Abstract
Aims
The aim of the study was to investigate the combined impact of genetic polymorphisms in key pharmacokinetic genes on plasma concentrations and clinical outcomes of cyclophosphamide (CPA) in Chinese patients with systemic lupus erythematosus (SLE).
Methods
One hundred and eighty nine Chinese SLE patients treated with CPA induction therapy (200 mg, every other day) were recruited and adverse reactions were recorded. After 4 weeks induction therapy, 128 lupus nephritis (LN) patients continued to CPA maintenance therapy (200–600 mg week–1) for 6 months, and their clinical outcomes were recorded. Blood samples were collected for CYP2C19, CYP2B6, GST and PXR polymorphism analysis, as well as CPA and its active metabolite (4‐hydroxycyclophosphamide (4‐OH‐CPA)) plasma concentration determination.
Results
Multiple linear regression analysis revealed that CYP2B6 ‐750 T > C (P < 0.001), −2320 T > C (P < 0.001), 15582C > T (P = 0.017), CYP2C19*2 (P < 0.001) and PXR 66034 T > C (P = 0.028) accounted for 47% of the variation in 4‐OH‐CPA plasma concentration. Among these variants, CYP2B6 ‐750 T > C and CYP2C19*2 were selected as the combination genetic marker because these two SNPs contributed the most to the inter‐individual variability in 4‐OH‐CPA concentration, accounting for 23.6% and 21.5% of the variation, respectively. Extensive metabolizers (EMs) (CYP2B6 ‐750TT, CYP2C19*1*1) had significantly higher median 4‐OH‐CPA plasma concentrations (34.8, 11.0 and 6.6 ng ml‐1 for EMs, intermediate metabolizers (IMs) and poor metabolizers (PMs), P < 0.0001), higher risks of leukocytopenia (OR = 7.538, 95% CI 2.951, 19.256, P < 0.0001) and gastrointestinal toxicity (OR = 7.579, 95% CI 2.934, 19.578, P < 0.0001), as well as shorter median time to achieve complete remission (13.2, 18.3 and 23.3 weeks for EMs, IMs and PMs, respectively, P = 0.026) in LN patients than PMs (CYP2B6 ‐750CC, CYP2C19*2*2) and IMs.
Conclusions
Our findings have indicated that genetic markers of drug metabolizing enzymes could predict the 4‐hydroxylation, adverse reactions and clinical efficacy of CPA. This is a necessary first step towards building clinical tools that will help assess clinical benefit and risk before undergoing CPA treatment in Chinese SLE patients.
Keywords: cyclophosphamide, CYP2B6, CYP2C19, lupus, pharmacogenetics
What is Already Known About this Subject
Genetic polymorphisms of CYP2B6 and CYP2C19 are known to influence cyclophosphamide (CPA) efficacy and side effects.
CYP3A4 is another enzyme that can metabolize CPA. Polymorphism is rare in the Chinese population although activity and expression are highly varied. Recent studies indicated that PXR plays an important role in CYP3A4 regulation and the change of PXR function can affect CYP3A4 expression.
The contribution of genetic polymorphism to the variability in CPA efficacy and side effects in Caucasians has been studied. However, the effect of genetic polymorphism on CPA efficacy and side effects in Chinese SLE patients remains to be investigated.
What this Study Adds
This research investigated the combined effect of key metabolizing enzyme variants on CPA plasma concentration, efficacy and side effects in Chinese SLE patients.
This study developed a multiple regression model including CYP2B6, CYP2C19 and PXR genotypes that explained 47.9% of the individual variability in CPA 4‐hydroxylation. This is the first study on the effect of PXR genetic polymorphisms on CPA pharmacokinetics and side effects. PXR 66034 T > C was responsible for 3.7% of the inter‐individual variation of 4‐OH‐CPA plasma concentration.
This is the first determination of the frequency of SNPs in the CYP2B6 non‐coding region in Chinese population.
Introduction
Cyclophosphamide (CPA) is widely prescribed for the treatment of cancer and auto‐immune diseases such as systemic lupus erythematosus (SLE) 1. Like all cytotoxic agents, the toxic metabolites of CPA enter normal tissues including the GI tract and bone marrow, where they induce host organ injuries in many patients 2. CPA‐based regimens for SLE patients often cause short term toxicity, such as myelosuppression, gastrointestinal (GI) symptoms (e.g. vomiting and diarrhoea) and infection due to marked suppression of the immune system. The usual dose‐limiting toxicity for CPA is myelosuppression 3.
CPA is a prodrug, which requires the activation via 4‐hydroxycyclophosphamide (4‐OH‐CPA) to phosphoramide mustard. The 4‐hydroxylation of CPA is catalyzed by cytochrome P450 (CYP) enzymes, while the formation of phosphoramide mustard is non‐enzymatic. Phosphoramide mustard is the alkylating agent responsible for therapeutic effects and toxicity such as immunosuppression, which usually manifests as myelosuppression, GI symptoms (e.g. vomiting and diarrhoea) and infection 3. Several isozymes of P450 are reported to be involved in the 4‐hydroxylation of CPA, including CYP2B6 4, CYP3A4/5 5, CYP2C19 6, 7 and CYP2C9 6. Conjugation of phosphoramide mustard and other CPA metabolites with glutathione is catalyzed by glutathione S‐transferases (GST) 3, which is the detoxification mechanism of CPA metabolites.
The CPA metabolizing enzymes are genetically polymorphic and are known to have variant alleles with diminished 8, 9, 10 or enhanced 4 metabolic activity of the expressed proteins. Considerable inter‐individual variation in 4‐OH‐CPA plasma concentrations has also been demonstrated. However the role of pharmacogenetics is still controversial and it is unclear which metabolizing genes are important. For example, it has been reported that CYP2C19*2 was related to decreased 4‐OH‐CPA formation 11 especially at a CPA dose lower than 1000 mg m–2. 7 Thereby, CYP2C19*2 was associated with worse therapeutic response in lupus nephritis (LN) patients 12 but decreased risks for ovarian toxicities in SLE patients 13, 14. In addition, some mutations of CYP2B6, such as CYP2B6 ‐750 T > C, −2320 T > C, could reduce 4‐OH‐CPA formation and were associated with decreased incidence of adverse reactions 15 or worse therapeutic effects (CYP2B6 1459 C > T (*5)) 12. On the other hand, other mutations such as CYP2B6 516G > T were correlated with higher 4‐OH‐CPA concentration 16. We also reported the correlation of GSTP1 105I > V mutation with the increased risk of leukocytopenia in SLE patients with pulsed CPA treatment 3. Nonetheless, the lack of statistically significant associations between genotypes and CPA pharmacokinetics/response has also been reported 17.
Until now, most CPA pharmacogenomic studies have focused on the separate effect of certain genotypes. Only one study used an in vitro‐in vivo method to determine the combined impact of CYP2C19 and CYP2B6 in a small sample size population 18. Moreover, our previous studies have revealed that CYP2B6, CYP2C19 and GST genes were polymorphic in the Chinese population and the frequency of the mutations was significantly different from other ethnic groups 19, 20. Since the frequency of SNPs in the CYP3A4 gene is sparse in the Chinese population 21, 22, the genetic polymorphisms of CYP3A4 are less likely to cause inter‐individual variation of CYP3A4 activity. However, we have recently shown that genetic polymorphisms of pregnane X receptor (PXR) are associated with CYP3A4 expression 23. Moreover, the combined effect of multiple genetic polymorphisms has not been studied in a larger sample size or in Chinese patients.
In this study, we aimed to investigate the impact of genetic polymorphisms of CYP2C19, CYP2B6, GST and PXR on CPA pharmacokinetics, efficacy and side effects. The genotypes closely correlated with 4‐OH‐CPA plasma concentrations and side effects of CPA were selected as a combination genetic marker to predict 4‐OH‐CPA concentration, efficacy and side effects of CPA.
