. doi: 10.1007/BF02816335. [DOI] [PubMed] [Google Scholar]
- Stander M. A., Bornscheuer U. T., Henke E., Steyn P. S. Screening of commercial hydrolases for the degradation of ochratoxin A. J. Agric. Food Chem. 2000;48:5736–5739. doi: 10.1021/jf000413j. [DOI] [PubMed] [Google Scholar]
- Stoev S. D., Djuvinov D., Mirtcheva T., Pavlov D., Mantle P. Studies on some feed additives giving partial protection against ochratoxin A toxicity in chicks. Toxicol Lett. 2002;135:33–50. doi: 10.1016/s0378-4274(02)00234-5. [DOI] [PubMed] [Google Scholar]
- Wache Y. J., Valat C., Postollec G., Bougeard S., Burel C., Oswald I. P., Fravalo P. Impact of deoxynivalenol on the intestinal microflora of pigs. Int. J. Mol. Sci. 2009;10:1–17. doi: 10.3390/ijms10010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Karlsson C., Olsson C., Adlerberth I., Wold A. E., Strachan D. P., Martricardi P. M., Aberg N., Perkin M. R., Tripodi S., et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J. Allergy Clin. Immunol. 2008;121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Wegst W., Lingens F. Bacterial degradation of ochratoxin A. FEMS Microbiol. Lett. 1983;17:341–344. [Google Scholar]
- Woodmansey E. J. Intestinal bacteria and ageing. J. Appl. Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- Zain M. E. Impact of mycotoxins on humans and animals. J. Saudi Chem. Soc. 2011;15:129–144. [Google Scholar]
- Zanic-Grubisic T., Zrinski R., Cepelak I., Petrik J., Radic B., Pepeljnjak S. Studies of ochratoxin A-induced inhibition of phenylalanine hydroxylase and its reversal by phenylalanine. Toxicol. Appl. Pharmacol. 2000;167:132–139. doi: 10.1006/taap.2000.8987. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhao Y., Zhang M., Pang X., Xu J., Kang C., Li M., Zhang C., Zhang Z., Zhang Y., et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.