Abstract
In this paper we report the set-up and results of the Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) organized in conjunction with the MICCAI 2012 and 2013 conferences. Twenty state-of-the-art tumor segmentation algorithms were applied to a set of 65 multi-contrast MR scans of low- and high-grade glioma patients—manually annotated by up to four raters—and to 65 comparable scans generated using tumor image simulation software. Quantitative evaluations revealed considerable disagreement between the human raters in segmenting various tumor sub-regions (Dice scores in the range 74%–85%), illustrating the difficulty of this task. We found that different algorithms worked best for different sub-regions (reaching performance comparable to human inter-rater variability), but that no single algorithm ranked in the top for all sub-regions simultaneously. Fusing several good algorithms using a hierarchical majority vote yielded segmentations that consistently ranked above all individual algorithms, indicating remaining opportunities for further methodological improvements. The BRATS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Index Terms: MRI, Brain, Oncology/tumor, Image segmentation, Benchmark
I. Introduction
Gliomas are the most frequent primary brain tumors in adults, presumably originating from glial cells and infiltrating the surrounding tissues [1]. Despite considerable advances in glioma research, patient diagnosis remains poor. The clinical population with the more aggressive form of the disease, classified as high-grade gliomas, have a median survival rate of two years or less and require immediate treatment [2], [3]. The slower growing low-grade variants, such as low-grade astrocytomas or oligodendrogliomas, come with a life expectancy of several years so aggressive treatment is often delayed as long as possible. For both groups, intensive neuroimaging protocols are used before and after treatment to evaluate the progression of the disease and the success of a chosen treatment strategy. In current clinical routine, as well as in clinical studies, the resulting images are evaluated either based on qualitative criteria only (indicating, for example, the presence of characteristic hyper-intense tissue appearance in contrast-enhanced T1-weighted MRI), or by relying on such rudimentary quantitative measures as the largest diameter visible from axial images of the lesion [4], [5].
By replacing the current basic assessments with highly accurate and reproducible measurements of the relevant tumor substructures, image processing routines that can automatically analyze brain tumor scans would be of enormous potential value for improved diagnosis, treatment planning, and follow-up of individual patients. However, developing automated brain tumor segmentation techniques is technically challenging, because lesion areas are only defined through intensity changes that are relative to surrounding normal tissue, and even manual segmentations by expert raters show significant variations when intensity gradients between adjacent structures are smooth or obscured by partial voluming or bias field artifacts. Furthermore, tumor structures vary considerably across patients in terms of size, extension, and localization, prohibiting the use of strong priors on shape and location that are important components in the segmentation of many other anatomical structures. Moreover, the so-called mass effect induced by the growing lesion may displace normal brain tissues, as do resection cavities that are present after treatment, thereby limiting the reliability of spatial prior knowledge for the healthy part of the brain. Finally, a large variety of imaging modalities can be used for mapping tumor-induced tissue changes, including T2 and FLAIR MRI (highlighting differences in tissue water relaxational properties), post-Gadolinium T1 MRI (showing pathological intratumoral take-up of contrast agents), perfusion and diffusion MRI (local water diffusion and blood flow), and MRSI (relative concentrations of selected metabolites), among others. Each of these modalities provides different types of biological information, and therefore poses somewhat different information processing tasks.
Because of its high clinical relevance and its challenging nature, the problem of computational brain tumor segmentation has attracted considerable attention during the past 20 years, resulting in a wealth of different algorithms for automated, semi-automated, and interactive segmentation of tumor structures (see [6] and [7] for good reviews). Virtually all of these methods, however, were validated on relatively small private datasets with varying metrics for performance quantification, making objective comparisons between methods highly challenging. Exacerbating this problem is the fact that different combinations of imaging modalities are often used in validation studies, and that there is no consistency in the tumor sub-compartments that are considered. As a consequence, it remains difficult to judge which image segmentation strategies may be worthwhile to pursue in clinical practice and research; what exactly the performance is of the best computer algorithms available today; and how well current automated algorithms perform in comparison with groups of human expert raters.
In order to gauge the current state-of-the-art in automated brain tumor segmentation and compare between different methods, we organized in 2012 and 2013 a Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) challenge in conjunction with the international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). For this purpose, we prepared and made available a unique dataset of MR scans of low- and high-grade glioma patients with repeat manual tumor delineations by several human experts, as well as realistically generated synthetic brain tumor datasets for which the ground truth segmentation is known. Each of 20 different tumor segmentation algorithms was optimized by their respective developers on a subset of this particular dataset, and subsequently run on the remaining images to test performance against the (hidden) manual delineations by the expert raters. In this paper we report the set-up and the results of this BRATS benchmark effort. We also describe the BRATS reference dataset and online validation tools, which we make publicly available as an ongoing benchmarking resource for future community efforts.
The paper is organized as follows. We briefly review the current state-of-the-art in automated tumor segmentation, and survey benchmark efforts in other biomedical image interpretation tasks, in Section II. We then describe the BRATS set-up and data, the manual annotation of tumor structures, and the evaluation process in Section III. Finally, we report and discuss the results of our comparisons in Sections IV and V, respectively. Section VI concludes the paper.
II. Prior Work
Algorithms for Brain Tumor Segmentation
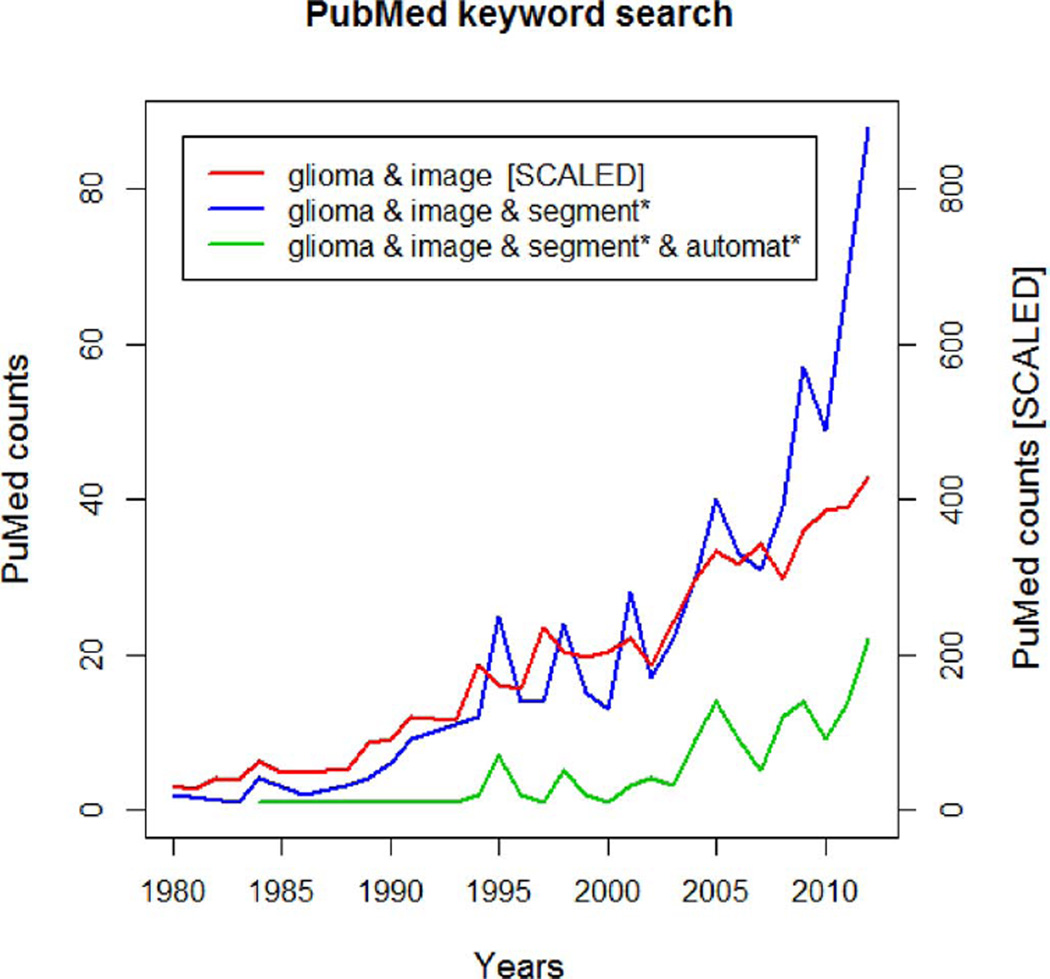
The number of clinical studies involving brain tumor quantification based on medical images has increased significantly over the past decades. Around a quarter of such studies relies on automated methods for tumor volumetry (Fig. 1). Most of the existing algorithms for brain tumor analysis focus on the segmentation of glial tumor, as recently reviewed in [6], [7]. Comparatively few methods deal with less frequent tumors such as meningioma [8]–[12] or specific glioma subtypes [13].
Fig. 1.
Results of PubMed searches for brain tumor (glioma) imaging (red), tumor quantification using image segmentation (blue), and automated tumor segmentation (green). While the tumor imaging literature has seen a nearly linear increase over the last 30 years, the number of publications involving tumor segmentation has grown more than linearly since 5–10 years. Around 25% of such publications refer to “automated” tumor segmentation.
Methodologically, many state-of-the-art algorithms for tumor segmentation are based on techniques originally developed for other structures or pathologies, most notably for automated white matter lesion segmentation that has reached considerable accuracy [14]. While many technologies have been tested for their applicability to brain tumor detection and segmentation—e.g., algorithms from image retrieval as an early example [9]—we can categorize most current tumor segmentation methods into one of two broad families. In the so-called generative probabilistic methods, explicit models of anatomy and appearance are combined to obtain automated segmentations, which offers the advantage that domain-specific prior knowledge can easily be incorporated. Discriminative approaches, on the other hand, directly learn the relationship between image intensities and segmentation labels without any domain knowledge, concentrating instead on specific (local) image features that appear relevant for the tumor segmentation task.
Generative models make use of detailed prior information about the appearance and spatial distribution of the different tissue types. They often exhibit good generalization to unseen images, and represent the state-of-the-art for many brain tissue segmentation tasks [15]–[21]. Encoding prior knowledge for a lesion, however, is difficult. Tumors may be modeled as outliers relative to the expected shape [22], [23] or image signal of healthy tissues [17], [24] which is similar to approaches for other brain lesions, such as Multiple Sklerosis [25], [26]. In [17], for instance, a criterion for detecting outliers is used to generate a tumor prior in a subsequent expectation-maximizations segmentation which treats tumor as an additional tissue class. Alternatively, the spatial prior for the tumor can be derived from the appearance of tumor-specific “bio-markers” [27], [28], or from using tumor growth models to infer the most likely localization of tumor structures for a given set of patient images [29]. All these models rely on registration for accurately aligning images and spatial priors, which is often problematic in the presence of large lesions or resection cavities. In order to overcome this difficulty, both joint registration and tumor segmentation [18], [30] and joint registration and estimation of tumor displacement [31] have been studied. A limitation of generative models is the significant effort required for transforming an arbitrary semantic interpretation of the image, for example, the set of expected tumor substructures a radiologist would like to have mapped in the image, into appropriate probabilistic models.
Discriminative models directly learn from (manually) annotated training images the characteristic differences in the appearance of lesions and other tissues. In order to be robust against imaging artifacts and intensity and shape variations, they typically require substantial amounts of training data [32]–[38]. As a first step, these methods typically extract dense, voxel-wise features from anatomical maps [35], [39] calculating, for example, local intensity differences [40]–[42], or intensity distributions from the wider spatial context of the individual voxel [39], [43], [44]. As a second step, these features are then fed into classification algorithms such as support vector machines [45] or decision trees [46] that learn boundaries between classes in the high-dimensional feature space, and return the desired tumor classification maps when applied to new data. One drawback of this approach is that, because of the explicit dependency on intensity features, segmentation is restricted to images acquired with the exact same imaging protocol as the one used for the training data. Even then, careful intensity calibration remains a crucial part of discriminative segmentation methods in general [47]–[49], and tumor segmentation is no exception to this rule.
A possible direction that avoids the calibration issues of discriminative approaches, as well as the limitations of generative models, is the development of joint generative-discriminative methods. These techniques use a generative method in a pre-processing step to generate stable input for a subsequent discriminative model that can be trained to predict more complex class labels [50], [51].
Most generative and discriminative segmentation approaches exploit spatial regularity, often with extensions along the temporal dimension for longitudinal tasks [52]–[54]. Local regularity of tissue labels can be encoded via boundary modeling for both generative [17], [55] and discriminative models [32], [33], [35], [55], [56], potentially enforcing non-local shape constraints [57]. Markov random field (MRF) priors encourage similarity among neighboring labels in the generative context [25], [37], [38]. Similarly, conditional random fields (CRFs) help enforce—or prohibit—the adjacency of specific labels and, hence, impose constraints considering the wider spatial context of voxels [36], [43]. While all these segmentation models act locally, more or less at the voxel level, other approaches consider prior knowledge about the relative location of tumor structures in a more global fashion. They learn, for example, the neighborhood relationships between such structures as edema, Gadolinium-enhancing tumor structures, or necrotic parts of the tumor through hierarchical models of super-voxel clusters [42], [58], or by relating image patterns with phenomenological tumor growth models adapted to patient scans [31].
While each of the discussed algorithms was compared empirically against an expert segmentation by its authors, it is difficult to draw conclusions about the relative performance of different methods. This is because datasets and pre-processing steps differ between studies, the image modalities considered, the annotated tumor structures, and the used evaluation scores all vary widely as well (Table I).
TABLE I.
Data Sets, MR Image Modalities, Evaluation Scores, and Even Tumor Types Used for Self-Reported Performances in the Brain Tumor Image Segmentation Literature Differ Widely. Shown Is a Selection of Algorithms Discussed Here and in [7]. Tumor Type Is Defined as G—Glioma (Unspecified), HG—High-Grade Glioma, LG—Low-Grade Glioma, M—Meningioma; “na” Indicates That No Information Is Reported. When Available the Number of Training and Testing Datasets Is Reported, Along With the Testing Mechanism: tt—Separate Training and Testing Datasets, cv—Cross-Validation
| Algorithm | MRI modalities |
Approach | Perform. score |
Tumor type |
trainining/testing (tt/cv) |
|---|---|---|---|---|---|
| Fletcher 2001 | T1 T2 PD | Fuzzy clustering w/ image retrieval |
Match (53–91%) |
na | 2/4 tt |
| Kaus 2001 | T1 | Template-moderated classification |
Accuracy (95%) |
LG, M | 10/10 tt |
| Ho 2002 | T1 T1c | Level-sets w/ region competition |
Jaccard (85–93%) |
G, M | na/5 tt |
| Prastawa 2004 | T2 | Generative model w/ outlier detection |
Jaccard (59–89%) |
G, M | na/3 tt |
| Corso 2008 | T1 T1c T2 FLAIR |
Weighted aggregation | Jaccard (62–69%) |
HG | 10/10 tt |
| Verma 2008 | T1 T1c T2 FLAIR DTI |
SVM | Accuracy (34–93%) |
HG | 14/14 cv |
| Wels 2008 | T1 T1c T2 | Discriminative model w/ CRF |
Jaccard (78%) |
G | 6/6 cv |
| Cobzas 2009 | T1c FLAIR | Level-set w/ CRF | Jaccard (50–75%) |
G | 6/6 tt |
| Wang 2009 | T1 | Fluid vector flow | Tanimoto (60%) |
na | 0/10 tt |
| Menze 2010 | T1 T1c T2 FLAIR |
Generative model w/ lesion class |
Dice (40–70%) |
G | 25/25 cv |
| Bauer 2011 | T1 T1c T2 FLAIR |
Hierarchical SVM w/ CRF |
Dice (77–84%) |
G | 10/10 cv |
Image Processing Benchmarks
Benchmarks that compare how well different learning algorithms perform in specific tasks have gained a prominent role in the machine learning community. In recent years, the idea of benchmarking has also gained popularity in the field of medical image analysis. Such benchmarks, sometimes referred to as “challenges,” all share the common characteristic that different groups optimize their own methods on a training dataset provided by the organizers, and then apply them in a structured way to a common, independent test dataset. This situation is different from many published comparisons, where one group applies different techniques to a dataset of their choice, which hampers a fair assessment as this group may not be equally knowledgeable about each method and invest more effort in optimizing some algorithms than others (see [59]).
Once benchmarks have been established, their test dataset often becomes a new standard in the field on how to evaluate future progress in the specific image processing task being tested. The annotation and evaluation protocols also may remain the same even when new data are added (to overcome the risk of over-fitting this one particular dataset that may take place after a while), or when related benchmarks are initiated. A key component in benchmarking is an online tool for automatically evaluating segmentations submitted by individual groups [60], as this allows the labels of the test set never to be made public. This helps ensure that any reported results are not influenced by unintentional overtraining of the method being tested, and that they are therefore truly representative of the method's segmentation performance in practice.
Recent examples of community benchmarks dealing with medical image segmentation and annotation include algorithms for artery centerline extraction [61], [62], vessel segmentation and stenosis grading [63], liver segmentation [64], [65], detection of microaneurysms in digital color fundus photographs [66], and extraction of airways from CT scans [67]. Rather few community-wide efforts have focused on segmentation algorithms applied to images of the brain (a current example deals with brain extraction (“masking”) [68]), although many of the validation frameworks that are used to compare different segmenters and segmentation algorithms, such as STAPLE [69], [70], have been developed for applications in brain imaging, or even brain tumor segmentation [71].
III. Set-Up of the Brats Benchmark
The BRATS benchmark was organized as two satellite challenge workshops in conjunction with the MICCAI 2012 and 2013 conferences. Here we describe the set-up of both challenges with the participating teams, the imaging data and the manual annotation process, as well as the validation procedures and online tools for comparing the different algorithms. The BRATS online tools continue to accept new submissions, allowing new groups to download the training and test data and submit their segmentations for automatic ranking with respect to all previous submissions.1 A common entry page to both benchmarks, as well as to the latest BRATS-related initiatives is www.braintumorsegmentation.org.2
A. The MICCAI 2012 and 2013 Benchmark Challenges
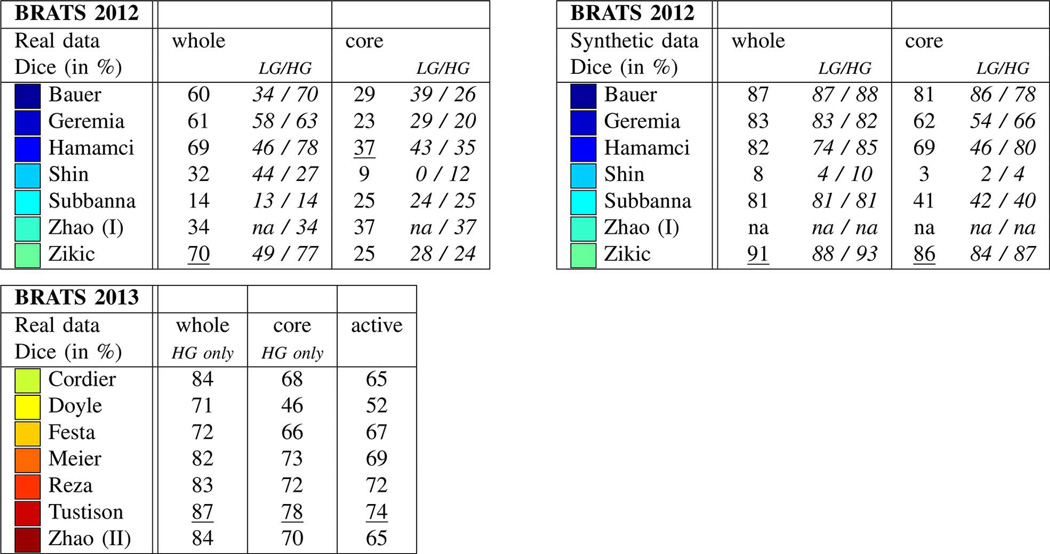
The first benchmark was organized on October 1, 2012 in Nice, France, in a workshop held as part of the MICCAI 2012 conference. During Spring 2012, participants were solicited through private e-mails as well as public e-mail lists and the MICCAI workshop announcements. Participants had to register with one of the online systems (cf. Section III-F) and could download annotated training data. They were asked to submit a four page summary of their algorithm, also reporting a cross-validated training error. Submissions were reviewed by the organizers and a final group of twelve participants were invited to contribute to the challenge. The training data the participants obtained in order to tune their algorithms consisted of multi-contrast MR scans of 10 low- and 20 high-grade glioma patients that had been manually annotated with two tumor labels (“edema” and “core,” cf. Section III-D) by a trained human expert. The training data also contained simulated images for 25 high-grade and 25 low-grade glioma subjects with the same two “ground truth” labels. In a subsequent “on-site challenge” at the MICCAI workshop, the teams were given a 12 h time period to evaluate previously unseen test images. The test images consisted of 11 high- and 4 low-grade real cases, as well as 10 high- and 5 low-grade simulated images. The resulting segmentations were then uploaded by each team to the online tools, which automatically computed performance scores for the two tumor structures. Of the twelve groups that participated in the benchmark, six submitted their results in time during the on-site challenge, and one group submitted their results shortly afterwards (Subbanna). During the plenary discussions it became apparent that using only two basic tumor classes was insufficient as the “core” label contained substructures with very different appearances in the different modalities. We therefore had all the training data re-annotated with four tumor labels, refining the initially rather broad “core” class by labels for necrotic, cystic and enhancing substructures. We asked all twelve workshop participants to update their algorithms to consider these new labels and to submit their segmentation results—on the same test data—to our evaluation platform in an “off-site” evaluation about six months after the event in Nice, and ten of them submitted updated results (Table II).