Methods
Study design
Patient enrolment, sample collection, treatment and follow‐up were performed according to the protocol registered at http://ClinicalTrials.gov (NCT01060410) and approved by the Human Investigation Ethics Committee at the School of Pharmaceutical Sciences at Sun Yat‐sen University, Guangzhou, China (No.200801). Participants were recruited from the Department of Rheumatology, the First Affiliated Hospital of Sun Yat‐sen University, Guangzhou, China. All patients had four or more revised American College of Rheumatology (ACR) criteria for SLE 24. Written informed consent was obtained from patients above 18 years old or their legal guardian for patients under 18 years. Eligibility criteria included age from 12 to 60 years old, HIV negative, aspartate aminotransferase, alanine aminotransferase, total bilirubin and serum creatinine below two times the upper limit of normal and SLE Disease Activity Index (SLEDAI) 25 ≥10. The SLEDAI of each patient was rated by physicians. Patients enrolled in the maintenance period had biopsy‐proven (within 6 months) lupus nephritis (LN) 26. Laboratory tests documented the presence of active nephritis, defined as proteinuria (protein excretion >1 g 24 h–1) or increased serum creatinine level (>1.3 mg dl–1) with active urinary sediment (any of >5 red blood cells/high‐power field, >5 white blood cells/high‐power field, or red blood cell casts in the absence of infection or other causes) in patients with class IV‐S or IV‐G and significant proteinuria (protein excretion >2 g 24 h–1) or increased serum creatinine level (>1.3 mg dl–1) in patients with class III or V 27. Exclusion criteria included taking non‐steroidal anti‐inflammatory drugs, taking or took methotrexate and azathioprine or herbal immuno‐modulators within 2 months, had a blood transfusion within 2 months, pregnant or breast‐feeding, had severe chronic heart, haematological (e.g. leucopenia, thrombocytopenia), renal, brain, liver or lung disease not due to SLE complications, alcoholic, taking or used phenobarbital, rifampicin or allopurinol within 2 months as these drugs may increase the toxicity of CPA due to drug interactions, had a recent vaccination and had received irradiation or cytotoxic therapy in the past 3 months.
Treatments included two parts, induction therapy and maintenance therapy. Patients who underwent induction therapy were intravenously injected (10 min injection) with CPA (Endoxan®, Baxter, China) at 200 mg dissolved in 30 ml normal saline, once every other day. They also received methylprednisolone 40–80 mg intravenously and hydroxychloroquine 0.4 g day–1 orally. CPA and 4‐OH‐CPA plasma concentrations and CPA short term toxicity were investigated in the induction period. After 1 month of induction therapy, patients diagnosed with LN continued maintenance therapy. The protocol of maintenance therapy was intravenous administration of CPA at 200–600 mg every week for 6 months. Patients treated with maintenance therapy were under the observation of short term efficacy of CPA.
Before treatment, blood samples (2 ml) were collected for DNA extraction and as blank plasma samples for CPA/4‐OH‐CPA concentration analysis. Another 2 ml blood sample was collected at 20 h (C 20 h) after the 4th administration of CPA induction therapy and the concentration of CPA and 4‐OH‐CPA was determined by LC‐MS/MS as previously described 28. The reason for choosing the blood sample 20 h (C 20 h) after the 4th administration is supported by previous studies from others 29 and our unpublished data. We have found that the plasma concentration of CPA and 4‐OH‐CPA at this time point was significantly correlated to their respective AUCs (data not shown).
Collection of clinical data
The adverse reaction to treatment was monitored and reported according to the National Cancer Institute Common Terminology Criteria for Adverse Events, version 3.0 (CTCAE v3.0) (http://ctep.cancer.gov/). During induction therapy, toxicities under observation included leukocytopenia, GI toxicity and infection. Leukocytopenia was selected as the indicator of myelosuppression and defined as the total count of white blood cells <4.0 × 109 l–1. The baseline white blood cell count before therapy was recorded. The nadirs of the counts after CPA administration were used to reflect the maximal myelosuppression. If the total white blood cells were less than 1.5 × 109 l–1, CPA therapy was discontinued. The GI toxicities were generally mild and thus were recorded only as positive/negative instead of the grades provided by CTCAE v3.0. The GI toxicity was defined as at least one of the following symptoms being observed: anorexia, diarrhoea (increase of >four stools per day over baseline), nausea and vomiting. Infection incidence included fever and coughs due to pneumonia. After CPA administration, the blood routine tests were taken weekly during the treatment. Patients were followed up for 4 weeks after the initiation of CPA induction therapy to observe acute toxicities. The evaluation of acute toxicities was performed by licensed clinicians.
Short term clinical outcomes were also observed in patients diagnosed with LN under CPA maintenance therapy. The primary end point was complete remission after 6 months of treatment. Secondary end points included response (defined as partial remission), changes in clinical parameters (including proteinuria, serum albumin, serum creatinine and serum C3 values) and adverse effects (including leukopenia, infections, gastrointestinal symptoms, amenorrhea, hair loss, liver function disorder, transient increase in serum creatinine level, etc). The clinical outcomes were defined based on a published study in LN patients 30.
Analysis of plasma CPA and 4‐OH‐CPA concentrations
Plasma concentrations of CPA and 4‐OH‐CPA were simultaneously determined with ifosfamide as the internal standard by an established LC‐MS/MS method 28. In brief, after protein precipitation with cold acetonitrile and stabilization of 4‐OH‐CPA by dansylhydrazine within 30 min after blood sample collection and extraction with ethyl acetate, separation was performed on a C18 3.5 μm 2.1 × 50 mm column with mobile phase of acetonitrile and water (50 : 50, v/v) with 0.1% formic acid at 200 μl min–1. Mass analysis and detection was performed on a Quattro micro™ triple quadruple mass spectrometer (Micromass, Notre Dame, UK) equipped with an electrospray ionization (ESI) source in the selected reaction monitoring (SRM) mode. The chromatographic run time was 3 min. The system control and data process were performed using Masslynx Version 4.0 software (Micromass, Notre Dame, UK). The lower limit of quantification for CPA and 4‐OH‐CPA were both 5 ng ml–1. The intra‐ and inter‐batch precision and accuracy were less than 15% for all quality control samples.
Analyses of genetic polymorphisms
Coded samples without any subject identification or clinical data were sent to the genotyping laboratory so that blinded analysis could be performed. Genomic DNA was extracted from peripheral leukocytes by the improved phenol‐chloroform extraction method as reported 31. All PCR reactions were carried out in a 25 μl volume containing 50 ng genomic DNA, 2.5 mm dNTPs, 10 mm of forward and reverse primers, 2.5 ml 10× Taq buffer and 0.75 U Taq DNA polymerase (Takara, Japan). Based on previous studies from us and others 19, 32, 33, the following SNPs with a higher prevalence in Asian population were selected: CYP2C19 681G > A (*2, rs4244285), CYP2C19 636G > A (*3, rs4986893), CYP2C19 4195C > T (*17, rs12248560), CYP2B6 ‐2320 T > C, CYP2B6 ‐750 T > C (rs4802101), CYP2B6 64C > T (rs8192709), CYP2B6 516G > T (rs3745274), CYP2B6 15582C > T, CYP2B6 785A > G (rs2279343), CYP2B6 1459C > T (rs3211371), PXR 66034 T > C (rs13059232), PXR 44477 T > C (rs1523130), PXR 45005C > T (rs3814055), PXR 69789A > G (rs7643645) and GSTP1(I105V, rs1695). Direct sequencing was used to determine PXR 66034 T > C, 44 477 T > C, 45005C > T and 69789A > G genotypes as described previously 33. After initial denaturation at 95 ºC for 5 min, the PCR amplification was performed by denaturating at 95 ºC for 30 s, annealing for 30 s, and extension at 72 ºC for 1 min for 30 cycles and a final extension at 72 ºC for 10 min. The annealing temperature for PXR 66034 T > C, 44 477 T > C (45005C > T) and 69789A > G was 59 ºC, 60 ºC and 59 ºC, respectively. Null alleles of GSTM1 and GSTT1 were genotyped by a previous reported multiplex PCR method 34. β‐Globin was co‐amplified and used as the internal control. The rest of SNPs under investigation were analyzed by PCR‐restriction fragment length polymorphism (PCR‐RFLP) methods as described previously 8, 15, 34, 35. Details on primer sequences, amplicon sizes and restriction enzymes for CYP2B6, CYP2C19, PXR and GST are summarized in Supplementary Table S1.
