Summary
For biological and statistical reasons it makes sense to combine information from variants at the level of the gene. One may wish to give more weight to variants which are rare and those that are more likely to affect function. A combined weighting scheme, implemented in the SCOREASSOC program, was applied to whole exome sequence data for 1392 subjects with schizophrenia and 982 with obesity from the UK10K project. Results conformed fairly well with null hypothesis expectations and no individual gene was strongly implicated. However, a number of the higher ranked genes appear plausible candidates as being involved in one or other phenotype and may warrant further investigation. These include MC4R, NLGN2, CRP, DONSON, GTF3A, IL36B, ADCYAP1R1, ARSA, DLG1, SIK2, SLAIN1, UBE2Q2, ZNF507, CRHR1, MUSK, NSF, SNORD115, GDF3 and HIBADH. Some individual variants in these genes have different frequencies between cohorts and could be genotyped in additional subjects. For other genes, there is a general excess of variants at many different sites so attempts at replication would be more difficult. Overall, the weighted burden test provides a convenient method for using sequence data to highlight genes of interest.
Keywords: Association, exome, burden test, DNA variant
Introduction
Although next generation sequencing has been used extensively for the study of rare Mendelian diseases, there is less experience of its application to large case‐control association studies of diseases with complex inheritance. Because of issues such as incomplete penetrance and allelic and locus heterogeneity, the task is to identify variants which are more frequent in cases rather than variants which are shared by all cases and not seen in any controls or in the general population. There are a number of approaches which can be used in an attempt to gain power by reducing the necessary correction for multiple testing. One general approach may be to restrict attention to variants which are judged to be “important” according to some criteria. These might relate to the predicted effect of the variant or to which gene it occurs in. The frequency of the variant might be used as a criterion; for example, if one is only interested in detecting variants with a large effect size then one may ignore common variants if the results of genomewide association studies have demonstrated that there are no common variants with a large effect size. Another approach is to group variants together and analyse them jointly. This may be done at the level of the gene, so that any correction only needs to be applied for the number of genes tested rather than the number of variants. Additionally, one may group genes into sets according to biological function and this can be viewed as a way of reducing multiple testing still further. Thus, if one is unable to conclusively implicate a gene then one may at least succeed in implicating a pathway.
A recent case‐control study of schizophrenia using whole exome sequence from over 5000 subjects provides a useful illustration of these approaches (Purcell et al., 2014). Attention was focussed on genes deemed a priori to be of interest based on previous GWAS, CNV and de novo SNV studies. Analyses were carried out with three different criteria for including variants based on their predicted effect on gene function. For each of these, three sets of allele frequency were used to include variants: singletons, up to 0.1% or up to 0.5% so that overall nine sets of analyses were carried out. Individual variants were tested and in addition two gene‐based tests were applied—a one‐sided burden test for increased rare variants in cases and the SNP‐set (sequence) kernel association test (SKAT), which tests for differences in case‐control allele frequencies in either direction (Wu et al., 2011). A polygenic burden test was applied to the predefined set of genes and also to subsets defined on the basis of biological function or through having been implicated by de novo SNVs. None of the variant and gene‐based tests achieved statistical significance but the study did demonstrate increased variant alleles in the gene sets.
As has been suggested previously (Madsen & Browning, 2009; Curtis, 2012), an alternative approach to carrying out repeated analyses with different sets of variants included according to effect and/or frequency is to carry out a combined analysis which accords different weights to different variants. Such an approach is implemented in the SCOREASSOC program, which provides a parabolic function to give higher weights to rarer alleles and which allows a functional weight to be specified which can be based on the predicted effect of the variant.