TABLE II.
Overview of the Algorithms Employed in 2012 and 2013. For a Full Description Please Refer to the Appendix and the Workshop Proceedings Available Online (see Section III-A). Three Non-Automatic Algorithms Required a Manual Initialization
| Method | Description | Fully automated |
|
|---|---|---|---|
| Integrated hierarchical random forest classification and CRF regularization | Yes | 2012 | |
| Spatial decision forests with intrinsic hierarchy [42] | Yes | ||
| “Tumorcut” method [72] | No | ||
| Generative lesion segmentation model [73] | Yes | ||
| Generative-discriminative model building on top of “Menze (G)” | Yes | ||
| Generative model with latent atlases and level sets | No | ||
| Hybrid clustering and classification by logistic regression | Yes | ||
| Hierarchical MRF approach with Gabor features | Yes | ||
| Learned MRF on supervoxels clusters | Yes | ||
| Context-sensitive features with a decision tree ensemble | Yes | ||
| Bit-grouping artificial immune network | Yes | 2013 | |
| Patch-based tissue segmentation approach | Yes | ||
| Hidden Markov fields and variational EM in a generative model | Yes | ||
| Random forest classifier using neighborhood and local context features | Yes | ||
| Semi-automatic segmentation using active contours | No | ||
| Appearance- and context-sensitive features with a random forest and CRF | Yes | ||
| Texture features and random forests | Yes | ||
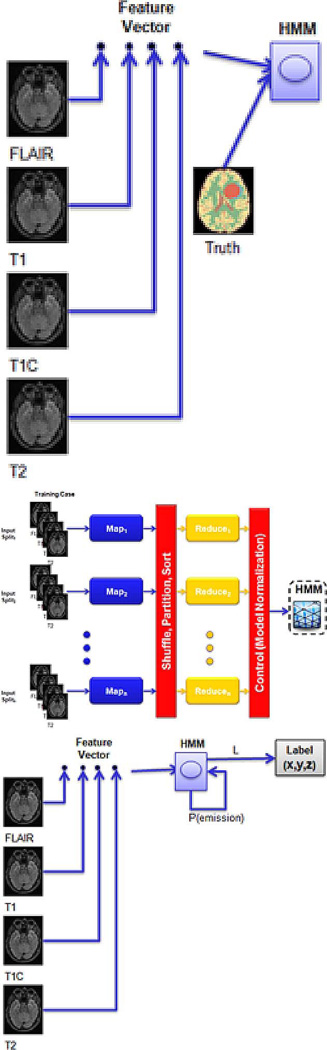
| “Map-Reduce Enabled” hidden Markov models | Yes | ||
| Random forest classifier using the open source ANTs/ANTsR packages | Yes | ||
| Like “Zhao (I)” with updated unary potential | Yes |
The second benchmark was organized on September 22, 2013 in Nagoya, Japan in conjunction with MICCAI 2013. Participants had to register with the online systems and were asked to describe their algorithm and report training scores during the summer, resulting in ten teams submitting short papers, all of which were invited to participate. The training data for the benchmark was identical to the real training data of the 2012 benchmark. No synthetic cases were evaluated in 2013, and therefore no synthetic training data was provided. The participating groups were asked to also submit results for the 2012 test dataset (with the updated labels) as well as to 10 new test datasets to the online system about four weeks before the event in Nagoya as part of an “off-site” leaderboard evaluation. The “on-site challenge” at the MICCAI 2013 workshop proceeded in a similar fashion to the 2012 edition: the participating teams were provided with 10 high-grade cases, which were previously unseen test images not included in the 2012 challenge, and were given a 12 h time period to upload their results for evaluation. Out of the ten groups participating in 2013 (Table II), seven groups submitted their results during the on-site challenge; the remaining three submitted their results shortly afterwards (Buendia, Guo, Taylor).
Altogether, we report three different test results from the two events: one summarizing the on-site 2012 evaluation with two tumor labels for a test set with 15 real cases (11 high-grade, four low-grade) and 15 synthetically generated images (10 high-grade, five low-grade); one summarizing the on-site 2013 evaluation with four tumor labels on a fresh set of 10 new real cases (all high-grade); and one from the off-site tests which ranks all 20 participating groups from both years, based on the 2012 real test data with the updated four labels. Our emphasis is on the last of the three tests.
B. Tumor Segmentation Algorithms Tested
Table II contains an overview of the methods used by the participating groups in both challenges. In 2012, four out of the twelve participants used generative models, one was a generative-discriminative approach, and five were discriminative; seven used some spatially regularizing model component. Two methods required manual initialization. The two automated segmentation methods that topped the list of competitors during the on-site challenge of the first benchmark used a discriminative probabilistic approach relying on a random forest classifier, boosting the popularity of this approach in the second year. As a result, in 2013 participants employed one generative model, one discriminative-generative model, and eight discriminative models out of which a total of four used random forests as the central learning algorithm; seven had a processing step that enforced spatial regularization. One method required manual initialization. A detailed description of each method is available in the workshop proceedings,3 as well as in the Appendix/Online Supporting Information.
C. Image Datasets
Clinical Image Data
The clinical image data consists of 65 multi-contrast MR scans from glioma patients, out of which 14 have been acquired from low-grade (histological diagnosis: astrocytomas or oligoastrocytomas) and 51 from high-grade (anaplastic astrocytomas and glioblastoma multiforme tumors) glioma patients. The images represent a mix of pre- and post-therapy brain scans, with two volumes showing resections. They were acquired at four different centers—Bern University, Debrecen University, Heidelberg University, and Massachusetts General Hospital—over the course of several years, using MR scanners from different vendors and with different field strengths (1.5T and 3T) and implementations of the imaging sequences (e.g., 2D or 3D). The image datasets used in the study all share the following four MRI contrasts (Fig. 2).
T1: T1-weighted, native image, sagittal or axial 2D acquisitions, with 1–6 mm slice thickness.
T1c: T1-weighted, contrast-enhanced (Gadolinium) image, with 3D acquisition and 1 mm isotropic voxel size for most patients.
T2: T2-weighted image, axial 2D acquisition, with 2–6 mm slice thickness.
FLAIR: T2-weighted FLAIR image, axial, coronal, or sagittal 2D acquisitions, 2–6 mm slice thickness.
To homogenize these data we co-registered each subject's image volumes rigidly to the T1c MRI, which had the highest spatial resolution in most cases, and resampled all images to 1 mm isotropic resolution in a standardized axial orientation with a linear interpolator. We used a rigid registration model with the mutual information similarity metric as it is implemented in ITK [74] (“VersorRigid3DTransform” with “MattesMutualInformation” similarity metric and three multi-resolution levels). No attempt was made to put the individual patients in a common reference space. All images were skull stripped [75] to guarantee anomymization of the patients.
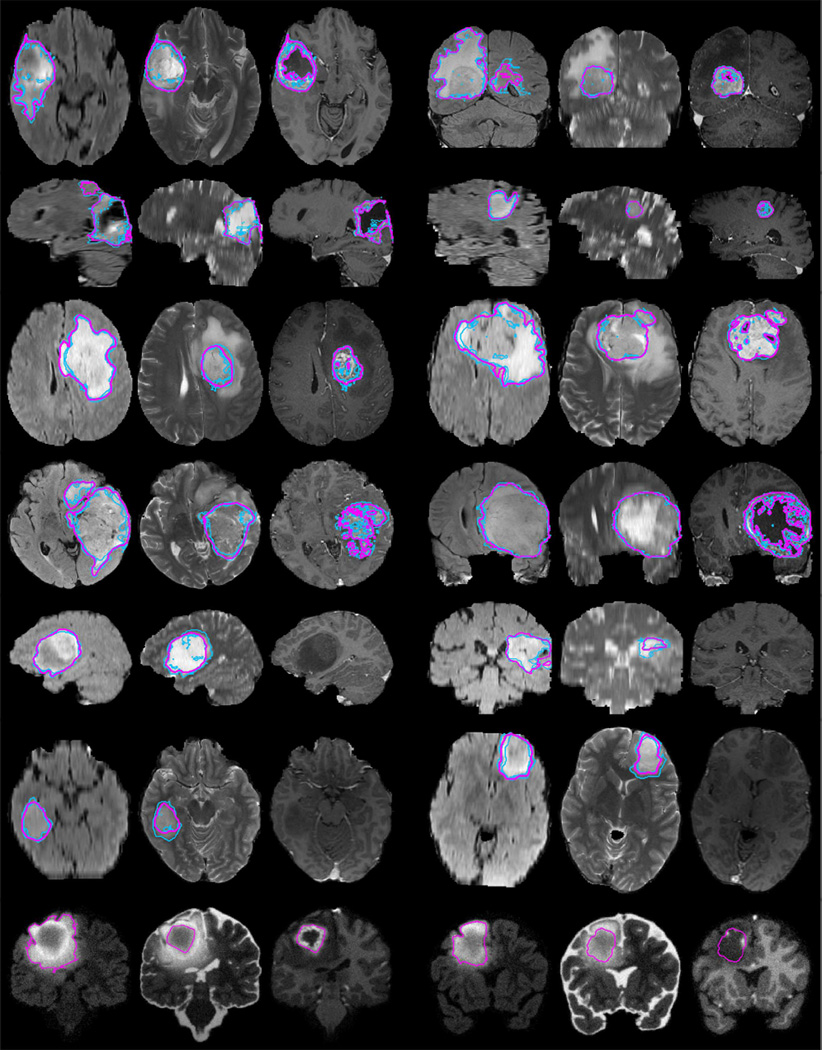
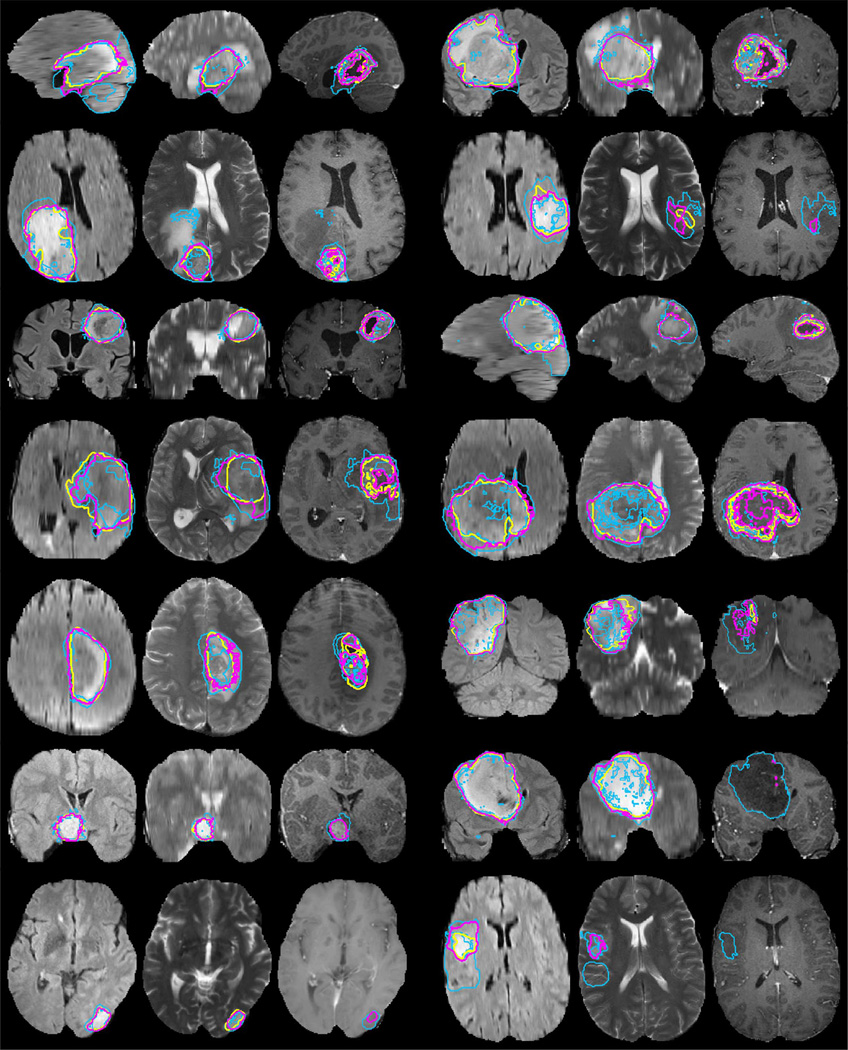
Fig. 2.
Examples from the BRATS training data, with tumor regions as inferred from the annotations of individual experts (blue lines) and consensus segmentation (magenta lines). Each row shows two cases of high-grade tumor (rows 1–4), low-grade tumor (rows 5–6), or synthetic cases (last row). Images vary between axial, sagittal, and transversal views, showing for each case: FLAIR with outlines of the whole tumor region (left); T2 with outlines of the core region (center); T1c with outlines of the active tumor region if present (right). Best viewed when zooming into the electronic version of the manuscript.
Synthetic Image Data
The synthetic data of the BRATS 2012 challenge consisted of simulated images for 35 high-grade and 30 low-grade gliomas that exhibit comparable tissue contrast properties and segmentation challenges as the clinical dataset (Fig. 2, last row). The same image modalities as for the real data were simulated, with similar 1 mm3 resolution. The images were generated using the TumorSim software,4 a cross-platform simulation tool that combines physical and statistical models to generate synthetic ground truth and synthesized MR images with tumor and edema [76]. It models infiltrating edema adjacent to tumors, local distortion of healthy tissue, and central contrast enhancement using the tumor growth model of Clatz et al. [77], combined with a routine for synthesizing texture similar to that of real MR images. We parameterized the algorithm according to the parameters proposed in [76], and applied it to anatomical maps of healthy subjects from the BrainWeb simulator [78], [79]. We synthesized image volumes and degraded them with different noise levels and intensity inhomogeneities, using Gaussian noise and polynomial bias fields with random coefficients.
D. Expert Annotation of Tumor Structures
While the simulated images came with “ground truth” information about the localization of the different tumor structures, the clinical images required manual annotations. We defined four types of intra-tumoral structures, namely “edema,” “non-enhancing (solid) core,” “necrotic (or fluid-filled) core,” and “non-enhancing core.” These tumor substructures meet specific radiological criteria and serve as identifiers for similarly-looking regions to be recognized through algorithms processing image information rather than offering a biological interpretation of the annotated image patterns. For example, “non-enhancing core” labels may also comprise normal enhancing vessel structures that are close to the tumor core, and “edema” may result from cytotoxic or vasogenic processes of the tumor, or from previous therapeutical interventions.
Tumor Structures and Annotation Protocol
We used the following protocol for annotating the different visual structures, where present, for both low- and high-grade cases (illustrated in Fig. 3).
The “edema” was segmented primarily from T2 images. FLAIR was used to cross-check the extension of the edema and discriminate it against ventricles and other fluid-filled structures. The initial “edema” segmentation in T2 and FLAIR contained the core structures that were then relabeled in subsequent steps [Fig. 3(A)].
As an aid to the segmentation of the other three tumor substructures, the so-called gross tumor core—including both enhancing and non-enhancing structures—was first segmented by evaluating hyper-intensities in T1c (for high-grade cases) together with the inhomogenous component of the hyper-intense lesion visible in T1 and the hypo-intense regions visible in T1 [Fig. 3(B)].
The “enhancing core” of the tumor was subsequently segmented by thresholding T1c intensities within the resulting gross tumor core, including the Gadolinium enhancing tumor rim and excluding the necrotic center and vessels. The appropriate intensity threshold was determined visually on a case-by-case basis [Fig. 3(C)].
The “necrotic (or fluid-filled) core” was defined as the tortuous, low intensity necrotic structures within the enhancing rim visible in T1c. The same label was also used for the very rare instances of hemorrhages in the BRATS data [Fig. 3(C)].
Finally, the “non-enhancing (solid) core” structures were defined as the remaining part of the gross tumor core, i.e., after subtraction of the “enhancing core” and the “necrotic (or fluid-filled) core” structures [Fig. 3(D)].
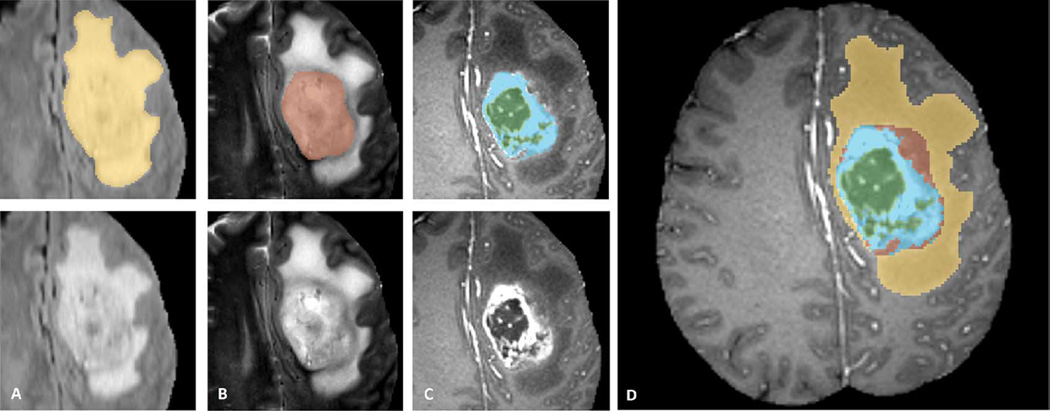
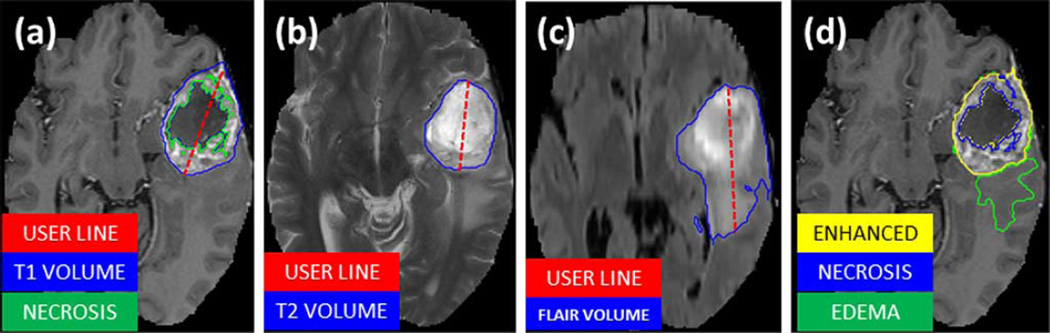
Fig. 3.
Manual annotation through expert raters. Shown are image patches with the tumor structures that are annotated in the different modalities (top left) and the final labels for the whole dataset (right). Image patches show from left to right: the whole tumor visible in FLAIR (A), the tumor core visible in T2 (B), the enhancing tumor structures visible in T1c (blue), surrounding the cystic/necrotic components of the core (green) (C). Segmentations are combined to generate the final labels of the tumor structures (D): edema (yellow), non-enhancing solid core (red), necrotic/cystic core (green), enhancing core(blue).
Following this protocol, the MRI scans were annotated by a trained team of radiologists and altogether seven radiographers in Bern, Debrecen and Boston. They outlined structures in every third axial slice, interpolated the segmentation using morphological operators (region growing), and visually inspected the results in order to perform further manual corrections, if necessary. All segmentations were performed using the 3D slicer software5, taking about 60 min per subject. As mentioned previously, the tumor labels used initially in the BRATS 2012 challenge contained only two classes for both high- and low-grade glioma cases: “edema,” which was defined similarly as the edema class above, and “core” representing the three core classes. The simulated data used in the 2012 challenge also had ground truth labels only for “edema” and “core.”
Algorithm 1. The hierarchical majority vote.
The number of raters/algorithms that assigned a given voxel to one of the four tumor structures is indicated by nedm, nnen, nnec, nenh; nall is the total number of raters/algorithms.
| label ← “nrm” | ▷ normal tissue |
| if (nedm + nnen + nnec + nenh) ≥ nall/2 then | |
| label ← “edm” | ▷ edema |
| if (nnen + nnec + nenh) ≥ nall/2 then | |
| label ← “nen” | ▷ non-enhancing core |
| if (nnec + nenh) ≥ nall/2 then | |
| label ← “nec” | ▷ necrotic core |
| if nenh ≥ nall/2 then | |
| label ← “enh” | ▷ enhancing core |
| end if | |
| end if | |
| end if | |
| end if |
Consensus Labels
In order to deal with ambiguities in individual tumor structure definitions, especially in infiltrative tumors for which clear boundaries are hard to define, we had all subjects annotated by several experts, and subsequently fused the results to obtain a single consensus segmentation for each subject. The 30 training cases were labeled by four different raters, and the test set from 2012 was annotated by three. The additional testing cases from 2013 were annotated by one rater. For the data sets with multiple annotations we fused the resulting label maps by assuming increasing “severity” of the disease from edema to non-enhancing (solid) core to necrotic (or fluid-filled) core to enhancing core, using a hierarchical majority voting scheme that assigns a voxel to the highest class to which at least half of the raters agree on (Algorithm 1). To illustrate this rule: a voxel that has been labeled as edema, edema, non-enhancing core, and necrotic core by the four annotators would be assigned to non-enhancing core structure as this is the most serious label that 50% of the experts agree on.
