Abstract
This article provides NODDI diffusion metrics in the brains of 52 healthy participants and computer simulation data to support compatibility of hybrid diffusion imaging (HYDI), “Hybrid diffusion imaging”[1] acquisition scheme in fitting neurite orientation dispersion and density imaging (NODDI) model, “NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain”[2]. HYDI is an extremely versatile diffusion magnetic resonance imaging (dMRI) technique that enables various analyzes methods using a single diffusion dataset. One of the diffusion data analysis methods is the NODDI computation, which models the brain tissue with three compartments: fast isotropic diffusion (e.g., cerebrospinal fluid), anisotropic hindered diffusion (e.g., extracellular space), and anisotropic restricted diffusion (e.g., intracellular space). The NODDI model produces microstructural metrics in the developing brain, aging brain or human brain with neurologic disorders. The first dataset provided here are the means and standard deviations of NODDI metrics in 48 white matter region-of-interest (ROI) averaging across 52 healthy participants. The second dataset provided here is the computer simulation with initial conditions guided by the first dataset as inputs and gold standard for model fitting. The computer simulation data provide a direct comparison of NODDI indices computed from the HYDI acquisition [1] to the NODDI indices computed from the originally proposed acquisition [2]. These data are related to the accompanying research article “Age Effects and Sex Differences in Human Brain White Matter of Young to Middle-Aged Adults: A DTI, NODDI, and q-Space Study” [3].
Keywords: NODDI, HYDI, Diffusion, Magnetic Resonance Imaging, White matter
Specifications Table
| Subject area | Neuroimaging |
| More specific subject area | Diffusion magnetic resonance imaging |
| Type of data | Figure, table |
| How data was acquired | Hybrid diffusion imaging (HYDI) at 3 T MRI scanner and computer simulation |
| Data format | Analyzed |
| Experimental factors | 48 white matter ROIs in the human brain and synthetic diffusion signals |
| Experimental features | Diffusion microstructural metrics |
| Data source location | University of Wisconsin - Madison |
| Data accessibility | Data is with this article |
Value of the data
-
•
The experimental data could serve as bench marker for studies on white matter microstructural measurements using either imaging or histologic approaches.
-
•
This simulation data could serve as a reference for future studies that use HYDI acquisition for NODDI computation.
-
•
This simulation approach could be applied on future studies to design acquisition schemes and investigate the schemes’ compatibility for diffusion compartment modeling.
1. Data
The experimental data (Table 1) include NODDI metrics (i.e., orientation dispersion index (ODI) and intercellular volume fraction (ICVF)) and other diffusion metrics for comparison such as DTI (i.e., axial diffusivity (AD), radial diffusivity (RD), mean diffusivity (MD), and factional anisotropy (FA)) and q-space analysis (i.e., zero-displacement probability (P0)). The means and standard deviations of diffusion metrics in 48 white matter ROIs were computed across 52 healthy participants with age between 18 and 72 years old (mean 39±14).
Table 1.
Mean and standard deviation (std) of the diffusion metrics in 48 white matter ROIs [4] across 52 subjects.