Statistical analysis
The allele and genotype frequency of the SNPs and linkage disequilibrium (LD) were calculated by an on‐line tool SHEsis 36 and the frequencies were compared with the expected frequency in the Chinese population from the HapMap reports 37 by χ2 test. The deviations from Hardy–Weinberg equilibrium were assessed by the goodness‐of‐fit χ2 test. The plasma concentrations of CPA and 4‐OH‐CPA were normally distributed. However, after division into different groups by genotypes, the plasma concentrations were not normally distributed in some groups. Therefore, comparison of CPA and 4‐OH‐CPA concentration relative to genotypes was performed by the Kruskal–Wallis H test followed by Dunn's multiple comparison tests to assess the difference between pairs of groups. Multiple linear regression analysis was conducted to analyze the contribution of different genetic polymorphisms to the variation of 4‐OH‐CPA plasma concentration at 20 h post‐injection. Variables were selected in a backward stepwise manner. To compare the incidence of adverse reactions, the subjects were divided into two groups according to their genotypes. In logistic regression analysis, the homozygous wild type genotypes were assigned into one group while heterozygous and homozygous mutant genotypes were combined into another group. The frequencies of adverse reaction between different genetic groups were compared by χ2 test or Fisher's exact test when appropriate. Binary logistic regression analysis was used to assess the association of adverse reactions and genotypes. The odds ratio (OR) and its 95% confidence interval (95% CI) showed the risks of adverse reactions in different genotypes. To evaluate the short term clinical response, cumulative probabilities of complete/partial remission and response were presented in Kaplan–Meier curves and survival analyses were based on log‐rank test. Time to first event was used for each end point. Patients missed or dropped during maintenance therapy were regarded as censored. The statistical analysis was performed using the Statistical Package for Social Science (ver 17.0; SPSS, Chicago, USA). Bonferroni correction for the P value 38 was conducted because the chances of a type I error will increase when multiple tests are performed in a single set of data. To analyze the relationship between genetic polymorphisms and adverse reaction of CPA, the multivariate logistics regression analysis contained three outcome measures (GI toxicities, infection and leukopenia) and 13 SNPs. Therefore, the Bonferroni‐corrected significance level was 0.05/(13*3) = 0.00128. P < 0.00128 was considered statistically significant in logistics analysis and χ2 test (Table 4). In the logistics analysis between genetic markers and 4‐OH‐CPA concentration, the adjusted P value was 0.05/(3*3) = 0.0056 because three outcome measures and three grouping measures (extensive metabolizers (EMs), intermediate metabolizers (IMs) and poor metabolizers (PMs)) were used. P < 0.0056 was considered significant. In other scenarios, P < 0.05 was considered statistically significant.
Table 4.
Comparison of adverse reaction frequencies among genotypes and logistics regression analysis for identification of factors associated with adverse reaction (n = 189)
| SNP | GI toxicity | Infection | Leukocytopenia | ||||
|---|---|---|---|---|---|---|---|
| n (%) | OR (95% CI) | n (%) | OR (95% CI) | n (%) | OR (95% CI) | ||
| CYP2B6 ‐2320 T > C | TT | 5 (18.5) | 0.841 (0.292, 2.423) | 8 (29.6) | 0.608 (0.244, 1.510) | 7 (25.9) | 1.065 (0.421, 2.694) |
| TC and CC | 26 (16.0) | 33 (20.4) | 44 (27.1) | ||||
| CYP2B6 ‐750 T > C | TT | 16 (33.3) | 0.238 (0.107, 0.523) † | 15 (31.1) | 0.497 (0.236, 1.047) | 21 (43.8) | 0.347(0.173, 0.699) † |
| TC and CC | 15 (10.6) * | 26 (18.4) | 30 (21.3) * | ||||
| CYP2B6 64C > T | CC | 29 (16.1) | 1.251 (0.277, 6.894) | 38 (21.1) | 1.848 (0.415, 8.234) | 50 (27.8) | 0.279 (0.032, 2.450) |
| CT and TT | 2 (22.2) | 3 (33.3) | 1 (11.1) | ||||
| CYP2B6 516G > T | GG | 16 (15.1) | 1.021 (0.417, 2.500) | 19 (17.9) | 1.733 (0.766, 3.920) | 29 (27.4) | 0.794 (0.377, 1.674) |
| GT and TT | 15 (18.1) | 22 (26.5) | 22 (26.5) | ||||
| CYP2B6 15582C > T | CC | 4 (11.8) | 1.429 (0.430, 4.753) | 5 (14.7) | 1.683 (0.564, 5.025) | 11 (32.4) | 0.713 (0.301, 1.688) |
| CT and TT | 27 (17.4) | 36 (23.2) | 40 (25.8) | ||||
| CYP2B6 785A > G | AA | 14 (14.7) | 1.216 (0.494, 2.997) | 20 (21.1) | 0.868 (0.380, 1.985) | 24 (25.3) | 1.272 (0.604, 2.681) |
| AG and GG | 17 (18.1) | 21 (22.3) | 27 (28.7) | ||||
| CYP2C19*2 | *1*1 | 26 (26.8) | 0.157 (0.057, 0.430) † | 31 (32.0) | 0.260 (0.119, 0.568) † | 40 (41.2) | 0.194 (0.092, 0.409) † |
| *1*2 and *2*2 | 5 (5.4) * | 10 (10.9) | 11 (12.0) | ||||
| CYP2C19*3 | *1*1 | 26 (15.8) | 1.459 (0.463) | 35 (21.2) | 1.232 (0.423, 3.589) | 41 (24.8) | 2.113 (0.281, 5.440) |
| *1*3 and *3*3 | 5 (20.8) | 6 (25.0) | 10 (41.7) | ||||
| GSTP1 105I > V | I/I | 18 (13.8) | 1.791 (0.835, 3.839) | 29 (22.3) | 0.828 (0.388, 1.768) | 32 (24.6) | 1.821 (0.953, 3.482) |
| I/V and V/V | 12 (21.4) | 11 (19.6) | 16 (28.6) | ||||
| PXR 66034 T > C | TT | 5 (17.2) | 0.931 (0.326, 2.664) | 5 (17.2) | 1.394 (0.496, 3.913) | 9 (31.0) | 0.791 (0.334, 1.837) |
| TC and CC | 26 (16.3) | 36 (22.5) | 42 (26.3) | ||||
| PXR 44477 T > C | TT | 21 (17.9) | 0.737 (0.325, 1.671) | 27 (23.1) | 0.805 (0.390, 1.661) | 30 (25.6) | 1.194 (0.620, 2.301) |
| TC and CC | 10 (13.9) | 14 (19.4) | 21 (29.2) | ||||
| PXR 45005C > T | CC | 1 (5.9) | 3.380 (0.432, 26.477) | 2 (11.8) | 2.199 (0.482, 10.035) | 3 (17.6) | 1.806 (0.497, 6.567) |
| CT and TT | 30 (17.4) | 39 (22.7) | 48 (27.9) | ||||
| PXR 69789A > G | AA | 11 (19.0) | 0.718 (0.232, 2.224) | 16 (26.2) | 0.464 (0.167, 1.291) | 16 (26.2) | 0.914 (0.368, 2.270) |
| AG and GG | 18 (14.1) | 23 (18.0) | 35 (27.3) | ||||
Bold values indicate P < 0.05.
P < 0.00128 by χ2 test or Fisher's exact test when appropriate (wild type vs. mutant).
P < 0.001 by binary logistic regression analysis.
Results
Patient characteristics
From September 2009 to June 2012, 214 Han Chinese patients with newly diagnosed SLE who met the inclusion criteria were included in the study. Among them 189 patients were assigned to CPA induction therapy. The others were discontinued (n = 8) or withdrew due to co‐administration (n = 13) and non‐standard medication (n = 4). After 4 weeks of induction therapy with CPA, 128 patients diagnosed with LN continued CPA maintenance therapy. Twelve of the 128 LN patients were lost during follow‐up. The algorithm for this study is shown in Figure 1.
Figure 1.