Materials and Methods
Data Used
In order to assess the performance of the weighted burden test in a real world example, it was applied to data produced by the UK10K project (The UK10K Consortium, 2015). Two cohorts of subjects were used, selected from the UK10K exomes arm. The OB cohort consisted of 982 subjects from the Severe Childhood Onset Obesity Project (Wheeler et al., 2013) and the SZ cohort consisted of 1392 subjects with schizophrenia recruited from five British centres. All subjects were British. Although a small proportion of schizophrenia subjects consisted of between two and five members of the same multiply affected pedigrees, for purposes of analysis all subjects were treated as if they were unrelated. The reason for using these two cohorts, rather than other subjects included in UK10K, was primarily that they represented two groups, each of which was phenotypically fairly homogeneous and which had similar geographical origins. The "case–case" design for association studies has the advantage that one does not require an additional set of controls but may have the disadvantage that if allele frequencies differ between the groups then one may not know which is the relevant phenotype (Curtis et al., 2011). As described elsewhere (The UK10K Consortium, 2015), the exome was targeted with the Agilent SureSelect 50Mb V3 exome library, followed by Illumina next generation sequencing with 75bp paired‐end reads. An average read depth of 79x was achieved in the bait regions. Variants were called with samtools/bcftools version 0.1.19‐3‐g4b70907. GATK Unified Genotyper (v1.6‐13‐g91f02df) was only used to recall at SNP sites discovered by samtools. This was to enable VQSR filtering of SNP calls. Three filters were applied to SNPs: LowQual, Description = "Low quality variant according to GATK (GATK)"; MinVQSLOD, Description = "Minimum VQSLOD score [SNPs:‐1.9667, truth sensitivity 99.48]"; SnpGap, Description = "SNP within INT bp around a gap to be filtered [10].” All SNP sites that did not fail these filters were marked as PASS. For the purpose of the current analyses, a number of additional constraints were applied to the downloaded VCF files to exclude some variants from analysis. Only single nucleotide variants (SNVs), not indels, were considered. Variants were excluded if they did not have a PASS in the information field, if there were more than five genotypes missing in either cohort or if the heterozygote count was smaller than both homozygote counts in both cohorts. At a subject level, variants were excluded if they had a genotype quality score less than 30.
Ethics Statement
This paper involves the analysis of data produced by UK10K. All subjects gave informed consent and each group received approval from the appropriate Research Ethics Committee in the United Kingdom.
Method of Analysis
Custom software was written to extract information for one gene at a time from the master VCF files. The variants from all transcripts of each refseq gene were extracted, using only variants called from within the targets. Variants were annotated using the hg19 reference sequence and where a variant had a different effect in different transcripts the one with the largest effect was used. The variants for each gene were analysed using the SCOREASSOC program. Rarer variants were accorded a higher weight than commoner ones, such that an extremely rare variant with minor allele frequency (MAF) close to 0 would be allocated a weight 10 times higher than a common one with MAF of 0.5, with variants of intermediate frequencies being allocated intermediate weights using a parabolic function (Madsen & Browning, 2009; Curtis, 2012). The allele of interest was considered to be the rarer allele, even if the reference allele was rarer than the alternate allele. If any variant had more than one alternate allele then these were grouped together so that the variant could be considered as biallelic. Weights were also allocated according to the effect of the variant. An arbitrary weighting scheme was devised, so that a variant producing a new stop codon within a coding region would be allocated 20 times the weight of an intergenic variant. Variants in coding regions were allocated a weight of 3 if they were synonymous and 10 if they were nonsynonymous. A full list of weights according to the effects of variants is presented in Table 1. The weights for each type of variant were chosen so that variants deemed more likely to have an effect on gene expression or protein function were allocated higher weights. Likewise, these weights were chosen to be of the same order of magnitude as those relating to the rarity of the variant. Thus, a variant might achieve a similar weight either through being very rare or through being likely to have a functional effect. Each variant was then allocated an overall weight, achieved by simply multiplying together the weight according to rarity and the weight according to effect. Thus rarer, more functional variants would be given a higher weight than common variants in noncoding regions. As described previously (Curtis, 2012), each subject would be assigned a score consisting of the sum of the weights of all the variant alleles possessed by that subject. An unpaired t test is used to test whether the average score for cases is higher than controls.
Table 1.
Scheme used to assign weights to each variant according to the predicted effect. (In the analyses described in this report, INDEL variants were not in fact used.)
| Predicted effect | Weight |
|---|---|
| NULL_CONSEQUENCE | 1 |
| INTERGENIC | 1 |
| DOWNSTREAM | 1 |
| INTRONIC | 3 |
| 3PRIME_UTR | 5 |
| SYNONYMOUS_CODING | 3 |
| UPSTREAM | 5 |
| 5PRIME_UTR | 5 |
| SPLICE_SITE | 5 |
| STOP_LOST | 5 |
| NON_SYNONYMOUS_CODING | 10 |
| CODINGINDEL | 15 |
| FRAMESHIFT_CODING | 20 |
| STOP_GAINED | 20 |
In the present study we wished to consider the results in two ways, firstly designating the SZ cohort as cases with the OB cohort as controls and then the other way round. All results were expressed as a signed log p value (SLP), this being the logarithm base 10 of the p value from the t test, being given a positive sign if there was an excess of rare, functional variants in SZ cases and a negative sign if the excess was in OB cases. Two sets of analyses were performed with different categories of variant. The broad category included all valid variants. The narrow category was restricted to splice site or nonsynonymous or stop variants and having MAF <0.1 in at least one of the cohorts.