|
Da (10−6mm2/s) |
Dr (10−6mm2/s) |
MD (10−6mm2/s) |
FA |
Po |
ODI |
ICVF |
FISO |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ROI | Mean | Std | Mean | Std | Mean | Std | Mean | Std | Mean | Std | Mean | Std | Mean | Std | Mean | Std |
| ACR-L | 840 | 40 | 392 | 4 | 541 | 36 | 0.470 | 0.033 | 0.463 | 0.042 | 0.292 | 0.023 | 0.655 | 0.057 | 0.149 | 0.033 |
| ACR-R | 848 | 37 | 395 | 4 | 546 | 34 | 0.467 | 0.035 | 0.457 | 0.039 | 0.285 | 0.024 | 0.656 | 0.055 | 0.160 | 0.029 |
| ALIC-L | 858 | 45 | 316 | 3 | 497 | 29 | 0.556 | 0.038 | 0.473 | 0.038 | 0.266 | 0.031 | 0.676 | 0.050 | 0.111 | 0.022 |
| ALIC-R | 876 | 51 | 322 | 3 | 506 | 32 | 0.556 | 0.037 | 0.461 | 0.038 | 0.260 | 0.026 | 0.682 | 0.053 | 0.124 | 0.025 |
| BCC | 1184 | 64 | 380 | 8 | 648 | 70 | 0.628 | 0.054 | 0.510 | 0.056 | 0.141 | 0.016 | 0.712 | 0.038 | 0.230 | 0.047 |
| CGC-L | 872 | 41 | 348 | 2 | 522 | 23 | 0.521 | 0.028 | 0.468 | 0.036 | 0.242 | 0.026 | 0.608 | 0.046 | 0.090 | 0.027 |
| CGC-R | 844 | 39 | 369 | 2 | 527 | 21 | 0.485 | 0.032 | 0.443 | 0.035 | 0.265 | 0.030 | 0.581 | 0.048 | 0.085 | 0.026 |
| CGH-L | 855 | 54 | 469 | 5 | 598 | 45 | 0.388 | 0.031 | 0.322 | 0.026 | 0.344 | 0.027 | 0.502 | 0.045 | 0.112 | 0.050 |
| CGH-R | 852 | 44 | 442 | 3 | 579 | 32 | 0.412 | 0.031 | 0.338 | 0.026 | 0.337 | 0.025 | 0.507 | 0.043 | 0.094 | 0.041 |
| CP-L | 1007 | 57 | 269 | 3 | 515 | 38 | 0.682 | 0.038 | 0.621 | 0.073 | 0.190 | 0.017 | 0.768 | 0.041 | 0.148 | 0.027 |
| CP-R | 1049 | 63 | 293 | 5 | 545 | 48 | 0.671 | 0.043 | 0.590 | 0.067 | 0.187 | 0.017 | 0.775 | 0.042 | 0.171 | 0.031 |
| CST-L | 883 | 83 | 332 | 7 | 516 | 72 | 0.584 | 0.042 | 0.686 | 0.134 | 0.208 | 0.019 | 0.769 | 0.053 | 0.181 | 0.049 |
| CST-R | 887 | 85 | 333 | 7 | 517 | 68 | 0.587 | 0.040 | 0.685 | 0.144 | 0.208 | 0.023 | 0.772 | 0.053 | 0.184 | 0.047 |
| EC-L | 835 | 45 | 399 | 5 | 544 | 44 | 0.455 | 0.034 | 0.379 | 0.033 | 0.320 | 0.026 | 0.531 | 0.037 | 0.075 | 0.037 |
| EC-R | 844 | 39 | 410 | 4 | 555 | 35 | 0.444 | 0.031 | 0.364 | 0.028 | 0.317 | 0.021 | 0.528 | 0.034 | 0.087 | 0.032 |
| Fx | 1716 | 141 | 830 | 16 | 1126 | 153 | 0.477 | 0.055 | 0.231 | 0.050 | 0.168 | 0.033 | 0.770 | 0.077 | 0.622 | 0.091 |
| Fx-ST-L | 1025 | 73 | 448 | 7 | 641 | 71 | 0.510 | 0.040 | 0.386 | 0.057 | 0.236 | 0.020 | 0.593 | 0.045 | 0.214 | 0.065 |
| Fx-ST-R | 1092 | 73 | 500 | 7 | 697 | 68 | 0.486 | 0.038 | 0.347 | 0.051 | 0.244 | 0.024 | 0.579 | 0.042 | 0.262 | 0.063 |
| GCC | 1325 | 93 | 530 | 8 | 795 | 83 | 0.575 | 0.033 | 0.411 | 0.045 | 0.180 | 0.018 | 0.722 | 0.047 | 0.336 | 0.054 |
| ICP-L | 817 | 46 | 340 | 3 | 499 | 27 | 0.526 | 0.038 | 0.569 | 0.032 | 0.248 | 0.028 | 0.683 | 0.038 | 0.125 | 0.039 |
| ICP-R | 810 | 46 | 322 | 3 | 484 | 27 | 0.539 | 0.039 | 0.590 | 0.037 | 0.247 | 0.030 | 0.683 | 0.038 | 0.105 | 0.032 |