We chose this hierarchical majority vote to include prior knowledge about the structure and the ranking of the labels. A direct application of other multi-class fusion schemes that do not consider relations between the class labels, such as the STAPLE algorithm [69], lead to implausible fusion results where, for example, edema and normal voxels formed regions that were surrounded by “core” structures.
E. Evaluation Metrics and Ranking
Tumor Regions Used for Validation
The tumor structures represent the visual information of the images, and we provided the participants with the corresponding multi-class labels to train their algorithms. For evaluating the performance of the segmentation algorithms, however, we grouped the different structures into three mutually inclusive tumor regions that better represent the clinical application tasks, for example, in tumor volumetry. We obtain
the “whole” tumor region (including all four tumor structures),
the tumor “core” region (including all tumor structures except “edema”),
and the “active” tumor region (only containing the “enhancing core” structures that are unique to high-grade cases).
Examples of all three regions are shown in Fig. 2. By evaluating multiple binary segmentation tasks, we also avoid the problem of specifying misclassification costs for trading false assignments in between, for example, edema and necrotic core structures or enhancing core and normal tissue, which cannot easily be solved in a global manner.
Performance Scores
For each of the three tumor regions we obtained a binary map with algorithmic predictions P ∈ {0, 1} and the experts' consensus truth T ∈ {0, 1}, and we calculated the well-known Dice score
where Λ is the logical AND operator, |·| is the size of the set (i.e., the number of voxels belonging to it), and P1 and T1 represent the set of voxels where P = 1 and T = 1, respectively (Fig. 4). The Dice score normalizes the number of true positives to the average size of the two segmented areas. It is identical to the F score (the harmonic mean of the precision recall curve) and can be transformed monotonously to the Jaccard score.
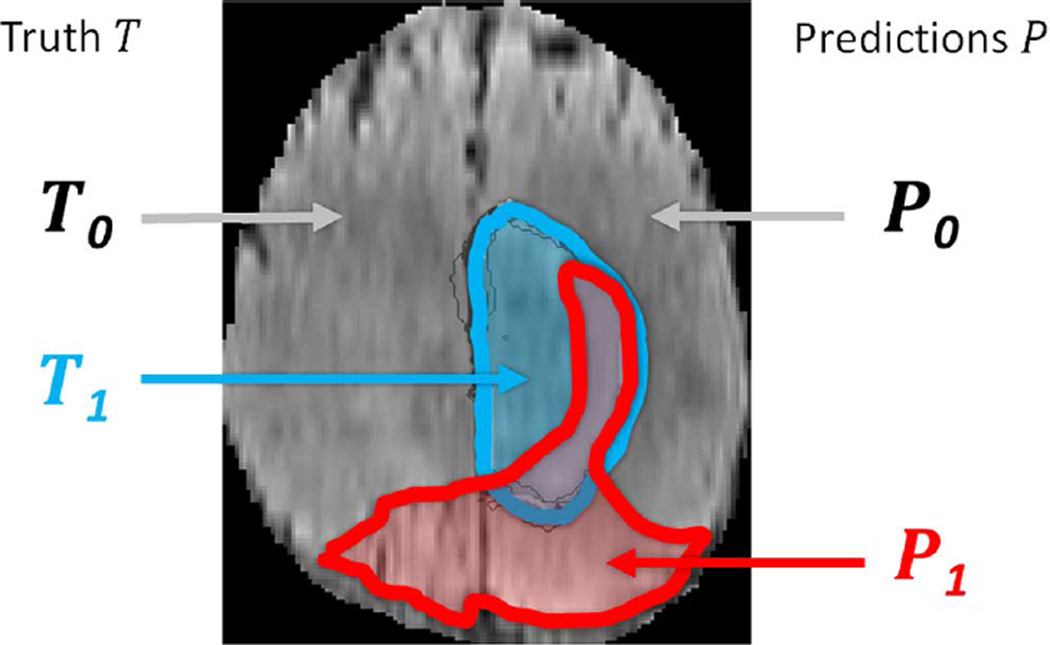
Fig. 4.
Regions used for calculating Dice score, sensitivity, specificity, and robust Hausdorff score. Region T1 is the true lesion area (outline blue), T0 is the remaining normal area. P1 is the area that is predicted to be lesion by—for example—an algorithm (outlined red), and P0 is predicted to be normal. P1 has some overlap with T1 in the right lateral part of the lesion, corresponding to the area referred to as P1 Λ T1 in the definition of the Dice score (Eq. III.E).
We also calculated the so-called sensitivity (true positive rate) and specificity (true negative rate)
where P0 and T0 represent voxels where P = 0 and T = 0, respectively.
Dice score, sensitivity, and specificity are measures of voxel-wise overlap of the segmented regions. A different class of scores evaluates the distance between segmentation boundaries, i.e., the surface distance. A prominent example is the Hausdorff distance calculating for all points p on the surface ∂P1 of a given volume P1 the shortest least-squares distance d(p, t) to points t on the surface ∂T1 of the other given volume T1, and vice versa, finally returning the maximum value over all d
Returning the maximum over all surface distances, however, makes the Hausdorff measure very susceptible to small outlying subregions in either P1 or T1. In our evaluation of the “active tumor” region, for example, both P1 or T1 may consist of multiple small areas or nonconvex structures with high surface-to-area ratio. In the evaluation of the “whole tumor,” predictions with few false positive regions—that do not substantially affect the overall quality of the segmentation as they could be removed with an appropriate postprocessing—might also have a drastic impact on the overall Hausdorff score. To this end we used a robust version of the Hausdorff measure—reporting not the maximal surface distance between P1 and T1, but the 95% quantile of it.
Significance Tests
In order to compare the performance of different methods across a set of images, we performed two types of significance tests on the distribution of their Dice scores. For the first test we identified the algorithm that performed best in terms of average Dice score for a given task, i.e., for the whole tumor region, tumor core region, or active tumor region. We then compared the distribution of the Dice scores of this “best” algorithm with the corresponding distributions of all other algorithms. In particular, we used a nonparametric Cox-Wilcoxon test, testing for significant differences at a 5% significance level, and recorded which of the alternative methods could not be distinguished from the “best” method this way.
In the same way we also compared the distribution of the inter-rater Dice scores, obtained by pooling the Dice scores across each pair of human raters and across subjects—with each subject contributing six scores if there are four raters, and three scores if there are three raters—to the distribution of the Dice scores calculated for each algorithm in a comparison with the consensus segmentation. We then recorded whenever the distribution of an algorithm could not be distinguished from the inter-rater distribution this way. We note that our inter-rater score somewhat overestimates variability as it is calculated from two manual annotations that may both be very eccentric. In the same way a comparison between a rater and the consensus label may somewhat underestimates variability, as the same manual annotations had contributed to the consensus label it now is compared against.
F. Online Evaluation Platforms
A central element of the BRATS benchmark is its online evaluation tool. We used two different platforms: the Virtual Skeleton Database (VSD), hosted at the University of Bern, and the Multimedia Digital Archiving System (MIDAS), hosted at Kitware [80]. On both systems participants can download annotated training and “blinded” test data, and upload their segmentations for the test cases. Each system automatically evaluates the performance of the uploaded label maps, and makes detailed—case by case—results available to the participant. Average scores for the different subgroups are also reported online, as well as a ranked comparison with previous results submitted for the same test sets.
The VSD6 provides an online repository system tailored to the needs of the medical research community. In addition to storing and exchanging medical image datasets, the VSD provides generic tools to process the most common image format types, includes a statistical shape modeling framework and an ontology-based searching capability. The hosted data is accessible to the community and collaborative research efforts. In addition, the VSD can be used to evaluate the submissions of competitors during and after a segmentation challenge. The BRATS data is publicly available at the VSD, allowing any team around the world to develop and test novel brain tumor segmentation algorithms. Ground truth segmentation files for the BRATS test data are hosted on the VSD but their download is protected through appropriate file permissions. The users upload their segmentation results through a web-interface, review the uploaded segmentation and then choose to start an automatic evaluation process. The VSD automatically identifies the ground truth corresponding to the uploaded segmentations. The evaluation of the different label overlap measures used to evaluate the quality of the segmentation (such as Dice scores) runs in the background and takes less than one minute per segmentation. Individual and overall results of the evaluation are automatically published on the VSD webpage and can be downloaded as a CSV file for further statistical analysis. Currently, the VSD has evaluated more than 10 000 segmentations and recorded over 100 registered BRATS users. We used it to host both the training and test data, and to perform the evaluations of the on-site challenges. Up-to-date ranking is available at the VSD for researchers to continuously monitor new developments and streamline improvements.
MIDAS7 is an open source toolkit that is designed to manage grand challenges. The toolkit contains a collection of server, client, and stand-alone tools for data archiving, analysis, and access. This system was used in parallel with VSD for hosting the BRATS training and test data in 2012, as well as managing submissions from participants and providing final scores using a collection of metrics. It has not been used any more for the 2013 BRATS challenge.
The software that generates the comparison metrics between ground truth and user submissions in both VSD and MIDAS is available as the open source COVALIC (Comparison and Validation of Image Computing) toolkit.8
IV. Results
In a first step we evaluate the variability between the segmentations of our experts in order to quantify the difficulty of the different segmentation tasks. Results of this evaluation also serve as a baseline we can use to compare our algorithms against in a second step. As combining several segmentations may potentially lead to consensus labels that are of higher quality than the individual segmentations, we perform an experiment that applies the hierarchical fusion algorithm to the automatic segmentations as a final step.
A. Inter-Rater Variability of Manual Segmentations
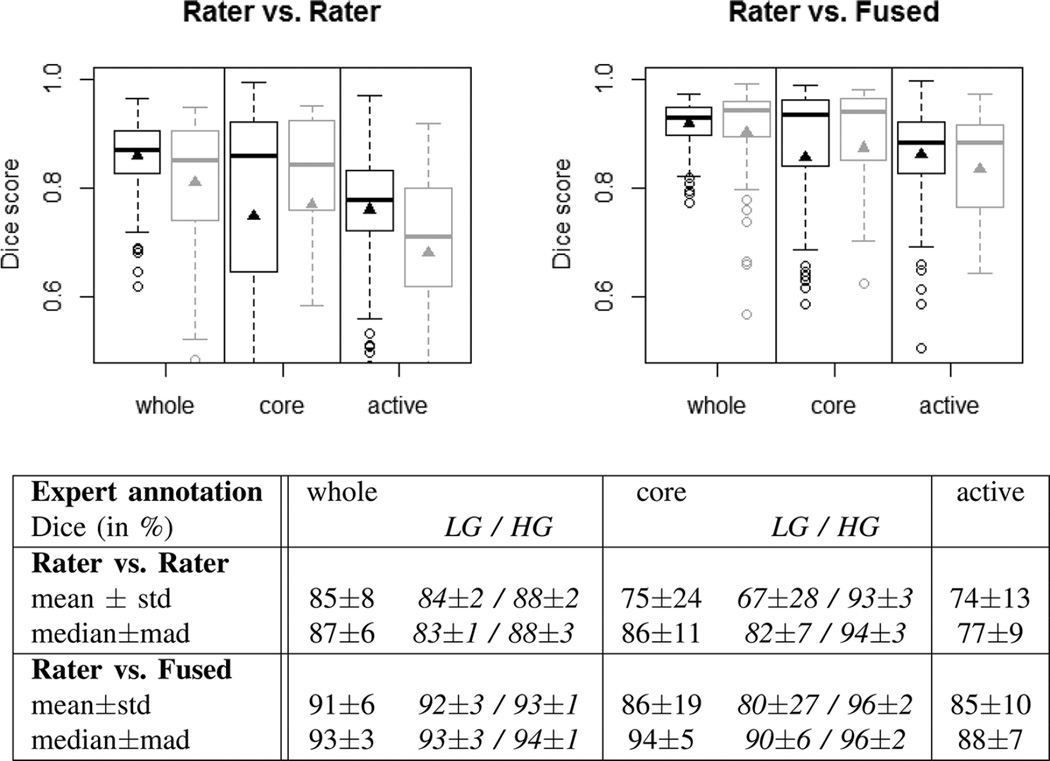
Fig. 5 analyzes the inter-rater variability in the four-label manual segmentations of the training scans (30 cases, four different raters), as well as of the final off-site test scans (15 cases, three raters). The results for the training and test datasets are overall very similar, although the inter-rater variability is a bit higher (lower Dice scores) in the test set, indicating that images in our training dataset were slightly easier to segment (Fig. 5, plots at the top). The scores obtained by comparing individual raters against the consensus segmentation provides an estimate of an upper limit for the performance of any algorithmic segmentation, indicating that segmenting the whole tumor region for both low- and high-grade and the tumor core region for high-grade is comparatively easy, while identifying the “core” in low-grade glioma and delineating the enhancing structures for high-grade cases is considerably more difficult (Fig. 5, table at the bottom). The comparison between an individual rater and the consensus segmentation, however, may be somewhat overly optimistic with respect to the upper limit of accuracy that can be obtained on the given datasets, as the consensus label is generated using the rater's segmentation it is compared against. So we use the inter-rater variation as an unbiased proxy that we compare with the algorithmic segmentations in the remainder. This sets the bar that has to be passed by an algorithm to Dice scores in the high 80% for the whole tumor region (median 87%), to scores in the high 80% for “core” region (median 94% for high-grade, median 82% for low-grade), and to average scores in the high 70% for “active” tumor region (median 77%) (Fig. 5, table at the bottom).
Fig. 5.
Dice scores of inter-rater variation (top left), and variation around the “fused” consensus label (top right). Shown are results for the “whole” tumor region (including all four tumor structures), the tumor “core” region (including enhancing, non-enhancing core, and necrotic structures), and the “active” tumor region (that features the T1c enhancing structures). Black boxplots show training data (30 cases); gray boxes show results for the test data (15 cases). Scores for “active” tumor region are calculated for high-grade cases only (15/11 cases). Boxes report quartiles including the median; whiskers and dots indicate outliers (some of which are below 0.5 Dice); and triangles report mean values. Table at the bottom shows quantitative values for the training and test datasets, including scores for low- and high-grade cases (LG/HG) separately; here “std” denotes standard deviation, and “mad” denotes median absolute deviance.
We note that on all datasets and in all three segmentation tasks the dispersion of the Dice score distributions is quite high, with standard deviations of 10% and more in particular for the most difficult tasks (tumor core in low-grade patients, active core in high-grade patients), underlining the relevance of comparing the distributions rather than comparing summary statistics such as the mean or the median and, for example, ranking measures thereof.
B. Performance of Individual Algorithms
On-Site Evaluation
Results from the on-site evaluations are reported in Fig. 6. Synthetic images were only evaluated in the 2012 challenge, and the winning algorithms on these images were developed by Bauer, Zikic, and Hamamci (Fig. 6, top right). The same methods also ranked top on the real data in the same year (Fig. 6, top left), performing particularly well for whole tumor and core segmentation. Here, Hamamci required some user interaction for an optimal initialization, while the methods by Bauer and Zikic were fully automatic. In the 2013 on-site challenge, the winning algorithms were those by Tustison, Meier, and Reza, with Tustison performing best in all three segmentation tasks (Fig. 6, bottom left).
Fig. 6.
On-site test results of the 2012 challenge (top left and right) and the 2013 challenge (bottom left), reporting average Dice scores. Test data for 2012 included both real and synthetic images, with a mix of low- and high-grade cases (LG/HG): 11/4 HG/LG cases for the real images and 10/5 HG/LG cases for the synthetic scans. All datasets from the 2012 on-site challenge featured “whole” and “core” region labels only. On-site test set for 2013 consisted of 10 real HG cases with four-class annotations, of which “whole,” “core,” “active” regions were evaluated (see text). Best results for each task are underlined. Top performing algorithms of the on-site challenge were Hamamci, Zikic, and Bauer in 2012; and Tustison, Meier, and Reza in 2013.
Overall, the performance scores from the on-site test in 2013 were higher than those in the previous off-site leader-board evaluation (compare Fig. 7, top with Fig. 6, bottom left). As the off-site test data contained the test cases from the previous year, one may argue that the images chosen for the 2013 on-site evaluation were somewhat easier to segment than the on-site test images in the previous—and one should be cautious about a direct comparison of on-site results from the two challenges.
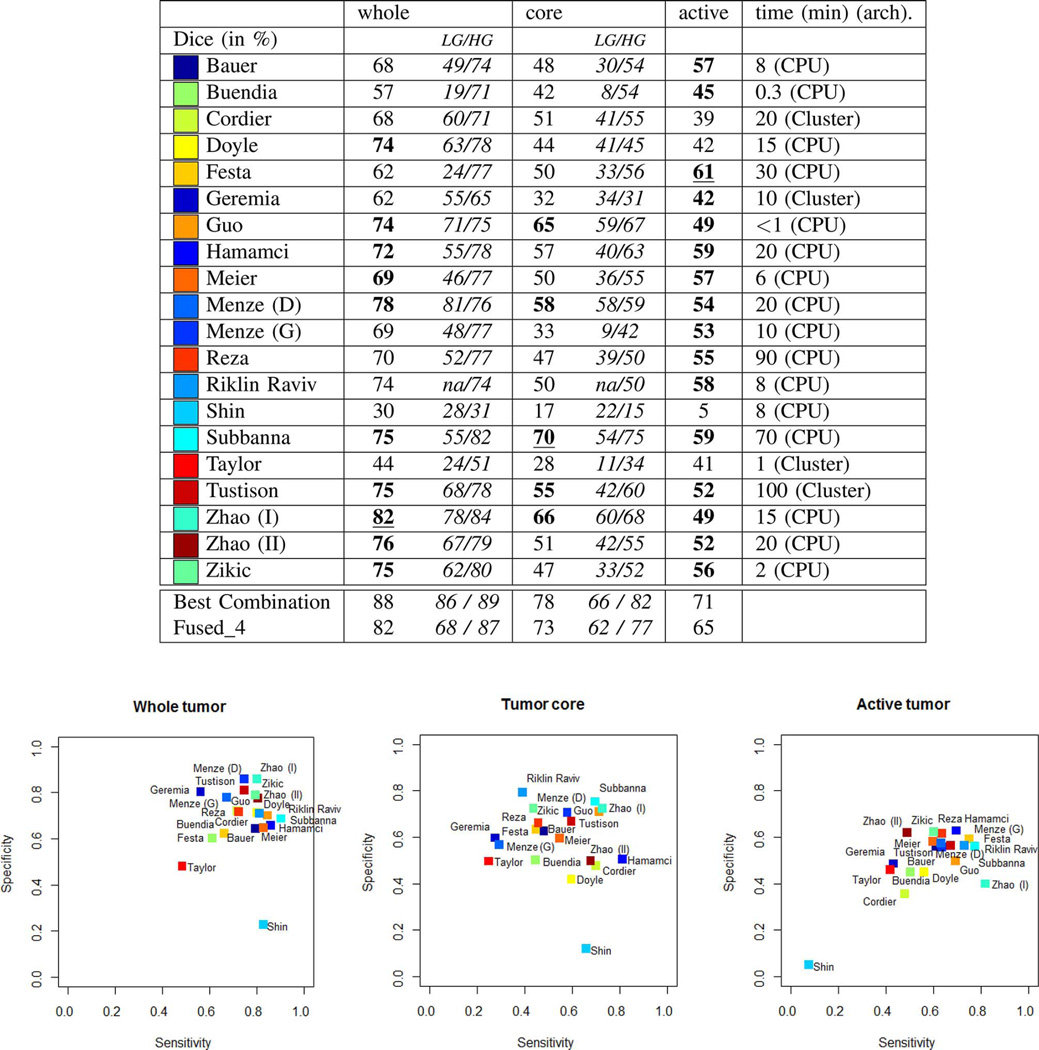
Fig. 7.
Average Dice scores from the “off-site” test, for all algorithms submitted during BRATS 2012 and 2013. The table at the top reports average Dice scores for “whole” lesion, tumor “core” region, and “active” core region, both for the low-grade (LG) and high-grade (HG) subsets combined and considered separately. Algorithms with the best average Dice score for the given task are underlined; those indicated in bold have a Dice score distribution on the test cases that is similar to the best (see also Fig. 8). “Best Combination” is the upper limit of the individual algorithmic segmentations (see text), “Fused_4” reports exemplary results when pooling results from Subbanna, Zhao (I), Menze (D), and Hamamci (see text). Reported average computation times per case are in minutes; an indication regarding CPU or Cluster based implementation is also provided. Plots at the bottom show the sensitivities and specificities of the corresponding algorithms. Colors encode the corresponding values of the different algorithms; written names have only approximate locations.