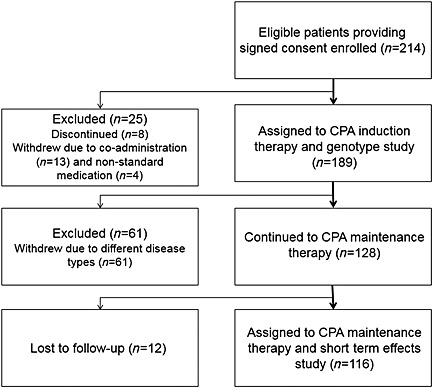
Study algorithm, including patient enrolment, study assignments, exclusion and outcomes
Table 1 summarizes the demographic and clinical characteristics of the recruited subjects in induction therapy and maintenance therapy. During the 4 weeks following the initiation of CPA induction treatment (accumulated dosage of 2–3 g), leukocytopenia, infection and GI toxicity were observed in 51 (27.0%), 41 (21.7%) and 31 (16.4%) patients, respectively. The adverse reaction incidence in LN patients was 21.6%, 22.6%, and 14.7% for leukocytopenia, infection and GI toxicity, respectively. The short term clinical outcomes including complete remission, partial remission and treatment failure in the maintenance therapy group were observed in 74 (63.8%), 32 (27.6%) and 10 (8.6%) patients, respectively.
Table 1.
Demographic and clinical characteristics of the included patients
| Induction therapy | Maintenance therapy | |
|---|---|---|
| n | 189 | 116 |
| Gender (male/female) | 43/146 | 21/95 |
| Age | 34 ± 14 years | 32 ± 13 years |
| Disease type | ||
| Lupus nephritis (LN) | 128 (67.7%) | 116 |
| LN and haematological damage | 26 (13.8%) | – |
| Haematological damage | 12 (6.3%) | – |
| Other types | 23 (12.2%) | – |
| Side effects | ||
| Leukocytopenia | 42 (21.9%) | 25 (21.6%) |
| Infection | 41 (21.4%) | 26 (22.6%) |
| Gastrointestinal toxicity | 31 (16.1%) | 17 (14.7%) |
| Outcome | ||
| Complete remission | 74 (63.8%) | |
| Partial remission | 32 (27.6%) | |
| Treatment failure | 10 (8.6%) |
Genotype analysis
The allelic frequencies and genotype distribution in SLE patients are shown in Table 2. All SNPs were in Hardy–Weinberg equilibrium. The allelic frequencies and genotype distribution were in accordance with the Han Chinese (Beijing) HapMap population 37. Within the PXR gene, a strong LD was observed between PXR 45005C > T and 44 477 T > C (|D’| = 1). A high LD was also shown between PXR 44477 T > C and 66 034 T > C (|D’| = 0.876). Within the CYP2B6 gene, there was a high LD (|D’| = 0.894) between 64C > T and ‐750 T > C.
Table 2.
Allelic and genotype frequency of the SNPs under investigation in Chinese SLE patients (n = 189)
| Gene | SNP | Genotype | Frequency (%) | Allele | Frequency (%) |
|---|---|---|---|---|---|
| CYP2B6 | −2320 T > C | TT | 27 (14.3) | T | 142 (37.6) |
| CT | 88 (46.6) | C | 236 (62.4) | ||
| CC | 74 (39.2) | ||||
| −750 T > C | TT | 48 (25.4) | T | 166 (43.9) | |
| CT | 70 (37.0) | C | 212 (56.0) | ||
| CC | 71 (37.6) | ||||
| 64C > T | CC | 180 (95.2) | C | 369 (97.6) | |
| CT | 9 (4.8) | T | 9 (2.4) | ||
| TT | 0 | ||||
| 516G > T | GG | 106 (56.1) | G | 281 (74.3) | |
| GT | 69 (36.5) | T | 97 (25.6) | ||
| TT | 14 (7.4) | ||||
| 15582C > T | CC | 34 (18.0) | C | 160 (42.3) | |
| CT | 92 (48.7) | T | 218 (57.7) | ||
| TT | 63 (33.3) | ||||
| 785A > G | AA | 95 (50.3) | A | 253 (66.9) | |
| AG | 63 (33.3) | G | 125 (33.0) | ||
| GG | 31 (16.4) | ||||
| 1459C > T | CC | 189 (100) | C | 378 (100) | |
| CT | 0 | T | 0 | ||
| TT | 0 | ||||
| CYP2C19 | *2 | *1*1 | 97 (51.3) | *1 | 260 (68.8) |
| *1*2 | 66 (34.9) | *2 | 118 (31.2) | ||
| *2*2 | 26 (13.8) | ||||
| *3 | *1*1 | 165 (87.3) | *1 | 354 (93.7) | |
| *1*3 | 24 (12.7) | *3 | 24 (6.3) | ||
| *3*3 | 0 | ||||
| *17 | *1*1 | 189 (100) | *1 | 378 (100) | |
| *1*17 | 0 | *17 | 0 | ||
| *17*17 | 0 | ||||
| GSTP1 | IIe/Val | I/I | 130 (68.8) | I | 316 (83.6) |
| I/V | 56 (30.7) | V | 62 (16.4) | ||
| VV | 3 (2.6) | ||||
| GSTM1 | + | 66 (46.8) | |||
| − | 75 (53.2) | ||||
| GSTT1 | + | 56 (39.7) | |||
| − | 85 (60.3) | ||||
| PXR | −25 913 C > T | TT | 117 (61.9) | T | 289 (76.5) |
| (rs1523130) | TC | 55 (29.1) | C | 89 (23.5) | |
| CC | 17 (9.0) | ||||
| −25385C > T | CC | 120 (63.5) | C | 292 (77.2) | |
| (rs3814055) | CT | 52 (27.5) | T | 86 (22.8) | |
| TT | 17 (9.0) | ||||
| −4356 T > C | TT | 29 (15.3) | T | 150 (39.7) | |
| (rs13059232) | TC | 92 (48.7) | C | 228 (60.3) | |
| CC | 68 (36.0) | ||||
| 579A > G | AA | 61 (32.3) | A | 201 (53.2) | |
| (rs7643645) | AG | 79 (41.8) | G | 177 (46.8) | |
| GG | 49 (25.9) |
Impact of genetic polymorphisms on plasma concentrations of CPA and 4‐OH‐CPA
The association between CPA and 4‐OH‐CPA plasma concentrations (C 20 h) and the genotypes of CYP2B6, CYP2C19, GST and PXR is shown in Supplementary Figures S1 and S2.
The C 20 h of 4‐OH‐CPA was significantly influenced by CYP2B6 ‐750 T > C and CYP2C19*2 (P < 0.001). As for CYP2B6 ‐750 T > C genotype, patients with CYP2B6 ‐750TT genotype (wildtype) had significantly higher 4‐OH‐CPA concentration (C 20 h median = 20.9 ng ml–1) than patients with TC and CC genotype (C 20 h median = 12.4 and 10.2 ng ml–1, respectively). Statistical difference was found between CYP2B6 TT and TC groups (P < 0.05), as well as between TT and CC groups (P < 0.05). Patients with CYP2C19*1*1 (wild type) genotype had much higher 4‐OH‐CPA concentrations than the patients with CYP2C19*1*2 or *2*2 (C 20 h median = 21.4, 9.8 and 6.6 ng ml–1 for *1*1, *1*2 and *2*2, respectively, P < 0.05). No significant relationship was observed between any of the other genotypes and 4‐OH‐CPA plasma concentration (Supplementary Figure S2). No significant relationship was observed between the genotypes and CPA plasma concentration (Supplementary Figure S1).
When multiple regression analysis was conducted to evaluate the impact of genotypes on 4‐OH‐CPA concentration, we found that CYP2C19*2, CYP2B6 ‐750 T > C, CYP2B6 15582C > T, CYP2B6 ‐2320 T > C and PXR 66034 T > C were responsible for 47.9% of the individual differences in 4‐OH‐CPA concentration (Table 3). Among these genotypes, CYP2C19*2 and CYP2B6 ‐750 T > C contributed the most, accounting for 23.6% and 21.5%, respectively, of the individual differences of 4‐OH‐CPA concentration. CYP2B6 15582C > T, CYP2B6 ‐2320 T > C and PXR 66034 T > C were minor but significant factors, which contributed to 4.4%, 5.9% and 3.7%, respectively, of the individual differences of 4‐OH‐CPA concentration.
Table 3.
Multiple linear regression model for 4‐OH‐CPA plasma concentration based on genotypes (n = 189)
| Variables | Beta‐standardized coefficients | P values | Partial r | Partial r 2 |
|---|---|---|---|---|
| CYP2C19*2 | −0.414 | <0.001 | −0.486 | 0.236 |
| CYP2B6 ‐750 T > C | −0.381 | <0.001 | −0.464 | 0.215 |
| CYP2B6 15582C > T | −0.182 | 0.017 | −0.241 | 0.044 |
| CYP2B6 ‐2320 T > C | −0.166 | <0.001 | −0.223 | 0.059 |
| PXR 66034 T > C | −0.138 | 0.028 | −0.190 | 0.037 |
General r 2 of the model =47.9%, the percentage of inter‐individual variability of 4‐OH‐CPA.