| MCP | 921 | 90 | 362 | 6 | 549 | 66 | 0.569 | 0.045 | 0.662 | 0.107 | 0.220 | 0.028 | 0.809 | 0.050 | 0.204 | 0.045 |
| ML-L | 884 | 64 | 288 | 3 | 487 | 22 | 0.615 | 0.048 | 0.575 | 0.035 | 0.180 | 0.024 | 0.642 | 0.029 | 0.097 | 0.028 |
| ML-R | 900 | 75 | 287 | 2 | 491 | 24 | 0.625 | 0.047 | 0.571 | 0.031 | 0.175 | 0.023 | 0.638 | 0.031 | 0.096 | 0.031 |
| PCR-L | 863 | 79 | 397 | 6 | 552 | 65 | 0.494 | 0.032 | 0.530 | 0.049 | 0.224 | 0.016 | 0.628 | 0.053 | 0.117 | 0.046 |
| PCR-R | 847 | 55 | 378 | 4 | 534 | 44 | 0.503 | 0.029 | 0.528 | 0.044 | 0.230 | 0.017 | 0.616 | 0.048 | 0.101 | 0.033 |
| PCT | 767 | 44 | 328 | 3 | 474 | 25 | 0.514 | 0.036 | 0.598 | 0.032 | 0.254 | 0.026 | 0.713 | 0.043 | 0.102 | 0.034 |
| PLIC-L | 860 | 32 | 259 | 2 | 460 | 22 | 0.643 | 0.027 | 0.646 | 0.032 | 0.209 | 0.018 | 0.743 | 0.036 | 0.105 | 0.020 |
| PLIC-R | 875 | 35 | 264 | 2 | 468 | 23 | 0.640 | 0.027 | 0.629 | 0.027 | 0.205 | 0.016 | 0.736 | 0.033 | 0.111 | 0.021 |
| PTR-L | 1072 | 99 | 418 | 9 | 636 | 89 | 0.580 | 0.038 | 0.488 | 0.051 | 0.161 | 0.022 | 0.634 | 0.053 | 0.191 | 0.064 |
| PTR-R | 962 | 64 | 332 | 5 | 542 | 48 | 0.608 | 0.036 | 0.548 | 0.063 | 0.180 | 0.022 | 0.656 | 0.057 | 0.132 | 0.035 |
| RLIC-L | 904 | 54 | 318 | 4 | 513 | 37 | 0.600 | 0.031 | 0.556 | 0.040 | 0.199 | 0.021 | 0.686 | 0.038 | 0.129 | 0.032 |
| RLIC-R | 905 | 44 | 328 | 3 | 520 | 29 | 0.588 | 0.030 | 0.540 | 0.037 | 0.211 | 0.019 | 0.681 | 0.040 | 0.135 | 0.031 |
| SCC | 1102 | 42 | 236 | 3 | 525 | 30 | 0.749 | 0.027 | 0.666 | 0.042 | 0.122 | 0.013 | 0.764 | 0.038 | 0.125 | 0.027 |
| SCP-L | 1316 | 106 | 534 | 8 | 795 | 82 | 0.579 | 0.033 | 0.454 | 0.036 | 0.154 | 0.026 | 0.724 | 0.041 | 0.329 | 0.057 |
| SCP-R | 1243 | 91 | 457 | 5 | 719 | 58 | 0.607 | 0.031 | 0.490 | 0.029 | 0.145 | 0.024 | 0.718 | 0.039 | 0.295 | 0.048 |
| SCR-L | 760 | 35 | 337 | 3 | 478 | 30 | 0.501 | 0.030 | 0.595 | 0.039 | 0.268 | 0.016 | 0.689 | 0.044 | 0.103 | 0.024 |
| SCR-R | 760 | 29 | 344 | 2 | 483 | 22 | 0.488 | 0.025 | 0.586 | 0.033 | 0.273 | 0.018 | 0.692 | 0.041 | 0.114 | 0.026 |
| SFO-L | 812 | 73 | 361 | 6 | 511 | 64 | 0.500 | 0.038 | 0.499 | 0.057 | 0.286 | 0.029 | 0.661 | 0.061 | 0.117 | 0.047 |
| SFO-R | 810 | 49 | 361 | 4 | 510 | 35 | 0.491 | 0.037 | 0.483 | 0.045 | 0.282 | 0.032 | 0.645 | 0.069 | 0.112 | 0.038 |
| SLF-L | 745 | 30 | 332 | 2 | 470 | 23 | 0.494 | 0.025 | 0.605 | 0.036 | 0.269 | 0.018 | 0.699 | 0.040 | 0.096 | 0.021 |
| SLF-R | 763 | 29 | 344 | 2 | 484 | 20 | 0.489 | 0.023 | 0.571 | 0.035 | 0.275 | 0.020 | 0.693 | 0.042 | 0.107 | 0.025 |
| SS-L | 967 | 73 | 419 | 5 | 602 | 52 | 0.511 | 0.029 | 0.427 | 0.037 | 0.225 | 0.026 | 0.594 | 0.041 | 0.172 | 0.047 |