Off-Site Evaluation
Results on the off-site evaluation (Figs. 7 and 8) allow us to compare algorithms from both challenges, and also to consider results from algorithms that did not converge within the given time limit of the on-site evaluation (e.g., Menze, Geremia, Riklin Raviv). We performed significance tests on the Dice score to identify which algorithms performed best or similar to the best one for each segmentation task (Fig. 7). We also performed significance tests on the Dice scores to identify which algorithms had a performance that is similar to the inter-rater variation that are indicated by stars on top of the box plots in Fig. 8. For “whole” tumor segmentation, Zhao (I) was the best method, followed by Menze (D), which performed the best on low-grade cases; Zhao (I), Menze (D), Tustison, and Doyle report results with Dice scores that were similar to the inter-rater variation. For tumor “core” segmentation, Subbanna performed best, followed by Zhao (I) that was best on low-grade cases; only Subbanna has Dice scores similar to the inter-rater scores. For “active” core segmentation Festa performs best; with the spread of the Dice scores being rather high for the “active” tumor segmentation task, we find a high number of algorithms (Festa, Hamamci, Subbanna, Riklin Raviv, Menze (D), Tustison) to have Dice scores that do not differ significantly from those recorded for the inter-rater variation. Sensitivity and specificity varied considerably between methods (Fig. 7, bottom).
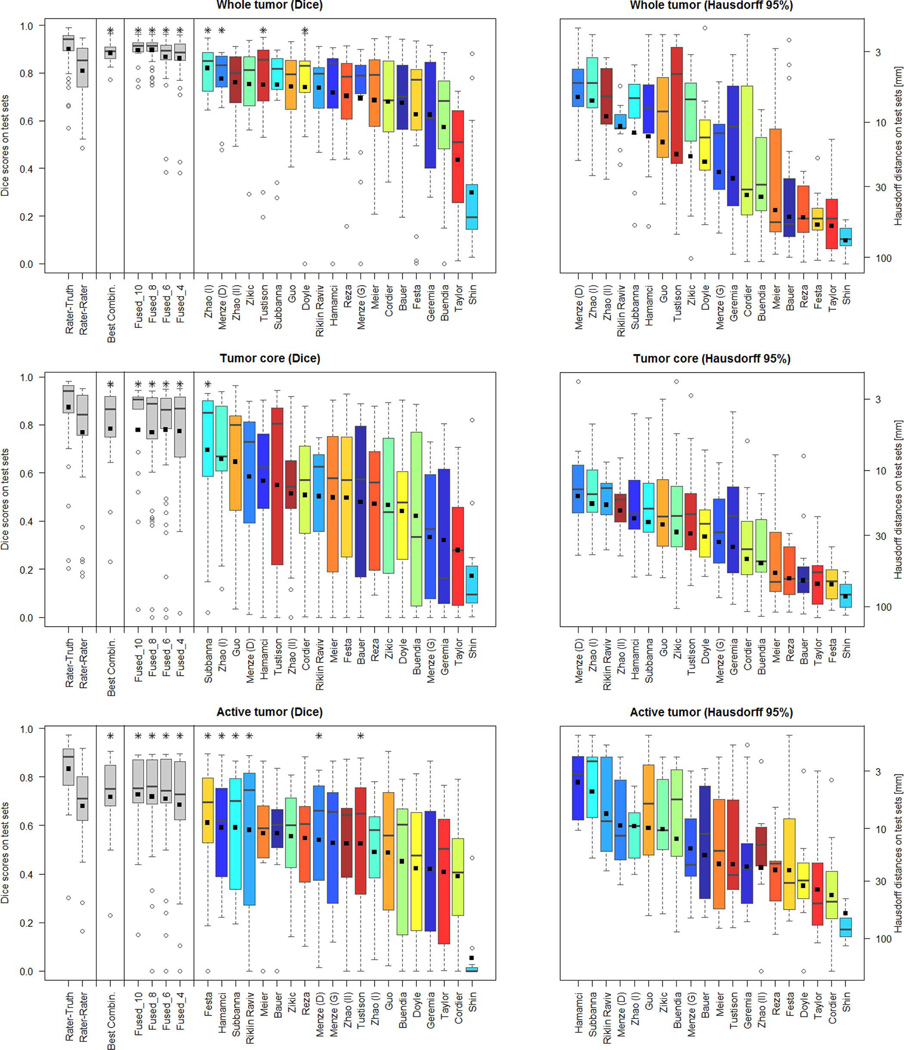
Fig. 8.
Dispersion of Dice and Hausdorff scores from the “off-site” test for the individual algorithms (color coded), and various fused algorithmic segmentations (gray), shown together with the expert results taken from Fig. 5 (also shown in gray). Boxplots show quartile ranges of the scores on the test datasets; whiskers and dots indicate outliers. Black squares indicate the mean score (for Dice also shown in the table of Fig. 7), which were used here to rank the methods. Also shown are results from four “Fused” algorithmic segmentations (see text for details), and the performance of the “Best Combination” as the upper limit of individual algorithmic performance. Methods with a star on top of the boxplot have Dice scores as high or higher than those from inter-rater variation. Hausdorff distances are reported on a logarithmic scale.
Using the Hausdorff distance metric we observe a ranking that is overall very similar (Fig. 7, boxes on the right), suggesting that the Dice scores indicate the general algorithmic performances sufficiently well. Inspecting segmentations of the one method that is an exception to this rule (Festa), we find it to segment the active region of the tumor very well for most volumes, but also to miss all voxels in the active region of three volumes (apparently removed from a very strong spatial regularization), with low Dice scores and Hausdorff distances of more than 50 mm. Averaged over all patients, this still leads to a very good Dice score, but the mean Hausdorff distance is unfavourably dominated by the three segmentations that failed.
C. Performance of Fused Algorithms
An Upper Limit of Algorithmic Performance
One can fuse algorithmic segmentations by identifying—for each test scan and each of the three segmentation tasks—the best segmentation generated by any of the given algorithms. This set of “optimal” segmentations (referred to as “Best Combination” in the remainder) has an average Dice score of about 90% for the “whole” tumor region, about 80% for the tumor “core” region, and about 70% for the “active” tumor region (Fig. 7, top), surpassing the scores obtained for inter-rater variation (Fig. 8). However, since fusing segmentations this way cannot be performed without actually knowing the ground truth, these values can only serve as a theoretical upper limit for the tumor segmentation algorithms being evaluated. The average Dice score of the algorithm performing best on the given task are about 10% below these numbers.
Hierarchical Majority Vote
In order to obtain a mechanism for fusing algorithmic segmentations in more practical settings, we first ranked the available algorithms according to their average Dice score across all cases and all three segmentation tasks, and then selected the best half. While this procedure guaranteed that we used meaningful segmentations for the subsequent pooling, we note that the resulting set included algorithms that performed well in one or two tasks, but performed clearly below average in the third one. Once the 10 best algorithms were identified this way, we sampled random subsets of 4, 6, and 8 of those algorithms, and fused them using the same hierarchical majority voting scheme as for combining expert annotations (Section III-D). We repeated this sampling and pooling procedure ten times. The results are shown in Fig. 8 (labeled “Fused_4,” “Fused_6,” and “Fused_8”), together with the pooled results for the full set of the 10 segmentations (named “Fused_10”). Exemplary segmentations for a Fused_4 sample are shown in Fig. 9—in this case, pooling the results from Subbanna, Zhao (I), Menze (D), and Hamamci. The corresponding Dice scores are reported in the table in Fig. 7.
Fig. 9.
Examples from the test data set, with consensus expert annotations (yellow) and consensus of four algorithmic labels overlaid (magenta). Blue lines indicate the individual segmentations of four different algorithms (Menze (D), Subbanna, Zhao (I), Hamamci). Each row shows two cases of high-grade tumor (rows 1–5) and low-grade tumor (rows 6–7). Three images are shown for each case: FLAIR (left), T2 (center), and T1c (right). Annotated are outlines of the whole tumor (shown in FLAIR), of the core region (shown in T2), and of active tumor region (shown in T1c, if applicable). Views vary between patients with axial, sagittal and transversal intersections with the tumor center. Note that clinical low-grade cases show image changes that have been interpreted by some of the experts as enhancements in T1c.
We found that results obtained by pooling four or more algorithms always outperformed those of the best individual algorithm for the given segmentation task. The hierarchical majority voting reduces the number of segmentations with poor Dice scores, leading to very robust predictions. It provides segmentations that are comparable to or better than the inter-rater Dice score, and it reaches the hypothetical limit of the “Best Combination” of case-wise algorithmic segmentations for all three tasks (Fig. 8).
V. Discussion
A. Overall Segmentation Performance
The synthetic data was segmented very well by most algorithms, reaching Dice scores on the synthetic data that were much higher than those for similar real cases (Fig. 6, top left), even surpassing the inter-rater accuracies. As the synthetic datasets have a high variability in tumor shape and location, but are less variable in intensity and less artifact-loaded than the real images, these results suggest that the algorithms used are capable of dealing well with variability in shape and location of the tumor segments, provided intensities can be calibrated in a reproducible fashion. As intensity-calibration of magnetic resonance images remains a challenging problem, a more explicit use of tumor shape information may still help to improve the performance, for example from simulated tumor shapes [81] or simulations that are adapted to the geometry of the given patients [31].
On the real data some of the automated methods reached performances similar to the inter-rater variation. The rather low scores for inter-rater variability (Dice scores in the range 74%–85%) indicate that the segmentation problem was difficult even for expert human raters. In general, most algorithms were capable of segmenting the “whole” region tumor quite well, with some algorithms reaching Dice scores of 80% and more (Zhao (I) has 82%). Segmenting the tumor “core” region worked surprisingly well for high-grade gliomas, and reasonably well for low-grade cases—considering the absence of enhancements in T1c that guide segmentations for high-grade tumors—with Dice scores in the high 60% (Subbanna has 70%). Segmenting small isolated areas of the “active” region in high-grade gliomas was the most difficult task, with the top algorithms reaching Dice scores in the high 50% (Festa has 61%). Hausdorff distances of the best algorithms are around 5–10 mm for the “whole” and the “active” tumor region, and about 20 mm for the tumor “core” region.
B. The Best Algorithm and Caveats
This benchmark cannot answer the question of what algorithm is overall “best” for glioma segmentation. We found that no single algorithm among the ones tested ranked in the top five for all three subtasks, although Hamamci, Subbanna, Menze (D), and Zhao (I) did so for two tasks (Fig. 8; considering Dice score). The results by Guo, Menze (D), Subbanna, Tustison, and Zhao (I) were comparable in all three tasks to those of the best method for respective task (indicated in bold in Fig. 7). Menze (D), Zhao (I), and Riklin Raviv led the ranking of the Hausdorff scores for two of the subtasks, and followed Hamamci and Subbanna for the third one.
Among the BRATS 2012 methods, we note that only Hamamci and Geremia performed comparably in the “off-site” and the “on-site” challenges, while the other algorithms performed significantly better in the “off-site” test than in the previous “on-site” evaluation. Several factors may have led to this discrepancy. Some of the groups had difficulties in submitting viable results during the “on-site” challenge and resolved them only for the “off-site” evaluation (Menze, Riklin Raviv). Others used algorithms during the “off-site” challenge that were significantly updated and reworked after the 2012 event (Subbanna, Shin). All 2012 participants had to adapt their algorithms to the new four-class labels and, if discriminative learning methods were used, to retrain their algorithms which also may have contributed to fluctuations in performance. Finally, we cannot rule out that some cross-checking between results of updated algorithms and available test images may have taken place in between the 2012 workshop and the 2013 “off-site” test.
There is another limitation regarding the direct comparison of “off-site” results between the 2012 and the 2013 workshop participants, as the test setting was inadvertently stricter for the latter group. In particular, the 2012 participants had several months to work with the test images and improve scores before the “off-site” evaluation took place—which, they were informed, would be used in a final ranking. In contrast, the 2013 groups were permitted access to those data only four weeks before their competition and were not aware that these images would be used for a broad comparison. It is therefore worth pointing out, once again, the algorithms that performed best on the on-site tests: these were the methods by Bauer, Zikic, and Hamamci in 2012, and Tustison's method in 2013.
C. “Winning” Algorithmic Properties
A majority of the top ranking algorithms relied on a discriminative learning approach, where low-level image features were generated in a first step, and a discriminative classifier was applied in a second step, transforming local features into class probabilities with MRF regularization to produce the final set of segmentations. Both Zikic and Menze (D) used the output of a generative model as input to a discriminative classifier in order to increase the robustness of intensity features. However, also other approaches that only used image intensities and standard normalization algorithms such as N4ITK [82] did surprisingly well. The spatial processing by Zhao (I), which considers information about tumor structure at a regional “super-voxel” level, did exceptionally well for “whole” tumor and tumor “core.” One may expect that performing such a non-local spatial regularization might also improve results of other methods. Most algorithms ranking in the lower half of the list used rather basic image features and did not employ a spatial regularization strategy, featuring small false positive outliers that decreased Dice score and increased the average Hausdorff distance.
Given the excellent results by the semi-automatic methods from Hamamci and Guo (and those by Riklin Raviv for the active tumor region), and because tumor segmentations will typically be looked at in the context of a clinical workflow anyway, it may be beneficial to take advantage of some user interaction, either in an initialization or in a postprocessing phase. In light of the clear benefit of fusing multiple automatic segmentations, demonstrated in Section IV-C, user interaction may also prove helpful in selecting the best segmentation maps for subsequent fusion.
The required computation time varied significantly among the participating algorithms, ranging from a few minutes to several hours. We observed that most of the computational burden related to feature detection and image registration sub-tasks. In addition, it was observed that a good understanding of the image resolution and amount of image subsampling can lead to a good trade-off between speed improvements and segmentation quality.
D. Fusing Automatic Segmentations
We note that fusing segmentations from different algorithms always performed better than the best individual algorithm applied to the same task. This observation aligns well with a common concept from ensemble learning, when a set of predictors that are unbiased but with high variability in the individual prediction, improve when their predictions are pooled [83]. In that case, averaging over multiple predictors reduces variance and, hence, reduces the prediction error. Subselecting only the best few segmentations, i.e., those with the least bias (or average misclassification) further improves results. In general there are two extrema: variance is maximal for single observations and minimal after fusing many, while bias is minimal for the one top-ranking algorithm and maximal when including a large number of (also lesser) predictions. For many applications, an optimum is reached in between these two extrema, depending on the bias and variance of the predictors that are fused. Optimizing the ensemble prediction by balancing variability reduction (fuse many predictors) and bias removal (fuse a few selected only) can be done on a test set representing the overall population, or for the individual image volume when partial annotation is available—for example from the limited user interaction mentioned above. Statistical methods that estimate and weight the performance of individual contributions—for example, based on appropriate multi-class extensions of STAPLE [69] and related probabilistic models [19], [84]—may also be used to trade bias and variance in an optimal fashion.
E. Limitations of the BRATS Benchmark
When designing the BRATS study, we made several choices that may have impacted the results and that could potentially have been improved. For example, we decided to homogenize the data by co-registering and reformatting each subject's image volumes using rigid registration and linear interpolation, as described in Section III-C. Although the registration itself was found to work well (as it was always between images acquired from the same subject and in the same acquisition session), it may have been advisable to use a more advanced interpolation method, because the image resolution differed significantly between sequences, patients, and centers. Furthermore, in order to build a consensus segmentation from multiple manual annotations, we devised a simple fusion rule that explicitly respects the known spatial and—with respect to the evolution of the disease— temporal relations between the tumor substructures, as more advanced fusion schemes were found to yield implausible results. These choices can certainly be criticized; however, we believe the major challenge for the segmentation algorithms was ultimately not interpolation or label fusion details, but rather the large spatial and structural variability of the tumors in the BRATS dataset, as well as the variability in image intensities arising from differences in imaging equipment and acquisition protocols.
Although we were able to identify several overall “winning” algorithmic properties (discussed in Section V-C), one general limitation of image analysis benchmarks is that it is often difficult to explain why a particular algorithm does well or—even more difficult—why it does not do well. This is because even the best algorithmic pipeline will fail if just one element is badly parameterized or implemented. Detecting such failures would require a meticulous study of each element of every processing pipeline—for a learning-based approach, for example, of the intensity normalization, the feature extraction, the classification algorithm, and the spatial regularization. Unfortunately, while this type of analysis is extremely valuable, it requires a careful experimental design that cannot easily be pursued post hoc on a heterogeneous set of algorithms contributed by different parties in a competitive benchmark such as BRATS.
Another limitation of the current study, which is also shared by other benchmarks, pertains to the selection of an appropriate overall evaluation metric that can be used to explicitly rank all competing algorithms. Although we reported separate results for sensitivity, specificity, and Hausdorff distance, we based our overall final ranking in different tumor regions on average Dice scores. As demonstrated by the results of the Festa method in “active tumor” segmentation, however, the exact choice of evaluation metric does sometimes affect the ranking results, as different metrics are sensitive to different types of segmentation errors.
Although the number of images included in the BRATS benchmark was large, the ranking of the segmentation algorithms reported here may still have been impacted by the high variability in brain tumors. As such, it will be desirable to further increase the number of training and test cases in future brain tumor segmentation benchmarks.
We wish to point out that all the individual segmentation results by all participants are publicly available,9 so that groups interested in brain tumor segmentation can perform their own internal evaluation, focusing specifically on what they consider most important. Looking at individual segmentations can also help understand better the advantages and drawbacks of the different algorithms under comparison, and we would strongly encourage taking advantage of this possibility. It is worth pointing out that the individual rater's manual segmentations of the training data are also available,10 so that groups that do not trust the consensus labels we provide, can generate their own training labels using a fusion method of their choice.
F. Lessons Learned
There are lessons that we learned from organizing BRATS 2012 and 2013 that may also be relevant for future benchmark organizers confronted with complex and expensive annotation tasks. First, it may be recommended to generate multiple annotations for the test data—rather than for the training set as we did here—as this is where the comparisons between experts and algorithms take place. Many algorithms will be able to overcome slight inconsistencies or errors in the training data that are present when only a single rater labels each case. At the same time, most algorithms will benefit from having larger training datasets and, hence, can be improved by annotating larger amounts of data even if this comes at the price of fewer annotations per image volume.
Second, while it may be useful to make unprocessed data available as well, we strongly recommend providing participants with maximally homogenized datasets—i.e., image volumes that are co-registered, interpolated to a standard resolution and normalized with respect to default intensity distributions— in order to ease participation, maximize the number of participants, and facilitate comparisons of the segmentation methods independently of preprocessing issues.
G. Future Work
Given that many of the algorithms that participated in this study offered good glioma segmentation quality, it would seem valuable to have their software implementations more easily accessible. Right now, only an implementation of Bauer and Meier's method is freely available,11 and Tustison's code12 The online MIDAS and VSD platforms that we used for BRATS may be extended to not only host and distribute data, but also to host and distribute such algorithms. Making the top algorithms available through appropriate infrastructures and interfaces—for example as developed for the VISCERAL benchmark13 [86], or as used in the commercial NITRC Amazon cloud service14—may help to make thoroughly benchmarked algorithms available to the wider clinical research community.
Since our results indicate that current automated glioma segmentation methods only reach the level of consensus-rater variation in the “whole” tumor case (Fig. 8), continued algorithmic development seems warranted. Other tumor substructures may also be relevant with respect to diagnosis and prognosis, and a more refined tumor model—with more than the four classes used in this study—may be helpful, in particular when additional image modalities are integrated into the evaluation. Finally, in clinical routine the change of tumor structures over time is often of primary relevance, something the current BRATS study did not address. Evaluating the accuracy of automated routines in longitudinal settings including both pre- and postoperative images, are important directions for future work along with further algorithmic developments.
VI. Summary and Conclusion
In this paper we presented the BRATS brain tumor segmentation benchmark. We generated the largest public dataset available for this task and evaluated a large number of state-of-the-art brain tumor segmentation methods. Our results indicate that, while brain tumor segmentation is difficult even for human raters, currently available algorithms can reach Dice scores of over 80% for whole tumor segmentation. Segmenting the tumor core region, and especially the active core region in high-grade gliomas, proved more challenging, with Dice scores reaching 70% and 60%, respectively. Of the algorithms tested, no single method performed best for all tumor regions considered. However, the errors of the best algorithms for each individual region fell within human inter-rater variability.
An important observation in this study is that fusing different segmenters boosts performance significantly. Decisions obtained by applying a hierarchical majority vote to fixed groups of algorithmic segmentations performed consistently, for every single segmentation task, better than the best individual segmentation algorithm. This suggests that, in addition to pushing the limits of individual tumor segmentation algorithms, future gains (and ultimately clinical implementations) may also be obtained by investigating how to implement and fuse several different algorithms, either by majority vote or by other fusion strategies.