Relationship between genetic polymorphisms and adverse reactions of CPA
To investigate the relationship between genetic polymorphisms and side effects including GI toxicity, infection and leukocytopenia, the frequency of side effects in different genotype pairs was compared. A binary logistics regression analysis was used to assess the risk of side effect in different genotypes. As shown in Table 4, of all the drug metabolizing enzymes examined in this study, only CYP2B6 ‐750 T > C and CYP2C19*2 were statistically associated with the adverse reactions of CPA.
As for CYP2B6 ‐750 T > C, the frequency of GI toxicity in patients carrying TT genotype (wild type) was 33.3%, significantly higher than that in patients carrying TC and CC genotypes (10.6%, P < 0.005). A similar tendency was observed in the percentage decrease of patients who developed leukocytopenia. 43.8% of patients with the TT genotype developed leukocytopenia compared with 21.3% of patients with the TC and CC genotype. The C carriers had a significantly lower leukocytopenia frequency (P < 0.005). Binary logistics regression analysis revealed that patients having at least one C allele of CYP2B6 ‐750 T > C were at lower risk of experiencing GI toxicity (OR 0.238, 95% CI 0.107, 0.523, P < 0.001) and leukocytopenia (OR 0.347, 95% CI 0.173, 0.699, P < 0.001) than other patients.
Similarly, binary logistics regression analysis indicated that patients possessing at least one dysfunctional allele of CYP2C19*2 were at lower risk of developing GI toxicity (OR 0.157, 95% CI 0.057, 0.430, P < 0.001), infection (OR 0.260, 95% CI 0.119, 0.568, P < 0.001) and leukocytopenia (OR 0.194, 95% CI 0.092, 0.409, P < 0.001) than other patients. Frequency of GI toxicity, infection and leukocytopenia in CYP2C19*2 carriers was 5.4%, 10.9% and 12.0% compared with 26.8%, 32.0% and 41.2% in CYP2C19*1*1 carriers, respectively. Frequencies of adverse reactions were significantly higher in CYP2C19*1*1 carriers than in *2 carriers (P < 0.001).
Determination of combination genetic marker and its association with 4‐OH‐CPA plasma concentration, clinical outcomes and adverse reactions of CPA
Multiple regression analysis indicated that both CYP2B6 ‐750 T > C and CYP2C19*2 were associated with the individual differences in 4‐OH‐CPA concentration. Furthermore, mutations in these two SNPs could increase the risk of experiencing adverse reactions to CPA. Therefore, CYP2B6 ‐750 T > C and CYP2C19*2 were selected as genetic markers for 4‐OH‐CPA plasma concentrations and toxicities. EMs were defined as CYP2C19*1*1 and CYP2B6 ‐750TT genotypes, while PMs were defined as CYP2C19*2*2 and CYP2B6 ‐750CC genotypes. Patients with other genotypes at these two sites were assigned into IMs.
The comparison of CPA and 4‐OH‐CPA concentrations (C 20 h) among EMs, IMs and PMs are presented in Figure 2. There were no significant associations found between different metabolizer categories and CPA concentration (Figure 2A). However, the differences in 4‐OH‐CPA concentration among different metabolizers were significant (Figure 2B, P <0.0001). The median concentration of 4‐OH‐CPA of EMs, IMs and PMs was 34.8 ng ml–1, 11.0 ng ml–1and 6.6 ng ml–1, respectively, and the differences between each of the two groups were significant (P < 0.05). There was significant gene–dose effect of 4‐OH‐CPA plasma concentration in the Chinese SLE subjects.
Figure 2.
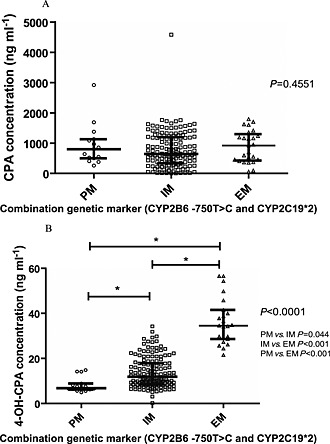
Scatterplots depicting the plasma concentration of cyclophosphamide (CPA) and 4‐hydroxycyclophosphamide (4‐OH‐CPA) classified by the combination genetic marker of CYP2C19*2 and CYP2B6 ‐750 T > C. (A) Scatter plot of CPA concentration grouped by genetic markers. (B) Scatter plot of 4‐OH‐CPA concentration grouped by genetic markers. Each dot represents the concentration data of one patient and the median value in each genotype is shown with a bar. The P values of Kruskal–Wallis H test are listed at the right of each plot. Extensive metabolizer (EM) was defined as CYP2C19*1*1 and CYP2B6 ‐750TT genotype while poor metabolizer (PM) was defined as CYP2C19*2*2 and CYP2B6 ‐750CC genotype. Patients with mixed genotypes at these two loci were defined as intermediate metabolizers (IM)* P < 0.05 (Dunn's multiple comparison tests between two groups).
To evaluate the risk of side effects in different genetic marker groups, logistic regression analysis was conducted. As summarized in Table 5, the risk of leukocytopenia in EMs was significantly higher than that in IMs (P < 0.0001, OR = 7.538, 95% CI 2.951, 19.256), while the risk in PMs was lower than that in IMs, though the difference was not statistically significant (P = 0.788, OR = 0.808, 95% CI 0.171, 3.826). EMs also had a higher risk of experiencing GI toxicity than IMs (P < 0.0001, OR = 7.579, 95% CI 2.934, 19.578), but no difference was observed between PMs and IMs (P = 0.999). However, no significant difference in infection incidence was found among PMs, IMs and EMs (P = 0.999). These results suggested that the genetic markers were significantly associated with the altered risk of leukocytopenia and GI toxicity.
Table 5.
The risks of side effects between different genetic marker groups by logistic regression analysis (n = 189)
| Side effect | Variable | Beta‐standardized coefficients | P value | OR | 95% CI | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Leukocytopenia | PM vs. IM | −0.214 | 0.788 | 0.808 | 0.171 | 3.826 |
| EM vs. IM | 2.020 | <0.0001 | 7.538 | 2.951 | 19.256 | |
| Infection | PM vs. IM | −19.929 | 0.999 | 0 | 0 | 0 |
| EM vs. IM | 0.646 | 0.179 | 1.907 | 0.744 | 4.886 | |
| GI toxicity | PM vs. IM | −19.265 | 0.999 | 0 | 0 | 0 |
| EM vs. IM | 2.025 | <0.0001 | 7.579 | 2.934 | 19.578 | |
EM, extensive metabolizer, patients with CYP2C19*1*1 and CYP2B6 ‐750TT;
IM, intermediate metabolizer, patients with any other combination of these genotypes other than EM and PM;
PM, poor metabolizer, patients with CYP2C19*2*2 and CYP2B6 ‐750CC.
Short term clinical outcomes of LN patients under CPA maintenance therapy were also investigated and categorized by different genetic markers. During the 6 month maintenance therapy, patients categorized as EMs had a significantly faster response time to achieve both complete and partial remission than IMs and PMs. As shown in Table 6 and Figure 3, the median response time to primary end point were 13.2, 18.3 and 23.3 weeks in EMs, IMs and PMs (P = 0.026), respectively. Similarly, the median response time to secondary end point in EMs, IMs and PMs was 8.6, 12.7 and 18.5 weeks (P = 0.006), respectively. The cumulative probability of complete and partial remission was statistically different among the three groups (Figure 3).
Table 6.
Kaplan–Meier estimates of response rate to primary and secondary end points in patients (n = 116)
| End point | Group | n | Median response time (weeks) | 95% CI | P value |
|---|---|---|---|---|---|
| Primary | PM | 10 | 23.3 | 21.5, 24.9 | |
| IM | 92 | 18.3 | 17.0, 19.7 | 0.026 | |
| EM | 14 | 13.2 | 9.3, 17.1 | ||
| Secondary | PM | 10 | 18.5 | 16.8, 22.3 | |
| IM | 92 | 12.7 | 11.3, 14.1 | 0.006 | |
| EM | 14 | 8.6 | 5.3, 11.9 |
The primary end point was complete remission after 6 months of treatment. Secondary end points included response (defined as partial remission), changes in clinical parameters (including proteinuria, serum albumin, serum creatinine and serum C3 values), and adverse effects (including leukopenia, infections, GI symptoms, amenorrhoea, hair loss, liver function disorder, transient increase in serum creatinine level, etc).