| SS-R | 951 | 57 | 387 | 3 | 575 | 36 | 0.536 | 0.028 | 0.447 | 0.039 | 0.237 | 0.028 | 0.635 | 0.049 | 0.167 | 0.032 |
| TAP-L | 2053 | 266 | 1328 | 25 | 1569 | 251 | 0.329 | 0.048 | 0.157 | 0.055 | 0.179 | 0.042 | 0.819 | 0.079 | 0.758 | 0.118 |
| TAP-R | 1788 | 190 | 1026 | 17 | 1280 | 176 | 0.414 | 0.054 | 0.230 | 0.064 | 0.143 | 0.028 | 0.759 | 0.089 | 0.624 | 0.123 |
| UNC-L | 864 | 66 | 438 | 6 | 580 | 56 | 0.419 | 0.048 | 0.329 | 0.037 | 0.306 | 0.052 | 0.455 | 0.033 | 0.061 | 0.047 |
| UNC-R | 926 | 76 | 464 | 8 | 618 | 72 | 0.425 | 0.055 | 0.303 | 0.045 | 0.296 | 0.053 | 0.474 | 0.054 | 0.116 | 0.103 |
The HYDI acquisition scheme includes 5 shells with different diffusion-weighting b-value and each shell has homogeneously distributed diffusion-weighting directions [1]. Details of the HYDI diffusion-encoding scheme are shown in Table 2. We simulated different combinations of HYDI shells to compute NODDI diffusion indices. The HYDI shell combination protocols are: p12, p123, p1234, and p12345 (Table 3). The p12 protocol comprises the first and second HYDI shell with b-value of 375 and 1500 s/mm2, respectively. The p123 protocol comprises the 1st, 2nd and 3rd HYDI shell. Similarly, p1234 comprises the 1st to 4th HYDI shell and p12345 has all 5 shells. The NODDI-p14 is the diffusion-encoding scheme recommended by [2]. The NODDI diffusion model yields microstructural indices including the intracellular volume fraction (ICVF), fiber orientation dispersion index (ODI), and free water volume fraction (FISO). The estimates of these microstructural indices will be compared between the HYDI protocols and the NODDI-p14.
Table 2.
HYDI shells, number of diffusion encoding directions (Ne), and the corrosponding b-values.
| HYDI shell | Ne | b-value (s/mm2) |
|---|---|---|
| 1 | 0 | |
| 1st | 6 | 375 |
| 2nd | 21 | 1500 |
| 3rd | 24 | 3375 |
| 4th | 24 | 6000 |
| 5th | 50 | 9375 |
| total | 126 |
Table 3.
Diffusion encoding protocols
| Protocol | b-value(s/mm2), (number of diffusion directions) |
|---|---|
| NODDI-p14 | b=711(30), b=2855(60) |
| p12 | b=375(6), b=1500(21) |
| p123 | b=375(6), b=1500(21), b=3375(24) |
| p1234 | b=375(6), b=1500(21), b=3375(24), b=6000(24) |
| p12345 | b=375(6), b=1500(21), b=3375(24), b=6000(24), b=9375(50) |
2. Experimental design, materials, and methods
The 48 white matter ROIs of the experimental data in Table 1 were defined through intersecting the white matter atlas, Johns Hopkins University (JHU) ICBM-DTI-81 [4] with the common white matter skeleton created from all of the subjects similar to [3]. The diffusion metrics of NODDI, DTI, and q-space were averaged within the ROIs and then across the 52 participants. The standard deviations reflect the variations of ROI means across the participants.