Acknowledgments
This research was supported by the NIH NCRR (P41-RR14075), the NIH NIBIB (R01EB013565), the Academy of Finland (133611), TEKES (ComBrain), the Lundbeck Foundation (R141-2013-13117), the Swiss Cancer League, the Swiss Institute for Computer Assisted Surgery (SICAS), the NIH NIBIB NAMIC (U54-EB005149), the NIH NCRR NAC (P41-RR13218), the NIH NIBIB NAC (P41-EB-015902), the NIH NCI (R15CA115464), the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images), the FCT and COMPETE (FCOM-01-0124-FEDER-022674), the MICAT Project (EU FP7 Marie Curie Grant No. PIRG-GA-2008-231052), the European Union Seventh Framework Programme under grant agreement no. 600841, the Swiss NSF project Computer Aided and Image Guided Medical Interventions (NCCR CO-ME), the Technische Universität München—Institute for Advanced Study (funded by the German Excellence Initiative and the European Union Seventh Framework Programme under Grant agreement 291763), the Marie Curie COFUND program of the European Union (Rudolf Mössbauer Tenure-Track Professorship to BHM).
Appendix
Here we reproduce a short summary of each algorithm used in BRATS 2012 and BRATS 2013, provided by its authors. A more detailed description of each method is available in the workshop proceedings.15
 Bauer, Wiest and Reyes (2012): Segmentation of Brain Tumor Images Based on Integrated Hierarchical Classification and Regularization
Bauer, Wiest and Reyes (2012): Segmentation of Brain Tumor Images Based on Integrated Hierarchical Classification and Regularization
Algorithm and Data
We are proposing a fully automatic method for brain tumor segmentation, which is based on classification with integrated hierarchical regularization [87]. It subcategorizes healthy tissues into CSF, WM, GM and pathologic tissues into necrotic, active, non-enhancing and edema compartment. The general idea is based on a previous approach presented in [43]. After pre-processing (denoising, bias-field correction, rescaling and histogram matching) [74], the segmentation task is modeled as an energy minimization problem in a conditional random field (CRF) formulation. The energy consists of the sum of the singleton potentials in the first term and the pairwise potentials in the second term of (1). The expression is minimized using [88] in a hierarchical way
| (1) |
The singleton potentials V(yi, xi) are computed according to (2), where ỹi is the label output from a classifier, xi is the feature vector and δ is the Kronecker-δ function
| (2) |
We use a decision forest as a classifier [89], which has the advantage of being able to handle multi-class problems and providing a probabilistic output [89]. The probabilistic output is used for the weighting factor p(ỹi|xi) in (2), in order to control the degree of spatial regularization. A 44-dimensional feature vector is used for the classifier, which combines the intensities in each modality with the first-order textures (mean, variance, skewness, kurtosis, energy, entropy) computed from local patches, statistics of intensity gradients in a local neighborhood and symmetry features across the mid-sagittal plane. The pairwise potentials W(yi, yj, xi, xj) account for the spatial regularization. In (3) ws(i, j) is a weighting function, which depends on the voxel spacing in each dimension. The term (1 − δ(yi, yj)) penalizes different labels of adjacent voxels, while the intensity term exp((−PCD(xi − xj))/(2·x̄) regulates the degree of smoothing based on the local intensity variation, where PCD is a pseudo-Chebyshev distance and x̄ is a generalized mean intensity. D(yi, yj) allows us to incorporate prior knowledge by penalizing different tissue adjacencies individually
| (3) |
Computation time for one dataset ranges from 4 to 12 min depending on the size of the images, most of the time is needed by the decision forest classifier.
Training and Testing
The classifier was trained using 5-fold cross-validation on the training dataset, with separate training for high- and low-grade as well as synthetic and patient data. The parameters of the algorithm were chosen empirically. We also compared the proposed approach to our previous method [43] which used SVMs as a classifier instead of decision forests and which had a less sophisticated regularization. With the new method, the computation time could be reduced by more than a factor of two and the accuracy was significantly improved. However, we still discovered difficulties with datasets that were very different from the training data, which hints at some problems of the supervised algorithm with generalization.
 Buendia, Taylor, Ryan and John (2013): A Grouping Artificial Immune Network for Segmentation of Tumor Images
Buendia, Taylor, Ryan and John (2013): A Grouping Artificial Immune Network for Segmentation of Tumor Images
Algorithm and Data
GAIN+ is an enhanced version of the original Grouping Artificial Immune Network that was developed for fully automated MRI brain segmentation. The model captures the main concepts by which the immune system recognizes pathogens and models the process in a numerical form. GAIN+ was adapted to support a variable number of input patterns for training and segmentation of tumors in MRI brain images. The model was demonstrated to operate with multi-spectral MR data with an increase in accuracy compared to the single spectrum case. The new input patterns include, in any combination, voxel intensities from 2D or 3D blocks or shapes of varying sizes customized to each MRI sequence (T1, T2, FLAIR, etc.), and also include feature and textural patterns such as mean and variance of selected block sizes, slice or radial distance, co-occurrence matrices, among others. Due to the representation of the voxel intensities as multi-bit values, it can be shown that not every bit carries the same entropy. That is, each bit does not contribute equally to the final interpretation of the data. The GAIN algorithm makes use of this fact to increase the speed of its operation. Bits are grouped into groups of size 2 bits. A new grouping approach was implemented based on the location of each bit within the input pattern, and the significance of the input features. Higher priority was given to higher order bits and overall to voxels at closer distance to the center voxel. This grouping approach runs in just a few seconds and the same grouping file can be used for all cases. Training takes an average of 1.3 min per input byte, thus, for example, an input pattern of 16 bytes takes an average of 21 min of training. Segmentation with post-processing of one case takes 20 s for the same input size. The preprocessing pipeline was designed to remove noise and inhomogeneities due to MR scanner bias fields, and match each spectrum's intensity histogram to the volumes used for training. Several post-processing options were added to the program, such as finding and extracting connected components, and performing dilation and erosion on those components.
Training and Testing
The original GAIN method was designed to train on a single case, and although GAIN+ has been adapted to train on multiple images, single case training performed best. We performed 20-fold cross validation on the real high-grade BRATS 2013 training set. GAIN+ performance was evaluated with the four BRATS 2013 labels: 1) Necrosis, 2) Edema, 3) Non-Enhancing tumor, and 4) Enhancing Tumor. In this case, GAIN+ was run with an input pattern of 16 bytes: 7 FLAIR + 7 TIC +1 T1 + 1 T2 voxels. The segmented images were uploaded to the BRATS 2013 Virtual Skeleton web site. The evaluation was done for three different tumor sub-compartments: 1) Region 1: Complete tumor (labels 1 + 2 + 3 + 4 for patient data), Dice: 0.73, 2) Region 2: Tumor core (labels 1 + 3 + 4 for patient data), Dice: 0.61, 3) Region 3: Enhancing tumor (label 4 for patient data), Dice: 0.64.
 Cordier, Menze, Delingette and Ayache (2013): Patch-Based Segmentation of Brain Tissues
Cordier, Menze, Delingette and Ayache (2013): Patch-Based Segmentation of Brain Tissues
Algorithm and Data
We describe a fully automated approach inspired by the human brain labelling method described in [90], and similar to multi-atlas label propagation methods [91]–[93]. A database of multi-channel local patches is first built from a set of training pathological cases. Then, given a test case, similar multi-channel patches are retrieved in the patch database, along with the corresponding labels. Finally, a classification map for the test case is inferred as a combination of the retrieved labels during a label fusion step [94]–[97].
To decrease the computation time, images are sub-sampled to 2-mm isotropic resolution [98]. A candidate tumor mask is defined by thresholding (50% percentile) a denoised [99] T2-weighted FLAIR image. Since the patch retrieval is driven by a sum-of-squared-differences (SSD), a global intensity alignment [98] is applied to the mean image intensity restricted to the candidate tumor mask. Training images are cropped along the Z-axis to focus on training patches surrounding the tumor.
Image features are the concatenation of 3 × 3 × 3 intensity patches extracted from four MR channels. Given a multi-channel patch query, the five nearest-neighbor patches are retrieved within each training patch database, each of which contributes to a weighted voting. We use exponential weights, based on patch similarity [90], [95], [97]; the decay parameter σ2 depends on the test case, and is set to the maximum of SSD between every voxel in the test case and every first-neighbor training patch. For each label, the weighted voting results in a probability-like map. Since the label regions are interlocked, label maps are hierarchically computed: first, complete tumor is distinguished from healthy tissues; then tumor core from edema; finally enhancing tumor from the rest of the core. At each step, weighted votes are rebalanced based on label frequencies, in order to penalize labels which would be more often picked if the patch retrieval were blind.
As post-processing, at most the two biggest connected components of the complete tumor are kept, the second one being kept only if its volume is greater than 20% of the volume of the first one. Classification maps are up-sampled to 1-mm isotropic resolution, and one iteration of Iterated Conditional Modes [100] smooths the result. On average, the segmentation total computation time is 20 min times the number of training cases.
Training and Testing
The most important parameters are manually set and consist of the patch size and the number of training cases. A range of values for the number of retrieved nearest-neighbor patches were tested, and the segmentation results were almost not affected. For the training data, the labelling is performed in a leave-one-out scheme, while for the test data, every relevant training case is used. Real cases are processed separately from simulated cases. For real low-grade test cases, the training dataset includes both high- and low-grade cases, while for real high-grade test cases, the training dataset only includes high-grade cases.
The algorithm shows a few shortcomings which would require the following steps to be refined.
The necrotic core is sometimes partially missed by the candidate tumor mask. Tumour detection could either be skipped at the expense of higher computational burden, or be more sensitive by using the T2-weighted image in addition to the FLAIR image.
For enhancing tumors, thin parts are often missed by the algorithm. This is visible on the probability maps and may be due to the sub-sampling step.
Tumor voxels can be misclassified as healthy tissues or edema, usually if the necrotic core is similar to the cerebrospinal fluid on the FLAIR channel. Enforcing the convexity of tumor connected components helps but the contours of the tumor compartments are not matched as closely. The regularization would be more relevant during the label fusion.
Shape criteria could help discard false positives in the occipital lobe and the cerebellum.
 Doyle, Vasseur, Dojat and Forbes (2013): Fully Automatic Brain Tumor Segmentation From Multiple MR Sequences Using Hidden Markov Fields and Variational EM
Doyle, Vasseur, Dojat and Forbes (2013): Fully Automatic Brain Tumor Segmentation From Multiple MR Sequences Using Hidden Markov Fields and Variational EM
Algorithm and Data
We propose an adaptive scheme for brain tumor segmentation using multiple MR sequences. Our approach is fully automatic and requires no training. The model parameters are instead estimated using a variational EM algorithm with MRF constraints and the inclusion of a priori probabilistic maps to provide a stable parameter trajectory during optimization.
We build on the standard hidden Markov field model by considering a more general formulation that is able to encode more complex interactions than the standard Potts model. In particular, we encode the possibility that certain tissue combinations in the neighborhood are penalized more than others, whereas the standard Potts model penalizes dissimilar neighboring classes equally, regardless of the tissues they represent.
A solution to the model is found using the Expectation Maximization (EM) framework [101] combined with variational approximation for tractability in the presence of Markov dependencies. In particular, we consider the so-called mean field principle that provides a deterministic way to deal with intractable Markov Random Field (MRF) models [102] and has proven to perform well in a number of applications.
We adopt a data model comprising of five normal tissue classes; white matter, grey matter, ventricular CSF, extraventricular CSF, and other. The glioma is modeled by a further four classes representing the diseased tissue state; edema, non-enhancing, enhancing and necrotic. In the absence of sufficient data to robustly and accurately estimate a full free interaction matrix B with the number of classes K = 9, further constraints are imposed on the B. The four glioma classes are considered a single structure, whose interaction with the normal tissue classes is not dependant on the specific glioma tissue state. Parameters are estimated using the variational EM algorithm, which provides a tractable solution for non trivial Markov models.
The deformable transform that describes the mapping between the International Consortium for Brain Mapping (ICBM) template and the data space is found using tools provided by the Insight Segmentation and Registration Toolkit (ITK). The transform is used to register the probabilistic tissue atlases to the MR sequences. An initial 5-class segmentation is performed, and the tumor region of interest (ROI) is detected by a simple morphological method comparing the segmentation result and the five tissue atlases. The prior probabilistic tissue atlas and the tumor ROI are incorporated a priori in the final segmentation algorithm via the singleton potential parameter in the MRF.
The computation time is 30 min per patient, giving an average Dice coefficient for high-grade and low-grade complete tumor volume of 0.84 and 0.81, respectively.
Training and Testing
The algorithm was tested on real-patient data from the BRATS 2012 and 2013 dataset. No training was performed; the initial labeling was random, and all model parameters were estimated iteratively.
 Festa, Pereira, Mariz, Sousa and Silva (2013): Automatic Brain Tumor Segmentation of Multi-Sequence MR Images Using Random Decision Forests
Festa, Pereira, Mariz, Sousa and Silva (2013): Automatic Brain Tumor Segmentation of Multi-Sequence MR Images Using Random Decision Forests
Algorithm and Data
The proposed algorithm is fully automated and uses all available MRI sequences. Three preprocessing steps were performed. The first aims for the bias field correction, with N4ITK method [103]. The second normalizes the intensity scale of each sequence to a chosen reference, by histogram matching using ITK [104]. Finally, since some FLAIR images were already limited to the volume of interest, all sequences from each subject were cropped to have the same brain volume. A random decision forest is used to classify each brain voxel, based on several features extracted from the training data. The main parameters in a decision forest are the number of trees and their depth, set to 50 and 25, respectively. Due to computational limitations, a maximum of 120 000 points per training subject were sampled. Half of these points are background and the other half are tumor and edema. The feature set includes: 1) MR sequences intensities and the difference between each two sequences; 2) neighborhood information with the mean, sum, median and intensity range of 3D cubic neighborhoods with edges of 3, 9, and 19 mm, centered in each voxel, from all MR sequences and the differences between sequences; 3) context information as the difference between each voxel and the mean value of 3 × 3 × 3 mm cubes, centered 3 mm from the voxel in six directions (two per axis), from all sequences; 4) texture information in all MR sequences, including edge density and local binary partition (signal and magnitude) extracted from 3 × 3 × 3 mm neighborhoods, and the Laws texture features [105] extracted from 2D 3 × 3 neighborhoods, in all three dimensions. Finally, a post processing step was performed assuming that very small isolated 3D regions, with less than seven voxels (value found empirically), of one label type should not exist. The total execution time is about 30 min for each test subject, mainly due to the features extraction, using the programming language Python on a computer with an Intel processor (i7-3930k, 3.2 GHz).
Training and Testing
Three datasets were available: “Training” (with corresponding ground truth), “LeaderBoard” and “Challenge.” The training step was done using all real data from the Training dataset, from both grades to increase the representation of some labels (like non-enhancing). The testing step was performed with all datasets. Leave-one-out cross-validation was used for the Training dataset. The features set, as well as the hyperparameters for the decision forest, were found using leave-one-out cross-validation of the Training dataset. To segment high-grade tumors, all images (used in training and testing stages) were normalized to a high-grade reference. Similarly, images were normalized to a low-grade reference when segmenting these tumors. The critical part of the proposed algorithm is the normalization, which influences the whole pipeline, especially with intensity related features used in a supervised classifier. A basic characterization of texture was used in the proposed algorithm and it seems to be helpful in the distinction of different tumor tissues. With a better texture characterization, it is expected to achieve further improvement in the segmentation of brain tumors.
 Geremia, Menze and Ayache (2012): Spatial Decision Forests for Glioma Segmentation in Multi-Channel MR Images
Geremia, Menze and Ayache (2012): Spatial Decision Forests for Glioma Segmentation in Multi-Channel MR Images
Medical imaging protocols produce large amounts of multi-modal volumetric images. The large size of the produced datasets contributes to the success of machine learning methods. These methods automatically learn from the data how to perform challenging task such as, for instance, semantic annotation. Although being a tremendous asset in theory, very large datasets are down-sampled to ensure tractability of the learning process. Moreover, the informative data is often submerged in overwhelming amounts of redundant data. Thus, most state of the art methods need to parse large amounts of uninformative data before reaching valuable data.
We present the “Spatially Adaptive Random Forests” (SARFs) [42] to overcome these issues in the context of volumetric medical images. SARFs automatically learn how to efficiently target valuable data. It avoids parsing and processing redundant data while focusing its computational power on critical image regions. We demonstrate its power to address multi-class glioma annotation in multi-modal brain MRIs.
SARF builds on three cutting-edge methods: 1) discriminative random forests, 2) an efficient multi-scale 3D image representation, and 3) structured labelling. Random forests demonstrated outstanding segmentation results in the context of brain lesion segmentation in MRIs and multi-organ segmentation in full body CT scans. Although real-time performance can be achieved during testing, training the forest is still time consuming due to the large amount of data that it needs to ingest.
In order to speed up training and testing, SARF relies on an efficient hierarchical representation of image volumes. The hierarchical representation is obtained by recursively applying an extended version of the SLIC algorithm to handle volumetric multi-modal images. The final result consists in a coarse to fine super-voxel hierarchical partition of the images similar to (cite bouman et al.).
Rather than merging the segmentations obtained from the different scales of the image, SARF iteratively refines the segmentation. This is made possible by carefully extrapolating the voxel-based ground truth to coarser scales. Additionally, SARF provides the ability of reasoning on semantically close classes by combining them in an hierarchical way (cite structured labelling). The resulting semantic tree together with the supervoxel hierarchy are powerful tools to efficiently parse and annotate the image volumes.
SARF makes use of these tools by integrating them into the random forest framework. During training, it learns the optimal image spatial sampling associated to the segmentation task. During testing, the algorithm quickly handles the background and focuses on challenging image regions to refine the segmentation. These properties were demonstrated together with promising results in the context of multi-class glioma segmentation in multi-modal brain MRIs.
 Guo, Schwartz and Zhao (2013): Semi-automatic Segmentation of Multimodal Brain Tumor Using Active Contours
Guo, Schwartz and Zhao (2013): Semi-automatic Segmentation of Multimodal Brain Tumor Using Active Contours
In this paper, we present a semi-automatic segmentation method for multimodal brain tumors. It requires only that a user manually draw a region of interest (ROI) roughly surrounding the tumor on a single image. The algorithm combines the image analysis techniques of region and edge-based active con-tours and level set approach, and has the advantages of easy initialization, quick segmentation, and efficient modification. The typical run-time for each case in the training dataset can be within 1 min.
 Hamamci and Unal (2012): Multimodal Brain Tumor Segmentation Using the “Tumor-Cut” Method
Hamamci and Unal (2012): Multimodal Brain Tumor Segmentation Using the “Tumor-Cut” Method
Algorithm and Data
As described in detail in the “Tumor-cut” article [72], the semi-automatic tumor segmentation method by Hamamci and Unal specifically targets the gross tumor volume (GTV) and the necrotic regions of the brain tumors on contrast enhanced T1-weighted MR images, requiring an initialization by drawing a line through the maximum diameter of the tumor as in the “Response Evaluation Criteria In Solid Tumors” (RECIST) guidelines [4]. For the BRATS challenge, the method was extended to multi-modal MRI to include also the labels for edema and non-enhanced regions. Hamamci and Unal's approach to fuse different MR modalities is to apply the original tumor-cut method to each channel seperately and then combine the segmented volumes by basic set operations based on the type of the modality. For each channel, a segmentation is initialized by drawing the maximum observable diameter of the tumor and performed independently (see Fig. 10). For FLAIR images, whole hyper-intense region is segmented as FLAIR volume (Vfl) and for T2 images only the core abnormality is segmented as T2 volume (Vt2). Tumor core is segmented on contrast enhanced T1 MRI (Vt1c) followed by the application of the necrotic segmentation method to segment the necrotic regions within the tumor core (Vnec). For the low-grade cases, Vt1c and Vnec are set to empty, because the tumors were not enhanced by the application of the contrast agent. Non-contrast enhanced T1 MR images were used neither for high- nor low-grade cases. For FLAIR segmentation, only the weight of the regularizer in the energy term for the level-set evolution is tuned to allow resulting tumor surfaces to have higher curvatures. Label for each class is determined by the following operations:
For each case, user interaction takes about 1–2 min and typical run time is around 10–30 min, depending on the size of the tumor, using a CPU. However, the parallel nature of the algorithm allows GPU implementation, which would reduce the processing time significantly.
Fig. 10.
Maximum diameter line drawn by the user to initialize the algorithm for CE-T1 (a), T2 (b), and Flair (c) modalities and the corresponding outputs, for a sample high-grade case. Manual labels overlayed on T1 for a sample slice (d).
Training and Testing
We observed that in one case only, we segmented an abnormal structure, which was not labeled as tumor by the experts. Although, this resulted a zero overlap score for the particular case, in fact, to allow user to choose what to segment is an advantage of the semi-automatic approach. In general, the T2 results did not provide useful information, as only a small portion of the tumors consist of the non-enhancing region and the segmentation results were not accurate due to the low contrast between tumor core and edema. The approach of Hamamci and Unal's algorithm was to apply their original algorithm independently to each modality. A combined algorithm that considers the multidimensional information from all available modalities have the potential to improve the results obtained.
 Meier, Bauer, Slotboom, Wiest and Reyes (2013): Appearance- and Context-sensitive Features for Brain Tumor Segmentation
Meier, Bauer, Slotboom, Wiest and Reyes (2013): Appearance- and Context-sensitive Features for Brain Tumor Segmentation
Algorithm and Data
In our approach, we regard image segmentation as a supervised classification problem. The present method is an improved version of the one proposed by Bauer et al. in [87] and can be subdivided into three main parts: a feature extraction yielding a voxel-wise feature vector, a classification step and a subsequent spatial regularization. Moreover, we preprocess the multimodal image data which encompasses noise-reduction, bias-field correction and intensity normalization.