Figure 3.
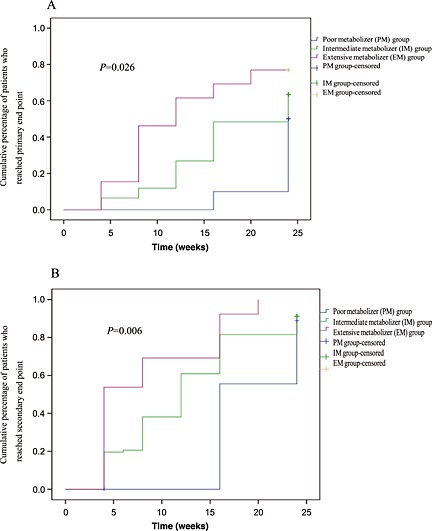
Kaplan–Meier estimates of response rate in different patients. (A) Primary end point patients. (B) Secondary end point of patients. The primary end point was complete remission after 6 months of treatment. Secondary end points included response (defined as partial remission), changes in clinical parameters (including proteinuria, serum albumin, serum creatinine, and serum C3 values), and adverse effects (including leukopenia, infections, gastrointestinal symptoms, amenorrhea, hair loss, liver function disorder, transient increase in serum creatinine level, etc)
Discussion
Cyclophosphamide is a prodrug that requires metabolic activation to 4‐OH‐CPA by cytochrome P450. Most of these enzymes have known variant alleles, and some have been shown to be associated with varied levels of protein expression or metabolic activity of the expressed proteins 8, 39, 40. The efficacy and side effects of CPA exhibit large inter‐individual variability, possibly due to differences in pharmacokinetics. Thus, we sought to study the inter‐individual variability in the plasma concentration of 4‐OH‐CPA, the short term clinical outcomes and side effects of CPA and their association with genetic polymorphisms of drug metabolizing enzymes in Chinese SLE patients. Genetic markers consisting of two genotypes were established and evaluated. To the best of our knowledge, this is the first research to study the effect of PXR genetic polymorphisms on CPA pharmacokinetics and side effects. Until now, no relevant research has been done in other ethnic groups. Moreover, it is also the first pharmacogenomic study of CPA in Chinese SLE patients.
Our previous studies revealed that CYP2B6, CYP2C19, GST and PXR genes were polymorphic in healthy Han Chinese, and the prevalence of these polymorphisms was different from other ethnic groups 19, 20, 23, 33, 41. Therefore, for Chinese patients, the genetic factors that cause varied response to CPA might differ from other ethnic groups. The prevalence of SLE in Chinese is much higher than that in Caucasians 42 Thus it is very important to investigate the genetic factors causing CPA response variation and to explore a plausible individualized CPA medication. In this study, we confirmed the impact of both CYP2C19 and CYP2B6 polymorphisms on CPA 4‐hydroxylation, short term outcomes and side effects. The multiregression analysis revealed the contribution of each genotype to the variation of 4‐OH‐CPA concentration (Table 3). We have investigated six SNPs of CYP2B6, three of them located in the coding region (exon), and the others located in the non‐coding region (5′‐flanking region or intron). In contrast to most previous studies 16, 43, SNPs located in the coding region have not been found to be related to the inter‐individual variation in CPA 4‐hydroxylation. However, SNPs located in the 5′‐flanking (promoter) region or intron showed significant association with decreased CPA 4‐hydroxylation. This finding is in accordance with a Japanese study carried out in cancer patients 15. In Caucasians, the frequencies of CYP2B6 ‐2320 T > C and CYP2B6 ‐750 T > C have been reported to be 30.0 and 56.6%, respectively 44. These frequencies in Japanese were reported as 51.9 and 68.4%, respectively 15. This is the first report on the frequency of – 2320 T > C and – 750 T > C (62.4 and 56.0%, respectively) in Chinese participants.
Most previous pharmacogenomic studies on CPA only considered the effects of single allele, while an in vitro‐in vivo approach conducted with 16 LN patients revealed that combined genotypes, instead of a single allele, were more accurate in predicting CPA intrinsic clearance 18. Our study with a larger sample size assessed the combined impact of genotypes. Among the genotypes under investigation, CYP2C19*2 and CYP2B6 ‐750 T > C showed the highest correlation with 4‐OH‐CPA concentration (r 2 = 0.236 and 0.215, respectively). Three other genotypes, including CYP2B6 15582C > T, CYP2B6 ‐2320 T > C and PXR 66034 T > C, were excluded from further analysis due to their relatively minor correlation with 4‐OH‐CPA concentration (r 2 = 4.4%, 5.9%, 3.7%, respectively). The combination genetic marker of CYP2C19*2 and CYP2B6 ‐750 T > C was significantly associated with 4‐OH‐CPA plasma concentration, short term outcomes and side effects.
We found in this study that PXR 66034 T > C was responsible for 3.7% of the inter‐individual variation of 4‐OH‐CPA plasma concentration (Table 3). This contribution was minor but significant. It is well known that dramatic variation in CYP3A4 activity and expression has been observed in Chinese, but the genetic polymorphisms of CYP3A4 are rare in Chinese. The most common CYP3A4 alleles in the Chinese population are *1 (wild‐type), *5, *6, *18 and *21 with frequency of 97%, 0.5%, 1%, 1% and 0.5%, respectively 22. Therefore, genetic polymorphism of CYP3A4 is unlikely to be a major cause of CYP3A4 inter‐individual variation in CPA metabolism and outcome in Chinese. Recent studies indicated that PXR plays an important role in CYP3A4 regulation and change of PXR function could affect CYP3A4 expression 45. Our previous study in healthy Chinese indicated that the PXR gene had different polymorphic characteristics from other ethnic groups 46. Furthermore, CPA can induce CYP2B6 expression, possibly via PXR activation 47. Hence the importance of PXR in CYP3A4 and CYP2B6 expression may explain our finding that PXR 66034 T > C was in part responsible for the inter‐individual variation in 4‐OH‐CPA concentration, which had a negative correlation with 4‐OH‐CPA (partial r = −0.190, Table 3). This is consistent with other reports that rifampicin‐mediated induction in CYP3A4 activity was higher in the PXR 66034TT genotype (wild type) than in the CC genotype 45.
In the current study, the protocol of CPA treatment included 1 month induction therapy (200 mg every other day) and 6 months maintenance therapy (200–600 mg week–1). Plasma concentrations of 4‐OH‐CPA and toxicity were investigated in the induction period, while short term efficacy was observed in the maintenance period. As compared with pulse therapy of CPA, the current protocol reduced the incidence of side effects. The occurrence rates for leukocytopenia, infection and GI toxicity were 21.9%, 21.4% and 16.1%, respectively, in the current studied cohort. In our previous study, the incidence of these side effects in SLE patients treated with pulse CPA were 40.2%, 15.7% and 31.4% for leukocytopenia, infection and GI toxicity, respectively 3. As LN is the major pathological type of SLE that results in a suitable sample size and excludes confounding clinical observations from other pathological types, patients diagnosed with LN were selected to continue maintenance therapy and observed for short term efficacy.
CPA is prescribed for various indications, such as cancer and auto‐immune disease. In the current study, we only focused on the subjects with SLE. CPA is commonly prescribed with other anti‐cancer agents and shares similar side effects with other co‐medications, such as leukocytopenia, GI toxicity, hair loss, mucositis, etc. However, co‐administration of other immunosuppressants in SLE treatment is not common. Moreover, common co‐medication in SLE treatment, such as glucocorticoids, hydroxychloroquine and NSAIDs, neither interferes with CPA pharmacokinetically nor shares side effects. Therefore, our study may be less affected by such confounding factors and can reflect the effect of CPA metabolizing enzymes more accurately.
Besides the CPA short term efficacy and side effects such as leukocytopenia and GI toxicity, the long term therapeutic response and toxicities might also be related to genotypes. The long term therapeutic response to the CPA treatment of SLE patients and side effects including reproductive toxicity will be the focus of our continuing observations in this cohort of patients.