The tissue diffusion properties were simulated using SynthMeas.m provided in the NODDI Matlab toolbox (http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab). According to the experimental data in Table 1, we designed the ground truth and initial inputs for computer simulation: ICVF = [0.2, 0.4, 0.5, 0.8]; kappa = [0, 0.25, 1, 4, 16]; and FISO=0. Similar design can also be found in [2]. The kappa is the Watson distribution parameter that determines the estimation of ODI [5]. The intracellular axial diffusivity is set as 1.7×10−3 mm2/s, and the fast free isotropic diffusivity representing cerebrospinal fluid is set as 3×10−3 mm2/s similar to free water diffusion. There were 30 random trials at each of the 250 different fiber orientations uniformly distributed on a unit sphere (Fig. 1). Five signal-to-noise ratios (SNR) defined at b-value =0 s/mm2 was simulated: SNRb0=[20, 30, 40, 50, Infinity]. The 4 b0 SNRs yield corresponding SNRs across the 50 diffusion directions in the outermost shell, SNRb=9375=[2.22±0.48, 3.71±0.81, 4.45±0.97, 5.56±1.21], respectively. Therefore, SNRb0=20 yields the outermost-shell SNRb=9375 (2.22±0.48) that is closest to the reported measurement (i.e., 2.85±0.71) in the human brain published in [1].
Fig. 1.
The vertices represent the fiber orientations on the unit spherical surface. There are 250 uniformly distributed vertices in this figure.
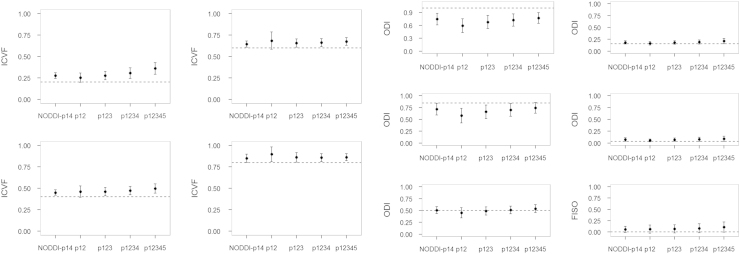
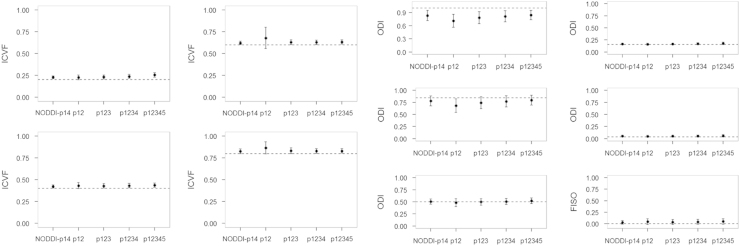
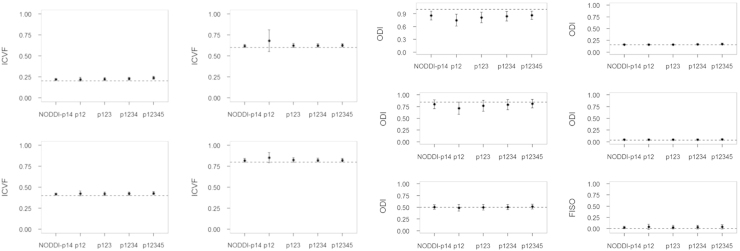
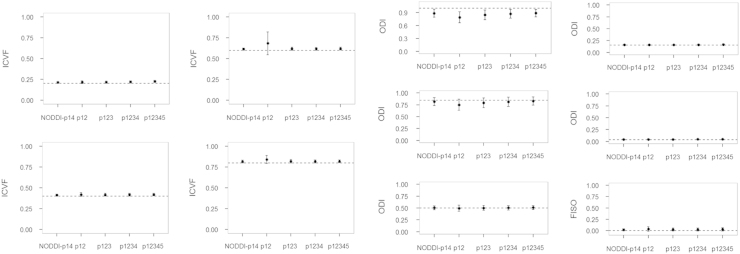
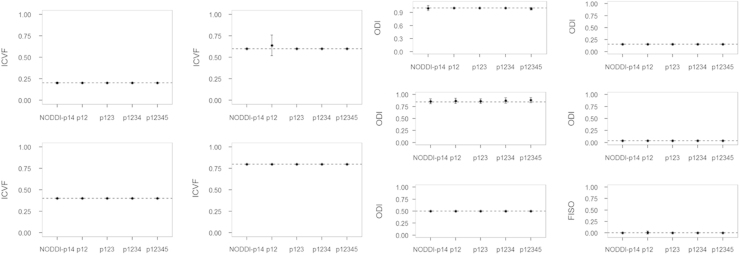
The simulation results are shown in Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6. For all of the ICVF and kappa conditions, the NODDI-p14 and HYDI p12345 schemes did not differ significantly from each other (p-value > 0.05). At SNRb0=20, HYDI p12345 overestimates low ICVFs more often than NODDI-p14, but both are comparable at a high ICVF (>0.5), which is a more realistic value for white matter listed in Table 1. Both schemes underestimate high ODIs, but work fine at a low ODI (<=0.5), which is again more realistic for white matter listed in Table 1. As the SNR increases, both NODDI-p14 and HYDI eventually reach the ground truth. Between the combinations of HYDI shells, p12 tends to underestimate ODI in high ODI tissue model at low SNR and also overestimate ICVF in high ICVF. Other combinations p123, p1234, and p12345 were comparable across simulated tissue properties and SNRs.