The major difference to [87] is that for every voxel we extract a 257-dimensional feature vector composed of appearance-sensitive (multimodal intensities and intensity differences, first-order textures and gradient textures) and context-sensitive features (atlas-normalized coordinates, multiscale symmetry features and multi-/monomodal ray features). As a classifier we employ a classification forest. The predicted class label is defined according to the MAP-rule applied on the posterior probability output from the classification forest. The implementation of the classification forest is based on the Sherwood library [46]. The regularization is conducted in a hierarchical manner as proposed in [106]. It is realized as an energy minimization problem of a conditional random field, which is defined on a grid graph representing the image. The probabilistic output of the classification forest is further used to define the unary potentials, which model the affiliation of a voxel to a possible tissue class. Pairwise potentials model the coherence between neighboring voxels and are used to incorporate tissue dependencies and to account for anisotropic voxel dimensions. For solving the energy minimization problem we relied on the Fast-PD algorithm proposed in [88].
Our method is fully automatic with a testing time of 2–12 min per subject depending on the size of the image volume, where the feature extraction consumes most of the time.
Training and Testing
Relevant parameters of the classification forest (depth, number of candidate weak learners and thresholds per node) are set according to a gridsearch. The model is trained either on high-grade or low-grade cases only. For preliminary results and training phase before the competition the method has been evaluated on the high-grade or low-grade cases of the BRATS2013 training set using 5-fold cross validation.
We observed that depending on the image data false positives in the infratentorial part of the brain might appear. Moreover, the discrimination between edema and non-enhancing tumor seems to be the most challenging one. We plan to employ additional image features to overcome these problems and to further improve the current accuracy.
 Menze, Van Leemput, Lashkari, Weber, Ayache and Golland (2012): Segmenting Glioma in Multi-Modal Images Using a Generative Model for Brain Lesion Segmentation
Menze, Van Leemput, Lashkari, Weber, Ayache and Golland (2012): Segmenting Glioma in Multi-Modal Images Using a Generative Model for Brain Lesion Segmentation
We evaluate a fully automated method for channel-specific tumor segmentation in multi-dimensional images proposed by us in [73] that extends the general “EM segmention” algorithm for situations when specific spatial structures cannot be described sufficiently through population priors. The method represents a tumor appearance model for multi-dimensional sequences that provides channel-specific segmentation of the tumor. Its generative model shares information about the spatial location of the lesion among channels while making full use of the highly specific multi-modal signal of the healthy tissue classes for segmenting normal tissues in the brain. In addition to tissue types, the model includes a latent variable for each voxel encoding the probability of observing tumor at that voxel, based on the ideas from [55], [56].
Approach amends physiological tissue atlas with personalized lesion prior.
During segmentation information on tumor localization is traded between modalities via latent prior. Results in an individual segmentation in every modality.
Outperforms both univariate and multivariate EM segmentation and is capable of considering channel-specific constraint on hypo- or hypo intensity of the lesion with respect to the intensities of normal tissues in the same image.
To initialize our algorithm we segment the volume into the three healthy and an outlier class using a freely available implementation of the EM segmentation with bias correction [25]. Outliers are defined as being more than three standard deviations away from the centroid of any of the three normal tissue classes. We apply our algorithm to the bias field corrected volumes returned from this EM segmenter and initialize intensity parameters with values estimated in the initial segmentation. We initialize the latent atlas α to 0.7 time the local prior for the presence of gray or white matter. For a semantic interpretation that is in line with the class definitions of the segmentation challenge, Channels-specific segmentations returned by our algorithm are transformed to Edema and Core classes. We label voxels that show tumor specific changes in the T2 channel as edema, and voxels that show hyper-intense tumor specific changes as tumor core. A discriminative classifier filters all tumor segments removing those that are most likely to be false positives, primarily evaluating shape and location of the tumor regions returned from the generative model.
 Menze, Geremia, Ayache and Szekely (2012): Segmenting Glioma in Multi-Modal Images Using a Generative-Discriminative Model for Brain Lesion Segmentation
Menze, Geremia, Ayache and Szekely (2012): Segmenting Glioma in Multi-Modal Images Using a Generative-Discriminative Model for Brain Lesion Segmentation
The present discriminative model [73] (described above) returns probability maps for the healthy tissues, and probability maps for the presences of characteristic hypo- or hyper-intense changes in each of the image volumes. While this provides highly specific information about different pathophysiological processes induced by the tumor, the analysis of the multimodal image sequence may still require to highlight specific structures of the lesion—such as edema, the location of the active or necrotic core of the tumor, “hot spots” of modified angiogenesis or metabolism—that cannot directly be associated with any of these basic parameter maps returned. As a consequence, we propose to use the probabilistic output of the generative model, together with few structural features that are derived from the same probabilistic maps, as input to a classifier modeling the posterior of the desired pixel classes. In this we follow the approach proposed by [40] that prove useful for identifying white matter lesion in multiple input volumes. The building blocks of this discriminative approach are the input features, the parametrization of the random forest classifier used, and the final post-processing routines.
The approach combines advantageous properties from both types of learning algorithms: First, it extracts tumor related image features in a robust fashion that is invariant to relative intensity changes by relying on a generative model encoding prior knowledge on expected physiology and pathophysiological changes. Second, it transforms image features extracted from the generative model—representing tumor probabilities in the different image channels—to an arbitrary image representation desired by the human interpreter through an efficient classification method that is capable of dealing with high-dimensional input data and that returns the desired class probabilities. In the following, we shortly describe the generative model from [73], and input features and additional regularization methods used similar to our earlier discriminative model from [40].
As input feature describing the image in i voxel we use the probabilities p(ki) for the K = 3 tissue classes . We also use the tumor probability for each channel C = 4 , and the C = 4 image intensities after calibrating them with a global factor that has been estimated from gray and white matter tissue . From these data we derive two types of features: the “long range features” that calculate differences of local image intensities for all three types of input features , and a distance feature that calculates the geodesic distance of each voxel i to characteristic tumor areas. We choose random forests as our discriminative model as it uses labeled samples as input and returns class probabilities. For the normal classes (that are not available from the manual annotation of the challenge dataset) we infer the maximum a posterior estimates of the generative model and use them as label during training. Random forests learn many decision trees from bootstrapped samples of the training data, and at each split in the tree they only evaluate a random subspaces to find the best split. To minimize correlation in the training data, and also to speed up training, we draw no more 2000 samples from each of the ≈ 106 voxels in each of the 25 dataset. We train an ensemble with 300 randomized decision trees, and choose a subspace dimensionality of 10. We use the random forest implementation from Breiman and Cutler. To improve segmentation, we use a Markov random field (MRF) imposing a smoothness constraint on the class labels. We optimize the function imposing costs when assigning different labels in a six neighborhood on the cross-validated predictions on the training data.
 Reza and Iftekharuddin (2013): Multi-Class Abnormal Brain Tissue Segmentation Using Texture Features
Reza and Iftekharuddin (2013): Multi-Class Abnormal Brain Tissue Segmentation Using Texture Features
Algorithm and Data
In this work, we propose fully automated multi-class abnormal brain tissue segmentation in multimodality brain MRI. Fig. 11 shows a generic flow diagram for our algorithm pipeline. Since BRATS-2013 dataset is already skull stripped and co-registered; the first step involves preprocessing of 2D MRI slices extracted from 3D volume for each patient. Intensity normalization and inhomogeneity correction are used as pre-processing steps. Then two primary sets of features are extracted from each preprocessed image. The first set includes non-local features such as pixel intensities (IT1, IT2, IFL, IT1c) and differences of intensities (d1 = IT1 − IT2, d2 = IT2 − IFL, d3 = IFL − IT1c) that represents global characteristics of brain tissues. To characterize the tumor surface variation, we employ our novel texture features such as fractal PTPSA [107], and mBm [108] as well as classical textons [109] as the second set of features. After extraction, all features are fused in a classical Random Forest [110] classifier. Once the labels are predicted simultaneously, we obtain a 3D volume image per patient for online evaluation.
Fig. 11.
Generic flow diagram of the proposed method.
Training and Testing
We performed 3-fold cross validation on training dataset to tune the parameters. Extensive experiments suggests employing all tumor samples and randomly selected equal number of non-tumor samples for training the RF classifier yields good training results. For a single patient it takes about an hour and half to complete the whole process as shown in Fig. 11 while 3-fold cross-validation takes only about 15 min. The most time consuming parts are preprocessing and feature extraction which are done offline. All results in this work are obtained using MATLAB 2011a on windows 64 bit 2.26 GHz Intel Xeon processor, with 12 GB RAM. We process HG and LG patients separately for both Leader Board and Challenge testing phases. There are a few leader board cases that show low scores. Our observation suggests that if the tumor tissue intensities are below the mean intensity of the image, the necrosis tissues are misclassified as non-tumor. Data redundancy in the samples and covariance among the features usually lower the classifier performance. In summary, our extensive experimental results with BRATS data confirm the efficacy of our texture-based methods for multi-class abnormal brain tissue segmentation.
 Riklin Raviv, Van Leemput and Menze (2012): Multi-Modal Brain Tumor Segmentation via Latent Atlases
Riklin Raviv, Van Leemput and Menze (2012): Multi-Modal Brain Tumor Segmentation via Latent Atlases
The work is based on a generative approach for patient-specific segmentation of brain tumors across different MR modalities much in the spirit of [55], [56]. The segmentation problem is solved via a statistically driven level-set framework. Specifically, image partitioning into regions of interest (tumor parts) and healthy brain parts are obtained via the joint evolution of four level-sets functions determined by the images gray level-distributions and a smoothness term. Manual initialization based on a few mouse clicks to determine the approximate tumor center and extent was used.
 Shin (2012): Hybrid Clustering and Logistic Regression for Multi-Modal Brain Tumor Segmentation
Shin (2012): Hybrid Clustering and Logistic Regression for Multi-Modal Brain Tumor Segmentation
Unsupervised learning approaches have potential for applications in medical image processing, as previously discussed [111]–[113]. Additionally, the approach can be extended readily to a previously unseen dataset avoiding the issues of overfitting that can occur in supervised learning methods, where overfitting has a larger influence in tumor segmentation when tumors have very heterogeneous characteristics. Unsupervised learning approaches were applied 1) in [112] for the previous two-class segmentation challenge, and 2) in [113] to detect multiple organs from a dataset where a few roughly labeled samples were available. These methods however were not directly applicable when the format of this challenge was changed to classify four-class labels.
The four-class segmentation problem was therefore approached with a supervised learning algorithm, used previously in [112], to segment the tumor-cores, trained with logistic regression. Four-dimensional patches (3 × 3 × 3 volume-patch ×4 channels) were used with second-order polynomial features as described in [112], as opposed to the three-dimensional patches (mpsl × mpsl 2-D image-patch ×T temporal-dimension) used previously in [113] to identify organs (but not for segmentation). This was because the dataset for this challenge was carefully registered with little motion, compared to the abdominal scans in [113] where the registration over the 40 volumes along the time-course was difficult as the region is usually affected by breathing motion. Deep neural networks with up to six layers were tried as well, pre-training the hidden-layers with stacked-autoencoder feature learning and subsequently fine-tuning them with the labeled samples in the training dataset. Neural network model was not used for the challenge however, because the improvement of classification accuracy was small (< ~0.05) relatively to the higher complexity compared to the logistic regression model.
Each channel of volumes was normalized separately, to try to learn the relation between the multi-channel intensity values, and to avoid any biases in the image intensities in different scans. The same type of classifier was used to classify all labels including the not-of-interest label (label:0), where they were trained only on the patient-dataset which has four-class labels, and applied to synthetic data which has only two labels. Two cross-validations were performed for the parameter adaptation, and no additional post-processing steps were applied to the patch-wise classification. It took about 5–10 min to segment a volume depending on the size of the whole head in the volume, as the classifier scans through all the non-zero entities.
The segmentation result is reasonably good, especially considering that only patch-wise classification was performed for the segmentation without any post-processing step, with a single (type of) classifier being used to segment all tumor classes and data-types (patient/synthetic). This demonstrates the application of a classification model applied to the segmentation of coarsely labeled tumors. Combining any post-processing steps might provide an immediate improvement on the final segmentation result, while application of unsupervised methods could be studied in the future for this four-class segmentation, e.g., segmenting label: x-vs-rest for all labels individually but similarly to [112]. Extending the classification model to a structured prediction model is an interesting avenue for future work for this model, while using a whole volume as an input to deep convolutional neural networks [114] might be worth investigating for the application of neural network models.
 Subbanna, Precup, Collins and Arbel (2012): Hierarchical Probabilistic Gabor and MRF Segmentation of Brain Tumours in MRI Volumes
Subbanna, Precup, Collins and Arbel (2012): Hierarchical Probabilistic Gabor and MRF Segmentation of Brain Tumours in MRI Volumes
The off-site classification results were produced by a fully automated hierarchical probabilistic framework for segmenting brain tumours from multispectral human brain MRIs using multiwindow Gabor filters and an adapted Markov Random Field (MRF) framework [115] (while the 2012 on-site results were produced by an earlier version of the work [116]).
Image pre-processing involves bias field correction using N3 [117], intra-subject multispectral volume registration [118], non-uniformity correction [119], and intensity normalization [104] The algorithm consists of two stages. At the first stage, the goal is to coarsely segment tumors (and associated sub-classes) from surrounding healthy tissues using texture features. During training, specialized Gabor functions are developed to optimally separate tumors from surrounding healthy tissues based on combined-space coefficients of tumors in multispectral brain MRIs [120]. A Bayesian classification framework is designed such that models for tumour/non-tumors are built during training, based on the combined space Gabor decomposition. During testing, a Bayesian classifier results in tumour/non-tumor probabilities and coarse tumor boundaries around regions with high tumor probabilities. Prior probabilities for healthy brain tissues are obtained by registering a healthy tissue prior atlas to regions outside tumor boundaries [118] The coarse boundaries are refined at the voxel level through a modified MRF framework that carefully separates different tumor subclasses from each other and from healthy tissues. This customized MRF differs from standard MRFs in that it is not simply a smoothing operator on priors. In addition to taking voxel intensities and class labels into account, it also models intensity differences between neighboring voxels in the likelihood model and considers transition probabilities between neighboring voxel classes. The second inference stage is shown to resolve local inhomogeneities and impose a smoothing constraint, while maintaining appropriate boundaries as supported by local intensity difference observations.
The method was trained and tested on the updated MICCAI 2012 BRATS Database, which included four tumor subclasses: necrotic core, edema, solid tumor, and enhanced tumor. The algorithm was trained and tested on clinical volumes, including low-grade and high-grade tumors. Classifiers were built separately for all categories. No other datasets were used for training or tuning. Online segmentation statistics (e.g., Dice overlap metrics) were provided. For training cases, the method was tested in a leave-one-out fashion. After training, the algorithm was tested on all test cases. On I7 Dell Optiplex machines, the training took a day, due to both convolution and simulated annealing algorithms used. Each volume took 70 min to classify, due to time consuming convolutions with different Gabor filters. For tumor core segmentation, the technique outperformed the top methods by about 30% in the clinical test cases in terms of Dice statistics, and had comparable performance with the highest performing methods in terms of segmentation of other tumour regions (in all statistics) for both training and test cases. In terms of shortcomings, the classifier is currently heavily dependent on the normalization step performing adequately, which caused a problem in at least one HG test case. In addition, should the classifier at the first stage fail to find tumours altogether, the second stage has difficulty recovering, as seen in an LG and HG case.
 Taylor, John, Buendia and Ryan (2013): Map-Reduce Enabled Hidden Markov Models for High Throughput Multimodal Brain Tumor Segmentation
Taylor, John, Buendia and Ryan (2013): Map-Reduce Enabled Hidden Markov Models for High Throughput Multimodal Brain Tumor Segmentation
We have developed a novel Map-Reduce enabled extension to hidden Markov models (HMMs) to enable high-throughput training and segmentation of tumors and edema in multimodal magnetic resonance images of the brain.
Preprocessing and Training
Preprocessing prepares the input MR spectra, T1, T1 with Gadolinium contrast-enhanced (T1C), T2, and FLAIR, for segmentation. The preprocessing pipeline has been designed to remove spatial inhomogeneities due to patient movement, remove image artifacts (skull, eyes) not related to the segmentation problem, remove inhomogeneities due to MR scanner bias fields, and match each spectrum's intensity histogram to the volumes used for training. Training the HMM (Fig. 12) involves extracting a feature vector for each voxel in the source case. We extract intensity voxels from FLAIR, T1, T1C, and T2 MR spectra. Neighboring voxels are added to the feature vector. The corresponding truth labels for the voxel neighborhood in the feature vector is utilized for supervised training of the HMM. Extending the HMM model to Map-Reduce (Fig. 12) involved adapting the HMM supervised learning algorithm to incrementally update based on individual feature vectors and coding a Mapper to perform feature extraction. In our current case, a single Mapper handles a single training case, extracting all of the feature vectors for the case and providing the vectors to the Reducer. The Reducer collects the feature vectors from all of the Mappers and incrementally updates the HMM model as new feature vectors are produced. A final Controller normalizes the probabilities in the HMM (initial, transition, emission) and stores the HMM to a file. The HMM was trained with the BRATS 2013 high-grade training data.
Fig. 12.
Left: Training the HMM model. Center: MapReduce model for HMM-based brain tumor segmentation. Right: Applying the HMM model for segmentation.
Segmentation and Results
Segmenting with the HMM (Fig. 3) involves extracting the feature vector for each voxel in the target case in the same manner as HMM training. Voxels from FLAIR, T1, T1C, and T2 in a neighborhood around the voxel of interest are organized into the feature vector and provided to the trained HMM model. The HMM model produces a predicted label for the feature vector. Postprocessing involved filtering out small objects and applying dilation and erosion operations on each segmented class. Our method has been evaluated on the BRATS2013 challenge dataset for high-grade glioma cases. We achieve an mean accuracy (Dice score) of [59.5]% for edema and [65.6]% for tumor in the real cases. The Map-Reduce enabled HMM is able to train on all cases simultaneously, performing 220% faster on an 8-node cluster than on a single node. Segmentation of a single patient case takes less than 1 min.
Limitations with the current algorithm include lack of support for spatial features, neighborhood-based textural features, and utilization of atlas-based priors, which have been shown to improve segmentation accuracy. We are currently working on a Decision Forest based extension to the HMM-Map Reduce algorithm to incorporate these features.
 Tustison, Wintermark, Durst and Avants (2013): ANTs and Árboles
Tustison, Wintermark, Durst and Avants (2013): ANTs and Árboles
Description
Given the success of random forest (RF)-based approaches in the BRATS 2012 challenge, we employed RFs to produce a completely automated, multi-modality brain segmentation framework. However, differences from related work include an expanded feature image set, concatenated RF modeling, and an open source implementation16 heavily dependent on the Advanced Normalization Tools (ANTs)17 repository including its R packaging (ANTsR).18 It is the latter open source aspect of our work which significantly motivated our participation in BRATS 2013 as it provides a reproducible and publicly available framework for performing such an important task in neuroimaging [121].
Algorithm and Data
The workflow for estimating tumor-based labeling from multi-modal images involves the following steps.
Symmetric multivariate template construction [122] using the data described in [123].
- Image preprocessing:
- Windowing intensities (quantiles [0.01, 0.99]);
- N4 bias correction [103];
- Rescaling intensity range to [0, 1].
- Stage 1 (GMM) processing:
- generation of feature images;
- construction of the Stage 1 RF model and probability images.
- Stage 2 processing:
- generation of single-modality MAP-MRF images using the Stage 1 RF probability images as spatial priors;
- construction of the Stage 2 RF model and labelings.
Refinement of Stage 2 labelings using a heuristically-derived binary morphological processing protocol.
We used the following feature images:
- Per modality (FLAIR, T1, T1C, T2)
-
—First-order neighborhood statistical images: mean, variance, skewness, and entropy. Neighborhood radius ∈ {1, 3}.
-
—GMM (stage 1) and MAP-MRF (stage 2) posteriors: CSF, gray matter, white matter, necrosis, edema, non-enhancing tumor and enhancing tumor (or a subset for the simulated data).
-
—GMM (stage 1) and MAP-MRF (stage 2) connected component geometry features: distance to tumor core label, volume, volume to surface area ratio, eccentricity, and elongation.
-
—Template-based: symmetric template difference and contralateral difference with Gaussian smoothing (σ = 4 mm).
-
—
Miscellaneous: normalized Euclidean distance based on cerebral mask, log Jacobian image, and (T1C - T1) difference image.
Prior cluster centers for specific tissue types learned from training data are used in the first stage to construct multiple GMM-based feature images [124]. The resulting spatial priors derived from application of the RF model for the first stage were used as input to an iterative n-tissue N4 ⇌ Atropos MAP-MRF segmentation protocol. These are used to create modified feature images for the second stage. ANTs registration [125] is also used to produce three sets of feature images: the log Jacobian image, intensity differences between each modality of each subject and the corresponding symmetric template, and contralateral differences.