Here we demonstrated the correlation of some CYP genotypes and CPA clinical efficacy and side effects. However it is necessary to test the activity of the enzymes involved in CPA metabolism in future studies to support further our conclusion.
In conclusion, our observation in Chinese patients with SLE indicated that the combination genetic marker of CYP2C19*2 and CYP2B6 ‐750 T > C genotypes was associated with 4‐OH‐CPA plasma concentration, short term efficacy and adverse reactions of CPA. The gene–dose effect of CPA 4‐hydroxylation observed in Chinese subjects is likely to have further pharmacogenetic implication when treating different ethnic groups, as the prevalence of EMs/IMs/PMs differs in different populations. The presence of increased function alleles (CYP2C19*1*1 and CYP2B6 ‐750TT) tended to result in higher 4‐OH‐CPA concentration, better short term outcomes and higher risks of leukocytopenia and GI toxicity. As such, we suggest that testing for the genotypes of CYP2C19*2 and CYP2B6 ‐750 T > C in Chinese patients might have a big potential of improving CPA dosing strategy in clinical practice.
Competing Interests
All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare no support from any organization for the submitted work, no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years and no other relationships or activities that could appear to have influenced the submitted work.
This study was supported by the grants from the National Natural Science Foundation of China (No. 81173131, 81072708, 81102515, 81201718, 31300774 and 81403019), the National Major Scientific and Technological Special Project for ‘Significant New Drugs Development’ during the Twelfth Five‐year Plan Period (No. 2012ZX09506001‐004), the Fundamental Research Funds for the Central Universities (No.2013ZM0067), and grant from Guangzhou Medical University Scientific Research Program for Young Scientists (No. 2013A39).
Author contributions
WS and SG made substantial contributions to the conception and design, analysis and interpretation of data, drafting of the manuscript, and were responsible for all experiments. XY and LL were the clinical investigators of the trial and responsible for the medical care of trial participants, communication with the research ethics committee, protocol, informed consent, data integrity and reporting. JL and ZC contributed to the genetic polymorphism analysis. YZ and LC contributed to the plasma concentration analysis. XW and MH contributed to the conception and design, and critically revised the manuscript for important intellectual content. All authors have given final approval of the manuscript.
Supporting information
Table S1 The primers, amplicons and restriction enzymes of PCR‐RFLP.
Figure S1 Box plots of cyclophosphamide (CPA) plasma concentration at 20 h after the 4th injection in Chinese SLE patients with different genotypes. The median value in each genotype is shown with a bar. The P values of Kruskal–Wallis H test are listed at the right of each plot. * P <0.05 (Dunn's multiple comparison tests between two groups).
Figure S2 Box plots of 4‐hydroxycyclophosphamide (4‐OH‐CPA) plasma concentration at 20 h after the 4th injection in Chinese SLE patients with different genotypes. The median value in each genotype is shown with a bar. The P values of Kruskal–Wallis H test are listed at the right of each plot. * P <0.05 (Dunn's multiple comparison tests between two groups).
Supporting info item
Shu, W. , Guan, S. , Yang, X. , Liang, L. , Li, J. , Chen, Z. , Zhang, Y. , Chen, L. , Wang, X. , and Huang, M. (2016) Genetic markers in CYP2C19 and CYP2B6 for prediction of cyclophosphamide's 4‐hydroxylation, efficacy and side effects in Chinese patients with systemic lupus erythematosus. Br J Clin Pharmacol, 81: 327–340. doi: 10.1111/bcp.12800.
W. Shu and S. Guan contributed equally to this work.
References
- 1. Petri M. Cyclophosphamide: new approaches for systemic lupus erythematosus. Lupus 2004; 13: 366–71. [DOI] [PubMed] [Google Scholar]
- 2. Pendse S, Ginsburg E, Singh AK. Strategies for preservation of ovarian and testicular function after immunosuppression. Am J Kidney Dis 2004; 43: 772–81. [DOI] [PubMed] [Google Scholar]
- 3. Zhong S, Huang M, Yang X, Liang L, Wang Y, Romkes M, Duan W, Chan E, Zhou SF. Relationship of glutathione S‐transferase genotypes with side‐effects of pulsed cyclophosphamide therapy in patients with systemic lupus erythematosus. Br J Clin Pharmacol 2006; 62: 457–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie HJ, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, Rane A. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 2003; 3: 53–61. [DOI] [PubMed] [Google Scholar]
- 5. Roy P, Yu LJ, Crespi CL, Waxman DJ. Development of a substrate‐activity based approach to identify the major human liver P‐450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA‐expressed activities and liver microsomal P‐450 profiles. Drug Metab Dispos 1999; 27: 655–66. [PubMed] [Google Scholar]
- 6. Griskevicius L, Yasar U, Sandberg M, Hidestrand M, Eliasson E, Tybring G, Hassan M, Dahl ML. Bioactivation of cyclophosphamide: the role of polymorphic CYP2C enzymes. Eur J Clin Pharmacol 2003; 59: 103–9. [DOI] [PubMed] [Google Scholar]
- 7. Timm R, Kaiser R, Lotsch J, Heider U, Sezer O, Weisz K, Montemurro M, Roots I, Cascorbi I. Association of cyclophosphamide pharmacokinetics to polymorphic cytochrome P450 2C19. Pharmacogenomics J 2005; 5: 365–73. [DOI] [PubMed] [Google Scholar]
- 8. Lang T, Klein K, Fischer J, Nussler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 2001; 11: 399–415. [DOI] [PubMed] [Google Scholar]
- 9. de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S‐mephenytoin metabolism in humans. J Biol Chem 1994; 269: 15419–22. [PubMed] [Google Scholar]
- 10. Brosen K, de Morais SM, Meyer UA, Goldstein JA. A multifamily study on the relationship between CYP2C19 genotype and s‐mephenytoin oxidation phenotype. Pharmacogenetics 1995; 5: 312–7. [DOI] [PubMed] [Google Scholar]
- 11. Afsar NA, Ufer M, Haenisch S, Remmler C, Mateen A, Usman A, Ahmed KZ, Ahmad HR, Cascorbi I. Relationship of drug metabolizing enzyme genotype to plasma levels as well as myelotoxicity of cyclophosphamide in breast cancer patients. Eur J Clin Pharmacol 2012; 68: 389–95. [DOI] [PubMed] [Google Scholar]
- 12. Takada K, Arefayene M, Desta Z, Yarboro CH, Boumpas DT, Balow JE, Flockhart DA, Illei GG. Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum 2004; 50: 2202–10. [DOI] [PubMed] [Google Scholar]
- 13. Singh G, Saxena N, Aggarwal A, Misra R. Cytochrome P450 polymorphism as a predictor of ovarian toxicity to pulse cyclophosphamide in systemic lupus erythematosus. J Rheumatol 2007; 34: 731–3. [PubMed] [Google Scholar]
- 14. Ngamjanyaporn P, Thakkinstian A, Verasertniyom O, Chatchaipun P, Vanichapuntu M, Nantiruj K, Totemchokchyakarn K, Attia J, Janwityanujit S. Pharmacogenetics of cyclophosphamide and CYP2C19 polymorphism in Thai systemic lupus erythematosus. Rheumatol Int 2011; 31: 1215–8. [DOI] [PubMed] [Google Scholar]
- 15. Nakajima M, Komagata S, Fujiki Y, Kanada Y, Ebi H, Itoh K, Mukai H, Yokoi T, Minami H. Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genomics 2007; 17: 431–45. [DOI] [PubMed] [Google Scholar]
- 16. Xie H, Griskevicius L, Stahle L, Hassan Z, Yasar U, Rane A, Broberg U, Kimby E, Hassan M. Pharmacogenetics of cyclophosphamide in patients with hematological malignancies. Eur J Pharmaceut Sci 2006; 27: 54–61. [DOI] [PubMed] [Google Scholar]