Fig. 2.
Simulated results for SNR=20. Dashed lines denote the ground truth of simulated diffusion properties (i.e., ICVF, ODI and FISO). The dot denotes the mean of the specific diffusion property estimated using NODDI-p14 and HYDI (p12, p123, p1234, and p12345) schemes. The error bar denotes the standard deviation across the 30 random trials, 250 fiber directions and the rest diffusion properties.
Fig. 3.
Simulated results for SNR=30. Dashed lines denote the ground truth of simulated diffusion properties (i.e., ICVF, ODI and FISO). The dot denotes the mean of the specific diffusion property estimated using NODDI-p14 and HYDI (p12, p123, p1234, and p12345) schemes. The error bar denotes the standard deviation across the 30 random trials, 250 fiber directions and the rest diffusion properties.
Fig. 4.
Simulated results for SNR=40. Dashed lines denote the ground truth of simulated diffusion properties (i.e., ICVF, ODI and FISO). The dot denotes the mean of the specific diffusion property estimated using NODDI-p14 and HYDI (p12, p123, p1234, and p12345) schemes. The error bar denotes the standard deviation across the 30 random trials, 250 fiber directions and the rest diffusion properties.
Fig. 5.
Simulated results for SNR=50. Dashed lines denote the ground truth of simulated diffusion properties (i.e., ICVF, ODI and FISO). The dot denotes the mean of the specific diffusion property estimated using NODDI-p14 and HYDI (p12, p123, p1234, and p12345) schemes. The error bar denotes the standard deviation across the 30 random trials, 250 fiber directions and the rest diffusion properties.
Fig. 6.
Simulated results for SNR = infinity (i.e., noise=0). Dashed lines denote the ground truth of simulated diffusion properties (i.e., ICVF, ODI and FISO). The dot denotes the mean of the specific diffusion property estimated using NODDI-p14 and HYDI (p12, p123, p1234, and p12345) schemes. The error bar denotes the standard deviation across the 30 random trials, 250 fiber directions and the rest diffusion properties.
Acknowledgments
The authors would like to thank Dr. Hui (Gary) Zhang for helpful discussions. This work was funded in part by NIH R21 NS075791.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.dib.2016.03.063.
Appendix A. Supplementary material
Supplementary material
References
- 1.Wu Y.C., Alexander A.L. Hybrid diffusion imaging. Neuroimage. 2007;36(3):617–629. doi: 10.1016/j.neuroimage.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- 3.Kodiweera C. Age effects and sex differences in human brain white matter of young to middle-aged adults: a DTI, NODDI, and q-space study. NeuroImage. 2016;128:180–192. doi: 10.1016/j.neuroimage.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oishi K., Zilles K., Amunts K., Faria A., Jiang H., Li X., Akhter K., Hua K., Woods R., Toga A.W., Pike G.B., Rosa-Neto P., Evans A., Zhang J., Huang H., Miller M.I., vanZijl P.C., Mazziotta J., Moris S. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43(3):447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardia K.V., Jupp P.E. xxi. J. Wiley; Chichester; New York: 2000. Directional statistics; p. 429. (Wiley series in probability and statistics). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material