All processing was performed using the computational cluster at the University of Virginia.19 Timing measures (single-threaded) included ~1.5 h per subject for feature image creation with the bulk of time devoted to spatial normalization with the symmetric template. Model construction required ~2 h with prediction taking approximately 15 min per subject.
Training and Testing
Training was performed separately for both real and simulated data and high-grade versus low-grade tumor assessment resulting in four RF modeling/prediction pathways. Training was limited to the 80 evaluation datasets provided by the organizers with evaluation employing a leave-one-out strategy for each of the four groupings.
 Zhao and Corso (2012): Brain Tumor Segmentation With MRF on Supervoxels
Zhao and Corso (2012): Brain Tumor Segmentation With MRF on Supervoxels
Algorithm and Data
For each MRI case, we first perform over-segmentation, which results in a set of supervoxels. We then solve the voxel labeling problem directly on the supervoxels constraining all voxels within one supervoxel to have the same label.
Consider a Markov random field defined over the supervoxels 𝒮. A labeling f assigns a label fP ∈ L to each supervoxel P, where L = {N, E, nonET, ET, C, B}, necrosis, edema, non-enhancing tumor, enhancing tumor, cerebrospinal fluid and background (white matter and gray matter), respectively. The energy function
where 𝒮 is the set of supervoxels and N𝒮 is the set of adjacent supervoxels, captures the cost of a certain labeling f. We define the data term as DQ (fQ) = ∑q∈Q − log(P(I(q) | fQ)), where P(I(q) | fQ) is the node class likelihood estimated by a Gaussian mixture model and I(q) denotes the feature of voxel q, the intensities of q of four channels. We define the smoothness term to capture the edge presence along the common boundary of the two supervoxels
where α,β are two nonnegative parameter, and Np is the neighborhood of p. Edge(p) is defined as
| (4) |
where rq,p is a voxel, such that q and rq,p are symmetric about p.
Finally, we solve the supervoxel labeling energy minimization problem using graph cuts [126]–[128].
The computing time is about 20 min for each case with Matlab an Intel Core i7-3770K, 3.50 GHz processor and 16 GB memory system. The most time consuming part is over-segmentation and computing VP,Q in (4).
Because we use intensities directly as the feature, we compute the standard scores to put the data in the same scale.
Training and Testing
We made a two-fold cross-validation on high-grade and low-grade cases, respectively. We learn individual classifiers for the high-grade set and the low-grade set with the same algorithm. As most other supervised methods using intensities as the feature, the accuracy of our method depends on the standardization of intensities. Hence, our method may fail if the case has different distribution with other cases. In some cases, our method fails because the data is not good enough. For example, in some cases, extraction is not good enough to remove the whole skull (we did not try to make a better extraction), and in some other cases, we do not have the whole image on FLAIR channel. But our method also fails on some good cases.
To overcome this problem, we could make a rough segmentation first, get the normal part of the case (white matter, gray matter, CSF), and make the intensity standardization only with the normal part. We are working on such a method and may use it in BRATS 2013 if it works.
 Zhao, Sarikaya and Corso (2013): Automatic Brain Tumor Segmentation With MRF on Supervoxels
Zhao, Sarikaya and Corso (2013): Automatic Brain Tumor Segmentation With MRF on Supervoxels
Algorithm and Data
We normalize the data and estimate the likelihood of pixels by the registration of a 3D joint histogram. We first perform over-segmentation on each case, resulting in a set of supervoxels. We then solve the voxel labeling problem directly on the supervoxels with Markov random field. This algorithm do not need manual input.
Pre-Processing
For each channel of each MRI case, we first denoise with SUSAN [129]; then we compute the standardized z-scores (zero mean and unit covariance) to put the data in the same scale, which are the feature vectors we use.
Oversegmentation of the Image With Supervoxels
In order to obtain supervoxels of MRI scan images, we use SLIC 3D [130] which generates supervoxels by clustering voxels based on their color similarity and proximity in the image volume. RegionSize and regularizer, the two parameters of SLIC, are 10 and 0.1, respectively.
Segmentation With Graph Cuts on a Markov Random Field
Consider a Markov random field defined over the supervoxels 𝒮 with A labeling f
where N𝒮 is the set of adjacent supervoxels. We define the data term as DQ (fQ) = ∑q∈Q − log(P(I(q) | fQ)), where P(I(q) | fQ) is the node class likelihood estimated by histogram based method and I(q) denotes the feature of voxel q. two supervoxels:
where Np is the neighborhood of p. Edge(p) is defined as
where rq,p is a voxel, such that q and rq,p are symmetric about p.
Finally, we solve the labeling energy minimization problem using graph cuts [131].
In this step, the key parameters a and b 0.5
A. Histogram Based Likelihood Estimation
Given a testing image Imgx and a labeled training image Imgi, we estimate the likelihood Pri(I(p) | fp) for each voxel p ∈ Imgx with Algorithm 2.
Algorithm 2.
Likelihood Estimation
| Input: Imgx, labelled image Imgi |
| 1: Compute Ii, Ix with quantization |
| 2: Compute Hi, Hx, Ht,i, t ∈ L |
| 3: With Hi, Hx, compute |
| 4: With and Ht,i, compute the deformed Histogram, , t ∈ L |
| 5: |
We then integrate information from each training image as follows:
Running Time
The running time is about 0.7n + 4 min for each case, where n is the number of cases of training data. The most consuming part is 3D registration of histograms. This depends on the size of the histogram and the method for registration.
Training and Testing
We learned individual classifiers for each of the four sub-problems. For each sub-problem, we use a 2-fold cross validation for the parameters a and b in the smoothness term of MRF, however, we set (a) / (b) to (1) / (30) manually. We only use the training data from the BRATS challenge.
Shortcomings
The performance of the over-segmentation limits the accuracy of our method. To overcome this, we could make a voxel level labeling in the supervoxels along the boundary, after the supervoxel labeling.
 Zikic, Glocker, Konukoglu, Shotton, Criminisi, Ye, Demiralp, Thomas, Das, Jena, and Price (2012): Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
Zikic, Glocker, Konukoglu, Shotton, Criminisi, Ye, Demiralp, Thomas, Das, Jena, and Price (2012): Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
Description
This submission is based on a classification forest, which is used such as to produce context-sensitive predictions. The method is based on our work focusing on high-grade glioma [39], with further technical details available in [51]. The context sensitivity arises from two components in the framework. The first one is that the forest does not operate only on the original input images, but also on initial patient-specific probabilities p′ for each tissue class c. These probabilities are computed at test time for each patient as the posterior probability p′(c | I(x)) = p(I(x) | c)p(c), based on the likelihood p(I(x) | c) of the multi-channel intensity I(x) given c. p(I(x) | c) and p(c) are estimated based on the training dataset—the likelihood by a Gaussian mixture model, and the prior as a normalized empirical histogram. While the initial probabilities often give reasonable rough estimates, they are noisy and erroneous, due to use of local intensity information only. Presenting the initial estimates to the forest as additional input has the effect of removing the noise and correcting some misclassifications. The second context-inducing component is the use of context-sensitive features for the forest (similar to [132], [133]), which capture intensity characteristics around the point of interest. Due to the regularizing effect of the context-sensitive forest, we did not find it necessary to use an explicit energy-based regularization.
We use the following preprocessing. For each patient, all scans are affinely aligned to the T1 contrast scan. We perform inhomogeneity correction with [82]. Instead of the standard histogram equalization, we multiply the intensities in each scan, such that the mean value equals 1000.
Our approach is fully automatic. The segmentation takes 1–2 min per scan (excluding pre-processing). The training of one tree takes ca. 20 min on a single PC. The key parameters of the method are the number of trees per forest and the maximal tree depth. We use forests with 100 trees with maximal depth of 20 for all challenge submissions (except the 2-class training data, where 40 trees per forest were used). An analysis of the parameter settings can be found in [39].
Method and parameter tuning were performed by a leave-one-out cross-validation on the initial BRATS 2-class training data. The same settings were then used for all submissions. We learn individual classifiers for the four sub-tasks (real/high, real/low, sim/high, sim/low). Since we did not perform a cross-validation to modify any parameters for the 4-class setting, the error reported in the system for 4-class training is based on a classifier trained on all images, which explains the high score.
The largest potential for improvement seems to be to attempt to achieve better results for outlier patients with very low accuracy (cf. [51]). This might be done by using more training data, or by taking into account further information, e.g., whether the scan is pre or post surgery.
Footnotes
B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani, J. Kirby, Y. Burren, N. Porz, J. Slotboom, R. Wiest, L. Lanczi, E. Gerstner, M.-A. Weber, M. Prastawa, M. Reyes, and K. Van Leemput co-organized the benchmark; all others contributed results of their algorithms as indicated in the appendix.
Available online: www.braintumorsegmentation.org
BRATS 2013: hal.inria.fr/hal-00912934; BRATS 2012: hal.inria.fr/hal-00912935
www.virtualskeleton.ch/→BRATS 2013→“BRATS 2013 Individual Observer Ground-truth Data”
BRATS 2013: hal.inria.fr/hal-00912934; BRATS 2012: hal.inria.fr/hal-00912935
Contributions
B. H. Menze, A. Jakab, S. Bauer, M. Reyes, M. Prastawa, and K. Van Leemput organized BRATS 2012. B. H. Menze, M. Reyes, J. Kalpathy-Cramer, J. Kirby, and K. Farahani organized BRATS 2013. A. Jakab and B. H. Menze defined the annotation protocol. A. Jakab, Y. Burren, N. Porz, J. Slotboom, R. Wiest, L. Lanczi, and M.-A. Weber acquired and annotated the clinical images. M. Prastawa generated the synthetic images. M. Prastawa and S. Bauer pre-processed the images. M. Prastawa implemented the evaluation scripts. S. Bauer, M. Reyes, and M. Prastawa adapted and maintained the online evaluation tools. All other authors contributed results of their tumor segmentation algorithms as indicated in the Appendix. B. H. Menze analyzed the data. B. H. Menze and K. Van Leemput wrote the manuscript. B. H. Menze wrote the first draft.
Contributor Information
Bjoern H. Menze, Institute for Advanced Study and Department of Computer Science, Technische Universität München, Munich, Germany, and with the Computer Vision Laboratory, ETH, Zürich, Switzerland, and with the Asclepios Project, Inria, Sophia-Antipolis, France, and also with the Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139 USA
Andras Jakab, Computer Vision Laboratory, ETH, Zürich, Switzerland, and also with the University of Debrecen, Debrecen, Hungary.
Stefan Bauer, Institute for Surgical Technology and Biomechanics, University of Bern, Switzerland, and also with the Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, Switzerland.
Jayashree Kalpathy-Cramer, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston MA, USA.
Keyvan Farahani, Cancer Imaging Program, National Cancer Institute, National Institutes of Health, Bethesda MD, USA.
Justin Kirby, Cancer Imaging Program, National Cancer Institute, National Institutes of Health, Bethesda MD, USA.
Yuliya Burren, Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, Switzerland.
Nicole Porz, Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, Switzerland.
Johannes Slotboom, Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, Switzerland.
Roland Wiest, Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, Switzerland.
Levente Lanczi, University of Debrecen, Debrecen, Hungary.
Elizabeth Gerstner, Department of Neuro-oncology, Massachusetts General Hosptial, Harvard Medical School, Boston MA, USA.
Marc-André Weber, Diagnostic and Interventional Radiology, University Hospital, Heidelberg, Germany.
Tal Arbel, Centre for Intelligent Machines, McGill University, Canada.
Brian B. Avants, Penn Image Computing and Science Lab, Department of Radiology, University of Pennsylvania, Philadelphia PA, USA
Nicholas Ayache, Asclepios Project, INRIA, Sophia-Antipolis, France.
Patricia Buendia, INFOTECH Soft, Inc., Miami FL, USA.
D. Louis Collins, McConnell Brain Imaging Centre, McGill University, Canada.
Nicolas Cordier, Asclepios Project, INRIA, Sophia-Antipolis, France.
Jason J. Corso, Computer Science and Engineering, SUNY, Buffalo NY, USA
Antonio Criminisi, Microsoft Research, Cambridge, U.K..
Tilak Das, Cambridge University Hospitals, Cambridge, U.K..
Hervé Delingette, Asclepios Project, INRIA, Sophia-Antipolis, France.
Çağatay Demiralp, Computer Science Department, Stanford University, Stanford CA, USA.
Christopher R. Durst, Department of Radiology and Medical Imaging, University of Virginia, Charlottesville VA USA
Michel Dojat, INRIA Rhône-Alpes, Grenoble, France, and also with the INSERM, U836, Grenoble, France.
Senan Doyle, INRIA Rhône-Alpes, Grenoble, France, and also with the INSERM, U836, Grenoble, France.
Joana Festa, Department of Electronics, University Minho, Portugal.
Florence Forbes, INRIA Rhône-Alpes, Grenoble, France, and also with the INSERM, U836, Grenoble, France.
Ezequiel Geremia, Asclepios Project, INRIA, Sophia-Antipolis, France.
Ben Glocker, BioMedIA Group, Imperial College, London, U.K..
Polina Golland, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge MA USA.
Xiaotao Guo, Department of Radiology, Columbia University, New York, NY USA.
Andac Hamamci, Faculty of Engineering and Natural Sciences, Sabanci University, Istanbul, Turkey.
Khan M. Iftekharuddin, Vision Lab, Department of Electrical and Computer Engineering, Old Dominion University, Norfolk, VA USA
Raj Jena, Cambridge University Hospitals, Cambridge, U.K..
Nigel M. John, INFOTECH Soft, Inc., Miami, FL USA, and also with the Department of Electrical and Computer Engineering, University of Miami, Coral Gables FL USA
Ender Konukoglu, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston MA USA.
Danial Lashkari, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge MA USA.
José António Mariz, Life and Health Science Research Institute (ICVS), School of Health Sciences, University of Minho, Braga, Portugal, and also with the ICVS/3B's—PT Government Associate Laboratory, Braga/Guimaraes, Portugal.
Raphael Meier, Institute for Surgical Technology and Biomechanics, University of Bern, 3014 Bern, Switzerland.
Sérgio Pereira, Department of Electronics, University Minho, Portugal.
Doina Precup, School of Computer Science, McGill University, Canada. and also with the Electrical and Computer Engineering Department, Ben-Gurion University, Beer-Sheva, Israel.
Stephen J. Price, Cambridge University Hospitals, Cambridge, U.K.
Syed M. S. Reza, Vision Lab, Department of Electrical and Computer Engineering, Old Dominion University, Norfolk, VA USA
Michael Ryan, INFOTECH Soft, Inc., Miami FL, USA.
Duygu Sarikaya, Computer Science and Engineering, SUNY, Buffalo NY, USA.
Lawrence Schwartz, Department of Radiology, Columbia University, New York, NY USA.
Hoo-Chang Shin, Sutton, U.K..
Jamie Shotton, Microsoft Research, Cambridge, U.K..
Carlos A. Silva, Department of Electronics, University Minho, Portugal
Nuno Sousa, Life and Health Science Research Institute (ICVS), School of Health Sciences, University of Minho, Braga, Portugal, and also with the ICVS/3B's—PT Government Associate Laboratory, Braga/Guimaraes, Portugal.
Nagesh K. Subbanna, Centre for Intelligent Machines, McGill University, Canada
Gabor Szekely, Computer Vision Laboratory, ETH, Zürich, Switzerland.
Thomas J. Taylor, INFOTECH Soft, Inc., Miami FL, USA
Owen M. Thomas, Cambridge University Hospitals, Cambridge, U.K.
Nicholas J. Tustison, Department of Radiology and Medical Imaging, University of Virginia, Charlottesville VA USA
Gozde Unal, Faculty of Engineering and Natural Sciences, Sabanci University, Istanbul, Turkey.
Flor Vasseur, INRIA Rhône-Alpes, Grenoble, France, and also with the INSERM, U836, Grenoble, France.
Max Wintermark, Department of Radiology and Medical Imaging, University of Virginia, Charlottesville VA USA.
Dong Hye Ye, Electrical and Computer Engineering Department, Purdue University, USA.
Liang Zhao, Computer Science and Engineering, SUNY, Buffalo NY, USA.
Binsheng Zhao, Department of Radiology, Columbia University, New York, NY USA.
Darko Zikic, Microsoft Research, Cambridge, U.K..
Marcel Prastawa, GE Global Research, Niskayuna NY, USA.
Mauricio Reyes, Institute for Surgical Technology and Biomechanics, University of Bern, 3014 Bern, Switzerland.
Koen Van Leemput, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02129 USA, and with the Technical University of Denmark, Denmark, and also with Aalto University, Finland.