- 17. Ekhart C, Doodeman VD, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4‐hydroxycyclophosphamide. Pharmacogenet Genomics 2008; 18: 515–23. [DOI] [PubMed] [Google Scholar]
- 18. Helsby NA, Hui CY, Goldthorpe MA, Coller JK, Soh MC, Gow PJ, De Zoysa JZ, Tingle MD. The combined impact of CYP2C19 and CYP2B6 pharmacogenetics on cyclophosphamide bioactivation. Br J Clin Pharmacol 2010; 70: 844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan S, Huang M, Li X, Chen X, Chan E, Zhou SF. Intra‐ and inter‐ethnic differences in the allele frequencies of cytochrome P450 2B6 gene in Chinese . Pharm Res 2006; 23: 1983–90. [DOI] [PubMed] [Google Scholar]
- 20. Zhong SL, Zhou S, Huang M. A comparison of glutathione S‐transferase mutant frequencies in healthy Han and uygur Chinese . Eur J Drug Metab Pharmacokinet 2005; 30: 181–5. [DOI] [PubMed] [Google Scholar]
- 21. Lamba JK, Lin YS, Thummel K, Daly A, Watkins PB, Strom S, Zhang J, Schuetz EG. Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics 2002; 12: 121–32. [DOI] [PubMed] [Google Scholar]
- 22. Zhou Q, Yu X, Shu C, Cai Y, Gong W, Wang X, Wang DM, Hu S. Analysis of CYP3A4 genetic polymorphisms in Han Chinese . J Hum Genet 2011; 56: 415–22. [DOI] [PubMed] [Google Scholar]
- 23. Wang XD, Li JL, Su QB, Guan S, Chen J, Du J, He YW, Zeng J, Zhang JX, Chen X, Huang M, Zhou SF. Impact of the haplotypes of the human pregnane X receptor gene on the basal and St john's wort‐induced activity of cytochrome P450 3A4 enzyme. Br J Clin Pharmacol 2009; 67: 255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hochberg MC. Updating the American college of rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40: 1725. [DOI] [PubMed] [Google Scholar]
- 25. Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The committee on prognosis studies in SLE. Arthritis Rheum 1992; 35: 630–40. [DOI] [PubMed] [Google Scholar]
- 26. Weening JJ, D'Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, Balow JE, Bruijn JA, Cook T, Ferrario F, Fogo AB, Ginzler EM, Hebert L, Hill G, Hill P, Jennette JC, Kong NC, Lesavre P, Lockshin M, Looi LM, Makino H, Moura LA, Nagata M. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol 2004; 15: 241–50. [DOI] [PubMed] [Google Scholar]
- 27. Sinclair A, Appel G, Dooley MA, Ginzler E, Isenberg D, Jayne D, Wofsy D, Solomons N. Mycophenolate mofetil as induction and maintenance therapy for lupus nephritis: rationale and protocol for the randomized, controlled Aspreva Lupus Management Study (ALMS). Lupus 2007; 16: 972–80. [DOI] [PubMed] [Google Scholar]
- 28. Shu W, Wang X, Yang X, Liang L, Li J, Chen Z, Chen X, Jin J, Li R, Zhao L, Huang M. Simultaneous determination of cyclophosphamide and 4‐hydroxycyclophosphamide in human plasma by high‐performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry ‐ application to Chinese systemic lupus erythematosus patients. Clin Chem Lab Med 2011; 49: 2029–37. [DOI] [PubMed] [Google Scholar]
- 29. Egorin MJ, Forrest A, Belani CP, Ratain MJ, Abrams JS, Van Echo DA. A limited sampling strategy for cyclophosphamide pharmacokinetics. Cancer Res 1989; 49: 3129–33. [PubMed] [Google Scholar]
- 30. Chen W, Tang X, Liu Q, Fu P, Liu F, Liao Y, Yang Z, Zhang J, Chen J, Lou T, Fu J, Kong Y, Liu Z, Fan A, Rao S, Li Z, Yu X. Short‐term outcomes of induction therapy with tacrolimus versus cyclophosphamide for active lupus nephritis: a multicenter randomized clinical trial. Am J kidney Dis 2011; 57: 235–44. [DOI] [PubMed] [Google Scholar]
- 31. Loparev VN, Cartas MA, Monken CE, Velpandi A, Srinivasan A. An efficient and simple method of DNA extraction from whole blood and cell lines to identify infectious agents. J Virol Methods 1991; 34: 105–12. [DOI] [PubMed] [Google Scholar]
- 32. Xiao ZS, Goldstein JA, Xie HG, Blaisdell J, Wang W, Jiang CH, Yan FX, He N, Huang SL, Xu ZH, Zhou HH. Differences in the incidence of the CYP2C19 polymorphism affecting the S‐mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther 1997; 281: 604–9. [PubMed] [Google Scholar]
- 33. Wang XD, Li JL, Su QB, Deng XY, Lu Y, Chen J, Zhang JX, Zhao LZ, Zuo Z, Chan E, Chen X, Chowbay B, Xue CC, Huang M, Zhou SF. A pharmacogenetic study of pregnane X receptor (NR1I2) in Han Chinese . Curr Drug Metab 2007; 8: 778–86. [DOI] [PubMed] [Google Scholar]
- 34. Zhong SL, Zhou S, Chen X, Huang M. Rapid determination of common mutations in glutathione S‐transferase gene by PCR‐based methods in healthy Chinese . Clinica Chimica Acta 2006; 364: 205–8. [DOI] [PubMed] [Google Scholar]
- 35. Goldstein JA, Blaisdell J. Genetic tests which identify the principal defects in CYP2C19 responsible for the polymorphism in mephenytoin metabolism. Methods Enzymol 1996; 272: 210–8. [DOI] [PubMed] [Google Scholar]
- 36. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005; 15: 97–8. [DOI] [PubMed] [Google Scholar]
- 37. The International HapMap Project . Nature 2003; 426: 789–96. [DOI] [PubMed] [Google Scholar]
- 38. Bland JM, Altman DG. Multiple significance tests: the bonferroni method. BMJ 1995; 310: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 2001; 27: 383–91. [DOI] [PubMed] [Google Scholar]
- 40. Goldstein JA, de Morais SM. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 1994; 4: 285–99. [DOI] [PubMed] [Google Scholar]
- 41. Guan S, Huang M, Chan E, Chen X, Duan W, Zhou SF. Genetic polymorphisms of cytochrome P450 2B6 gene in Han Chinese . Eur J Pharmaceut Sci 2006; 29: 14–21. [DOI] [PubMed] [Google Scholar]
- 42. Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med 2008; 358: 929–39. [DOI] [PubMed] [Google Scholar]
- 43. Kirchheiner J, Klein C, Meineke I, Sasse J, Zanger UM, Murdter TE, Roots I, Brockmoller J. Bupropion and 4‐OH‐bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 2003; 13: 619–26. [DOI] [PubMed] [Google Scholar]
- 44. Zukunft J, Lang T, Richter T, Hirsch‐Ernst KI, Nussler AK, Klein K, Schwab M, Eichelbaum M, Zanger UM. A natural CYP2B6 TATA box polymorphism (−82 T– > C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol 2005; 67: 1772–82. [DOI] [PubMed] [Google Scholar]
- 45. Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron one of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos 2008; 36: 169–81. [DOI] [PubMed] [Google Scholar]
- 46. Wang XD, Deng XY, Chen J, Li JL, Chen X, Zhao LZ, Lu Y, Chowbay B, Su QB, Huang M, Zhou SF. Single nucleotide polymorphisms of the pregnane x receptor gene in Han Chinese and a comparison with other ethnic populations. Pharmacology 2008; 81: 350–4. [DOI] [PubMed] [Google Scholar]
- 47. Wang D, Li L, Fuhrman J, Ferguson S, Wang H. The role of constitutive androstane receptor in oxazaphosphorine‐mediated induction of drug‐metabolizing enzymes in human hepatocytes. Pharm Res 2011; 28: 2034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The primers, amplicons and restriction enzymes of PCR‐RFLP.
Figure S1 Box plots of cyclophosphamide (CPA) plasma concentration at 20 h after the 4th injection in Chinese SLE patients with different genotypes. The median value in each genotype is shown with a bar. The P values of Kruskal–Wallis H test are listed at the right of each plot. * P <0.05 (Dunn's multiple comparison tests between two groups).
Figure S2 Box plots of 4‐hydroxycyclophosphamide (4‐OH‐CPA) plasma concentration at 20 h after the 4th injection in Chinese SLE patients with different genotypes. The median value in each genotype is shown with a bar. The P values of Kruskal–Wallis H test are listed at the right of each plot. * P <0.05 (Dunn's multiple comparison tests between two groups).
Supporting info item