References
- 1.Holland EC. Progenitor cells and glioma formation. Curr. Opin. Neurol. 2001;14:683–688. doi: 10.1097/00019052-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol. Exp. Neurol. 2005 Jun;64(6):479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 3.Louis DH, Ohgaki H, Wiestler OD, Cavanee WK. WHO classification of tumours of the central nervous system WHO/IARC. Lyon, France: Tech. Rep; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenhauer E, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J Clin. Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 6.Angelini ED, Clatz O, Mandonnet E, Konukoglu E, Capelle L, Duffau H. Glioma dynamics and computational models: A review of segmentation, registration, and in silico growth algorithms and their clinical applications. Curr. Med. Imag. Rev. 2007;3:262–276. [Google Scholar]
- 7.Bauer S, Wiest R, Nolte L-P, Reyes M. A survey of MRI-based medical image analysis for brain tumor studies. Phys. Med. Biol. 2013 Jul;58(13):R97–R129. doi: 10.1088/0031-9155/58/13/R97. [DOI] [PubMed] [Google Scholar]
- 8.Kaus M, et al. Segmentation of meningiomas and low grade gliomas in MRI. Proc. MICCAI. 1999:1–10. [Google Scholar]
- 9.Fletcher-Heath LM, Hall LO, Goldgof DB, Murtagh FR. Automatic segmentation of non-enhancing brain tumors in magnetic resonance images. Artif. Intell. Med. 2001;21(1–3):43–63. doi: 10.1016/s0933-3657(00)00073-7. [DOI] [PubMed] [Google Scholar]
- 10.Tsai Y-F, Chiang I-J, Lee Y-C, Liao C-C, Wang K-L. Automatic MRI meningioma segmentation using estimation maximization. Proc. IEEE Eng. Med. Biol. Soc. 2005;3:3074–3077. doi: 10.1109/IEMBS.2005.1617124. [DOI] [PubMed] [Google Scholar]
- 11.Konukoglu E, et al. Monitoring slowly evolving tumors. Proc. ISBI. 2008:1–4. doi: 10.1109/ISBI.2008.4541120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuadra MB, Craene MD, Duay V, Macq B, Pollo C. and J.-P. Thiran,"Dense deformation field estimation for atlas-based segmentation of pathological MR brain images. Comput. Methods Programs Biomed. 2006;84:66–75. doi: 10.1016/j.cmpb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Weizman L, et al. Automatic segmentation, internal classification, and follow-up of optic pathway gliomas in MRI. Med. Image Anal. 2012;16(1):177–188. doi: 10.1016/j.media.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Styner M, et al. 3D segmentation in the clinic: A grand challenge II: MS lesion segmentation. MIDAS J. 2008:1–5. [Google Scholar]
- 15.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imag. 1999 Oct;18(10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- 16.Kaus MR, Warfield SK, Nabavi A, Black PM, Jolesz FA, Kikinis R. Automated segmentation of MR images of brain tumors. Radiology. 2001 Feb;218(2):586–591. doi: 10.1148/radiology.218.2.r01fe44586. [DOI] [PubMed] [Google Scholar]
- 17.Prastawa M, Bullitt E, Ho S, Gerig G. A brain tumor segmentation framework based on outlier detection. Med. Image Anal. 2004;8:275–283. doi: 10.1016/j.media.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Pohl KM, Fisher J, Levitt JJ, Shenton ME, Kikinis R, Grimson WEL, Wells WM. A unifying approach to registration, segmentation, and intensity correction. Proc. MICCAI. 2005:310–318. doi: 10.1007/11566465_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaster FO, Menze BH, Weber M-A, Hamprecht FA. Comparative validation of graphical models for learning tumor segmentations from noisy manual annotations. Proc. MICCAI-MCV. 2010 [Google Scholar]
- 20.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Zacharaki EI, Shen D, Davatzikos C. ORBIT: A multiresolution framework for deformable registration of brain tumor images. IEEE Trans. Med. Imag. 2008 Aug;27(8):1003–1017. doi: 10.1109/TMI.2008.916954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach Cuadra B, Pollo C, Bardera A, Cuisenaire O, Thiran JP. Atlas-based segmentation of pathological brain MR images using a model of lesion growth. IEEE Trans. Med. Imag. 2004 Oct;23(10):1301–1314. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 24.Gering D, Grimson W, Kikinis R. Recognizing deviations from normalcy for brain tumor segmentation. Proc. MICCAI 2002. 2002 Sep;2488:388–395. [Google Scholar]
- 25.Van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imag. 2001;20:677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 26.Seghier ML, Ramlackhansingh A, Crinion J, Leff AP, Price CJ. Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage. 2008;41(4):1253–1266. doi: 10.1016/j.neuroimage.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon N, Bullitt E, Van Leemput K, Gerig G. Model-based brain and tumor segmentation. Proc. ICPR. 2002:528–531. [Google Scholar]
- 28.Prastawa M, Bullitt E, Moon N, Leemput KV, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Acad. Radiol. 2003;10:1341–1348. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gooya A, et al. GLISTR: Glioma image segmentation and registration. IEEE Trans. Med. Imag. 2012 Oct;31(10):1941–1954. doi: 10.1109/TMI.2012.2210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parisot S, Duffau H, Chemouny S, Paragios N. Joint tumor segmentation and dense deformable registration of brain MR images. Proc. MICCAI. 2012:651–658. doi: 10.1007/978-3-642-33418-4_80. [DOI] [PubMed] [Google Scholar]
- 31.Gooya A, Biros G, Davatzikos C. Deformable registration of glioma images using EM algorithm and diffusion reaction modeling. IEEE Trans. Med. Imag. 2011 Feb;30(2):375–390. doi: 10.1109/TMI.2010.2078833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cobzas D, Birkbeck N, Schmidt M, Jagersand M, Murtha A. 3D variational brain tumor segmentation using a high dimensional feature set. Proc. ICCV. 2007:1–8. [Google Scholar]
- 33.Lefohn A, Cates J, Whitaker R. Interactive, GPU-based level sets for 3D brain tumor segmentation. Proc. MICCAI. 2003:564–572. [Google Scholar]
- 34.Verma R, et al. Multiparametric tissue characterization of brain neoplasms and their recurrence using pattern classification of MR images. Acad. Radiol. 2008 Aug;15(8):966–977. doi: 10.1016/j.acra.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho S, Bullitt E, Gerig G. Level-set evolution with region competition: Automatic 3D segmentation of brain tumors. Proc. ICPR. 2002:532–535. [Google Scholar]
- 36.Gorlitz L, Menze BH, Weber M-A, Kelm BM, Hamprecht FA. Semi-supervised tumor detection in magnetic resonance spectroscopic images using discriminative random fields. Proc. DAGM. 2007:224–233. ser. LNCS. [Google Scholar]
- 37.Lee C, Wang S, Murtha A, Greiner R. Segmenting brain tumors using pseudo conditional random fields. Proc. MICCAI. 2008:359–366. [PubMed] [Google Scholar]
- 38.Wels M, Carneiro G, Aplas A, Huber M, Hornegger J, Comaniciu D. A discriminative model-constrained graph cuts approach to fully automated pediatric brain tumor segmentation in 3D MRI. Proc. MICCAI. 2008:67–75. doi: 10.1007/978-3-540-85988-8_9. LNCS 5241. [DOI] [PubMed] [Google Scholar]
- 39.Zikic D, et al. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Proc. MICCAI. 2012 doi: 10.1007/978-3-642-33454-2_46. [DOI] [PubMed] [Google Scholar]
- 40.Geremia E, Menze BH, Clatz O, Konukoglu E, Criminisi A, Ayache N. Spatial decision forests for MS lesion segmentation in multichannel MR images. Proc. MICCAI. 2010 doi: 10.1007/978-3-642-15705-9_14. [DOI] [PubMed] [Google Scholar]
- 41.Geremia E, Clatz O, Menze BH, Konukoglu E, Criminisi A, Ayache N. Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. Neuroimage. 2011;57:378–390. doi: 10.1016/j.neuroimage.2011.03.080. [DOI] [PubMed] [Google Scholar]
- 42.Geremia E, Menze BH, Ayache N. Spatially adaptive random forests. Proc. IEEE Int. Symp. Biomed. Imag. 2013:1344–1347. [Google Scholar]
- 43.Bauer S, Nolte L-P, Reyes M. Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. Proc. MICCAI. 2011:354–361. doi: 10.1007/978-3-642-23626-6_44. [DOI] [PubMed] [Google Scholar]
- 44.Wu W, Chen AY, Zhao L, Corso JJ. Brain tumor detection and segmentation in a conditional random fields framework with pixel-pairwise affinity and superpixel-level features. Int. J. Comput. Assist. Radiol. Surg. 2013:1–13. doi: 10.1007/s11548-013-0922-7. [DOI] [PubMed] [Google Scholar]
- 45.Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20(3):273–297. [Google Scholar]
- 46.Criminisi A, Shotton J. Decision Forests for Computer Vision and Medical Image Analysis. Heidelberg, Germany: Springer; 2013. [Google Scholar]
- 47.Iglesias JE, Konukoglu E, Zikic D, Glocker B, Van Leemput K, Fischl B. Is synthesizing MRI contrast useful for inter-modality analysis? Proc. MICCAI. 2013:631–638. doi: 10.1007/978-3-642-40811-3_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy S, Carass A, Prince J. A compressed sensing approach for MR tissue contrast synthesis. Proc. IPMI. 2011:371–383. doi: 10.1007/978-3-642-22092-0_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy S, et al. Longitudinal intensity normalization in the presence of multiple sclerosis lesions. Proc. ISBI. 2013:1384–1387. doi: 10.1109/ISBI.2013.6556791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menze BH, Van Leemput K, Lashkari D, Weber M-A, Ayache N, Golland P. Segmenting glioma in multi-modal images using a generative model for brain lesion segmentation. Proc. MICCAI-BRATS. 2012:1–8. doi: 10.1007/978-3-642-15745-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zikic D, et al. Context-sensitive classification forests for segmentation of brain tumor tissues. Proc. MICCAI-BRATS. 2012:22–30. [Google Scholar]
- 52.Tarabalka Y, Charpiat G, Brucker L, Menze BH. Enforcing monotonous shape growth or shrinkage in video segmentation; Proc. Br. Mach. Vis. Conf; 2013. [DOI] [PubMed] [Google Scholar]
- 53.Tarabalka Y, Charpiat G, Brucker L, Menze BH. Spatio-temporal video segmentation with shape growth or shrinkage constraint. IEEE Trans. Image Process. 2014 Sep;23(9):3829–3840. doi: 10.1109/TIP.2014.2336544. [DOI] [PubMed] [Google Scholar]
- 54.Bauer S, Tessier J, Krieter O, Nolte L, Reyes M. Integrated spatio-temporal segmentation of longitudinal brain tumor imaging studies. Proc. MICCAI-MCV. 2013:122–130. [Google Scholar]
- 55.Riklin-Raviv T, et al. Joint segmentation via patient-specific latent anatomy model. Proc. MICCAI-PMMIA. 2009:244–255. [Google Scholar]
- 56.Riklin-Raviv T, Van Leemput K, Menze BH, Wells WM, 3rd, Golland P. Segmentation of image ensembles via latent atlases. Med. Image Anal. 2010;14:654–665. doi: 10.1016/j.media.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cobzas D, Schmidt M. Increased discrimination in level set methods with embedded conditional random fields; Proc. IEEE Conf. Comput. Vis. Pattern Recognit; 2009. Jun, pp. 328–335. [Google Scholar]
- 58.Corso JJ, et al. Efficient multilevel brain tumor segmentation with integrated Bayesian model classification. IEEE Trans. Med. Imag. 2008 May;27(5):629–640. doi: 10.1109/TMI.2007.912817. [DOI] [PubMed] [Google Scholar]
- 59.Tustison NJ, et al. Instrumentation bias in the use and evaluation of scientific software: Recommendations for reproducible practices in the computational sciences. Front. Neurosci. 2013;7:162. doi: 10.3389/fnins.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shattuck DW, Prasad G, Mirza M, Narr KL, Toga AW. Online resource for validation of brain segmentation methods. Neuroimage. 2009;45(2):431–439. doi: 10.1016/j.neuroimage.2008.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metz C, et al. 3D segmentation in the clinic: A grand challenge II-coronary artery tracking. Insight J. 2008;1(5):6. [Google Scholar]
- 62.Schaap M, et al. Standardized evaluation methodology and reference database for evaluating coronary artery centerline extraction algorithms. Med. Image Anal. 2009;13(5):701–714. doi: 10.1016/j.media.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hameeteman K, et al. Evaluation framework for carotid bifurcation lumen segmentation and stenosis grading. Med. Image Anal. 2011;15(4):477–488. doi: 10.1016/j.media.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Heimann T, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imag. 2009 Aug;28(8):1251–1265. doi: 10.1109/TMI.2009.2013851. [DOI] [PubMed] [Google Scholar]
- 65.Van Ginneken B, Heimann T, Styner M. 3D segmentation in the clinic: A grand challenge. 3D Segmentation Clinic: A Grand Challenge. 2007:7–15. [Google Scholar]
- 66.Niemeijer M, et al. Retinopathy online challenge: Automatic detection of microaneurysms in digital color fundus photographs. IEEE Trans. Med. Imag. 2010 Jan;29(1):185–195. doi: 10.1109/TMI.2009.2033909. [DOI] [PubMed] [Google Scholar]
- 67.Lo P, et al. Extraction of airways from CT (EXACT'09) IEEE Trans. Med. Imag. 2012 Nov;31(11):2093–2107. doi: 10.1109/TMI.2012.2209674. [DOI] [PubMed] [Google Scholar]
- 68.Eskildsen SF, et al. BEaST: Brain extraction based on nonlocal segmentation technique. Neuroimage. 2012;59(3):2362–2373. doi: 10.1016/j.neuroimage.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 69.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imag. 2004 Jul;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou KH, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 2004;11(2):178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Archip N, Jolesz FA, Warfield SK. A validation framework for brain tumor segmentation. Acad. Radiol. 2007;14(10):1242–1251. doi: 10.1016/j.acra.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 72.Hamamci A, Kucuk N, Karaman K, Engin K, Unal G. Tumorcut: Segmentation of brain tumors on contrast enhanced MR images for radiosurgery applications. IEEE Trans. Med. Imag. 2012 Mar;31(3):790–804. doi: 10.1109/TMI.2011.2181857. [DOI] [PubMed] [Google Scholar]
- 73.Menze BH, et al. A generative model for brain tumor segmentation in multi-modal images. Proc. MICCAI. 2010:151–159. doi: 10.1007/978-3-642-15745-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ibanez L, et al. The ITK Software Guide. Clifton Park, NY: Kitware; 2003. [Google Scholar]
- 75.Bauer S, Fejes T, Reyes M. A skull-stripping filter for ITK. Insight J. 2012:70–78. [Google Scholar]
- 76.Prastawa M, Bullitt E, Gerig G. Simulation of brain tumors in MR images for evaluation of segmentation efficacy. Med. Image Anal. 2009;13:297–311. doi: 10.1016/j.media.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clatz O, et al. Brain tumor growth simulation. Tech. Rep. 2004 [Google Scholar]
- 78.Cocosco CA, Kollokian V, Kwan RK-S, Pike GB, Evans AC. Brainweb: Online interface to a 3D MRI simulated brain database. NeuroImage. 1997:301–307. [Google Scholar]
- 79.Aubert-Broche B, Griffin M, Pike GB, Evans AC, Collins DL. Twenty new digital brain phantoms for creation of validation image data bases. IEEE Trans. Med. Imag. 2006 Nov;25(11):1410–1416. doi: 10.1109/TMI.2006.883453. [DOI] [PubMed] [Google Scholar]
- 80.Kistler M, Bonaretti S, Pfahrer M, Niklaus R, Büchler P. The virtual skeleton database: An open access repository for biomedical research and collaboration. J Med. Internet Res. 2013;15(11):e245. doi: 10.2196/jmir.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohamed A, Zacharakib EI, Shena D, Davatzikos C. Deformable registration of brain tumor images via a statistical model of tumor-induced deformation. Med. Image Anal. 2006;10:752–763. doi: 10.1016/j.media.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 82.Tustison N, Gee J. N4ITK: Nick's N3 ITK implementation for MRI bias field correction. Insight J. 2010:1–22. [Google Scholar]
- 83.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. 2nd. New York: Springer; 2009. [Google Scholar]
- 84.Bouix S, et al. On evaluating brain tissue classifiers without a ground truth. Neuroimage. 2007 Jul;36:1207–1224. doi: 10.1016/j.neuroimage.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porz N, et al. Multi-modalodal glioblastoma segmentation: Man versus machine. PLOS ONE. 2014;9:e96873. doi: 10.1371/journal.pone.0096873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanbury A, Müller H, Langs G, Menze BH. Cloud-based research infrastructure for evaluation on big data. In: Galis AEA, editor. The Future Internet— Future Internet Assembly. New York: Springer; 2013. pp. 104–114. [Google Scholar]
- 87.Bauer S, et al. Segmentation of brain tumor images based on integrated hierarchical classification and regularization. Proc. MICCAI-BRATS. 2012:32–38. [Google Scholar]
- 88.Komodakis N, Tziritas G, Paragios N. Performance vs computational efficiency for optimizing single and dynamic MRFs: Setting the state of the art with primal-dual strategies. Comput. Vis. Image Understand. 2008;112(1) [Google Scholar]
- 89.Criminisi A, Shotton J, Konukoglu E. Decision forests for classification, regression, density estimation, manifold learning and semi-supervised learning Microsoft. Tech. Rep. 2011 [Google Scholar]
- 90.Rousseau F, Habas PA, Studholme C. A supervised patch-based approach for human brain labeling. IEEE Trans. Med. Imag. 2011 Oct;30(10):1852–1862. doi: 10.1109/TMI.2011.2156806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cabezas M, Oliver A, Lladó X, Freixenet J, Cuadra MB. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Programs Biomed. 2011;104(3):e158–e177. doi: 10.1016/j.cmpb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Yushkevich PA. Multi-atlas segmentation without registration: A supervoxel-based approach. Proc. MICCAI. 2013:535–542. doi: 10.1007/978-3-642-40760-4_67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tong T, Wolz R, Coupé P, Hajnal JV, Rueckert D. Segmentation of MR images via discriminative dictionary learning and sparse coding: Application to hippocampus labeling. NeuroImage. 2013:320–328. doi: 10.1016/j.neuroimage.2013.02.069. [DOI] [PubMed] [Google Scholar]
- 94.Artaechevarria X, Munoz-Barrutia A, Ortiz-de Solorzano C. Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Trans. Med. Imag. 2009 Aug;28(8):1266–1277. doi: 10.1109/TMI.2009.2014372. [DOI] [PubMed] [Google Scholar]
- 95.Coupé P, et al. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 96.Wang H, Suh JW, Pluta J, Altinay M, Yushkevich P. Optimal weights for multi-atlas label fusion. Proc. Inf. Process. Med. Imag. 2011:73–84. doi: 10.1007/978-3-642-22092-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu G, et al. Minimizing joint risk of mislabeling for iterative patch-based label fusion. Proc. MICCAI. 2013:551–558. doi: 10.1007/978-3-642-40760-4_69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zikic D, et al. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Proc. MICCAI. 2012:369–376. doi: 10.1007/978-3-642-33454-2_46. [DOI] [PubMed] [Google Scholar]
- 99.Chan SH, Khoshabeh R, Gibson KB, Gill PE, Nguyen TQ. An augmented Lagrangian method for total variation video restoration. IEEE Trans. Image Process. 2011 Nov;20(11):3097–3111. doi: 10.1109/TIP.2011.2158229. [DOI] [PubMed] [Google Scholar]
- 100.Besag J. On the statistical analysis of dirty pictures. JR Stat. Soc. 1986:259–302. [Google Scholar]
- 101.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. JR Stat. Soc. 1977;39:1–38. [Google Scholar]
- 102.Celeux G, Forbes F, Peyrard N. EM procedures using mean field-like approximations for Markov model-based image segmentation. Pattern Recognit. 2003;36(1):131–144. [Google Scholar]
- 103.Tustison NJ, et al. N4ITK: Improved n3 bias correction. IEEE Trans. Med. Imag. 2010 Jun;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imag. 2000 Feb;19(2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 105.Laws KI. International Society for Optics and Photonics, Rapid texture identification; Proc. 24th Annu. Tech. Symp; 1980. pp. 376–381. [Google Scholar]
- 106.Bauer S, Nolte L-P, Reyes M. Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. Proc. MICCAI. 2011;14 doi: 10.1007/978-3-642-23626-6_44. [DOI] [PubMed] [Google Scholar]
- 107.Ahmed S, Iftekharuddin KM, Vossough A. Efficacy of texture, shape, and intensity feature fusion for posterior-fossa tumor segmentation in MRI. IEEE Trans. Inf. Technol. Biomed. 2011 Feb;15(2):206–213. doi: 10.1109/TITB.2011.2104376. [DOI] [PubMed] [Google Scholar]
- 108.Islam A, Reza SMS, Iftekharuddin KM. Multi-fractal texture estimation for detection and segmentation of brain tumors. IEEE Trans. Biomed. Eng. 2013 Nov;60(11):3204–3215. doi: 10.1109/TBME.2013.2271383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leung T, Malik J. Representing and recognizing the visual appearance of materials using three-dimensional textons. Int. J. Comput. Vis. 2001;43(1):29–44. [Google Scholar]
- 110.Breiman L. Random forests. Mach. Learn. J. 2001;45:5–32. [Google Scholar]
- 111.Shin H-C, Orton M, Collins DJ, Doran S, Leach MO. Autoencoder in time-series analysis for unsupervised tissues characterisation in a large unlabelled medical image dataset. Proc. IEEE 10th Int. Conf. Mach. Learn. Appl. Workshop. 2011;1:259–264. [Google Scholar]
- 112.Shin H-C. Hybrid clustering and logistic regression for multi-modal brain tumor segmentation. Proc. Workshops Challanges MICCAI. 2012 [Google Scholar]
- 113.Shin H-C, Orton MR, Collins DJ, Doran SJ, Leach MO. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. IEEE Trans. Pattern Anal. Mach. Intell. 2013 Aug;35(8):1930–1943. doi: 10.1109/TPAMI.2012.277. [DOI] [PubMed] [Google Scholar]
- 114.Krizhevsky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional neural networks. Proc. Neural Inf. Process. Syst. 2012:1106–1114. [Google Scholar]
- 115.Subbanna N, Precup D, Collins L, Arbel T. Hierarchical probabilistic Gabor and MRF segmentation of brain tumours in MRI volumes. Proc. MICCAI. 2013;8149:751–758. doi: 10.1007/978-3-642-40811-3_94. [DOI] [PubMed] [Google Scholar]
- 116.Subbanna N, Arbel T. Probabilistic Gabor and Markov random fields segmentation of brain tumours in MRI volumes. Proc. MICCAI Brain Tumor Segmentat. Challenge. 2012:28–31. doi: 10.1007/978-3-642-40811-3_94. [DOI] [PubMed] [Google Scholar]
- 117.Sled J, Pike G. Correction for B(1) and B(0) variations in quantitative T2 measurements using MRI. Mag. Reson. Med. 2000;43(4):589–593. doi: 10.1002/(sici)1522-2594(200004)43:4<589::aid-mrm14>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 118.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardised talairach space. J Comp. Assist. Tom. 1994;18:192–205. [PubMed] [Google Scholar]
- 119.Belaroussia B, Millesb J, Carmec S, Zhua H, Benoit-Cattina YM. Intensity non-uniformity correction in MRI: Existing methods and their validation. Med. Image Anal. 2006;10(2):234–246. doi: 10.1016/j.media.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 120.Subbanna N, Zeevi Y. Existence conditions for discrete noncanonical multiwindow Gabor schemes. IEEE Trans. Signal Process. 2007 Oct;55(10):5113–5117. [Google Scholar]
- 121.Ince DC, Hatton L, Graham-Cumming J. The case for open computer programs. Nature. 2012 Feb;482(7386):485–488. doi: 10.1038/nature10836. [DOI] [PubMed] [Google Scholar]
- 122.Avants BB, et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010 Feb;49(3):2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Landman BA, et al. Multi-parametric neuroimaging reproducibility: A 3-T resource study. Neuroimage. 2011 Feb;54(4):2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics. 2011 Dec;9(4):381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Avants BB, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011 Feb;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boykov Y, Kolmogorov V. An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. Pattern Anal. Mach. Intell. 2004 Sep;26(9):1124–1137. doi: 10.1109/TPAMI.2004.60. [DOI] [PubMed] [Google Scholar]
- 127.Kolmogorov V, Zabih R. What energy functions can be minimized via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2004 Dec;26(12):147–159. doi: 10.1109/TPAMI.2004.1262177. [DOI] [PubMed] [Google Scholar]
- 128.Boykov RY, Veksler O. Efficient approximate energy minimization via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2001 Dec;20(12):1222–1239. [Google Scholar]
- 129.Smith SM, Brady JM. Susan—A new approach to low level image processing. Int. J. Comput. Vis. 1997;23:45–78. [Google Scholar]
- 130.Achanta R, et al. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012 Nov;34(11):2274–2282. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 131.Boykov Y, Veksler O, Zabih R. Efficient approximate energy minimization via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2001 Nov;23(11):1222–1239. [Google Scholar]
- 132.Shotton J, et al. Real-time human pose recognition in parts from a single depth image; Proc. IEEE Conf. Comput. Vis. Pattern Recongnit; 2011. Jun, pp. 1297–1304. [Google Scholar]
- 133.Viola P, Jones MJ. Robust real-time face detection. Int. J. Comput. Vis. 2004;57(2):137–154. [Google Scholar]