The SLPs of individual genes were considered. Also, some genes were grouped into sets of a priori interest. For SZ, the same sets were used as described in Table S2 of the schizophrenia exome study, consisting of postsynaptic density (PSD), calcium channel, FMRP targets and SZ de novo (Purcell et al., 2014). For obesity, genes listed in OMIM were used, consisting of NR0B2, SDC3, POMC, GHRL, PPARG, UCP1, CART, ADRB2, PPARGC1B, SIM1, ENPP1, ADRB3, UCP3, AGRP, PYY, MC4R, LEP, LEPR and PCSK1.
Database Submission
Variants which appeared to be associated with the phenotypes studied were submitted to the Human Variation database at NCBI (http://www.ncbi.nlm.nih.gov/).
Results
Distribution of Test Statistic
There were 1,028,678 valid variants in 20,438 genes. The SLPs produced from the broad and narrow categories of variant were highly correlated, r 2 = 0.65. The Q:Q plots are displayed in Figure 1. These show that when there was an excess of rare, functional variants in SZ subjects and the SLP was positive then the values obtained conformed well with null hypothesis expectations. However when the excess was in the OB subjects then the line for the negative SLPs is steeper, indicating that the p value obtained is somewhat anticonservative. To explore the possibility that this might have happened because the scores were not normally distributed, Wilcoxon's signed rank test was applied instead of a t test but almost identical Q:Q plots were obtained. Taking a suitable threshold for “genomewide significance” as log(0.05×1/20438) = 5.6, only one gene, NSF, almost reached this with SLP = −5.5 in the broad analysis. However, this is fairly meaningless if one takes into account the nonconservative nature of the test for the negative SLPs and overall the results do not produce real evidence for the involvement of any particular gene.
Figure 1.
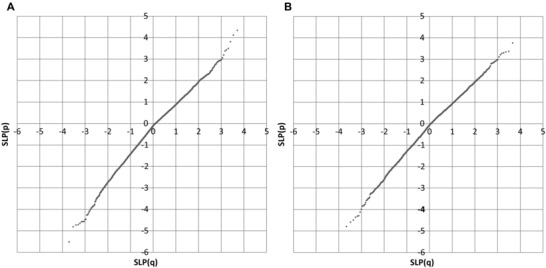
Q:Q plots for SLP obtained from SCOREASSOC compared to expected under null hypothesis. Positive SLPs indicate an excess of rare, functional variants in SZ subjects, negative SLPs indicate an excess in OB subjects. (A) Shows results using broad category of variants, (B) for narrow category.
Although the analyses did not highlight any genes reaching conventional criteria for statistical significance once appropriate allowance was made for the number of genes tested, the results could still be used to rank genes, with the idea being that genes contributing to risk of SZ or OB might tend to have the highest or lowest ranks respectively. Also, one might expect to see genes belonging to the predefined sets tending to have more extreme SLPs than the rest. However when this was formally tested there was no evidence for such enrichment. For each set, the genes within it had the same average SLP as the others not in the set. Nevertheless, ranking genes according to SLP did draw attention to some individual genes which arguably are of interest. The highest and the lowest ranked genes for the broad and narrow analyses are shown in Table 2. The full results for all genes are provided in Table S1.
Table 2.
Highest and lowest ranked genes with corresponding SLPs using SZ and OB definitions of caseness and including broad and narrow categories of variant
| Highest SLPs (SZ cases) | Lowest SLPs (OB cases) | ||||||
|---|---|---|---|---|---|---|---|
| Broad category | Narrow category | Broad category | Narrow category | ||||
| Symbol | SLP | Symbol | SLP | Symbol | SLP | Symbol | SLP |
| DFNA5 | 4.3 | MC4R | 3.8 | NSF | −5.5 | SRSF8 | −4.8 |
| GTF3A | 4.1 | DONSON | 3.4 | CCDC58 | −4.8 | KIAA0947 | −4.6 |
| MC4R | 3.8 | SLAIN1 | 3.3 | MAPT | −4.7 | SLC17A2 | −4.5 |
| NLGN2 | 3.5 | ARR3 | 3.3 | KIAA0947 | −4.7 | HIBCH | −4.4 |
| CRP | 3.4 | ZNF507 | 3.3 | HIST1H4A | −4.7 | SCEL | −4.3 |
| SCARNA11 | 3.4 | ADCYAP1R1 | 3.2 | STH | −4.6 | DUOX1 | −4.3 |
| TRIM77P | 3.2 | DFNA5 | 3.1 | SNORD115 | −4.6 | TGM2 | −4.1 |
| FHIT | 3.1 | NPHS1 | 3.0 | TGM2 | −4.5 | ATRX | −4.0 |
| FAM212B | 3.0 | SKIV2L | 3.0 | SRSF8 | −4.5 | MFSD1 | −3.9 |
| AGXT | 2.9 | ARSA | 2.9 | RPAIN | −4.3 | PXDN | −3.8 |
| IL36B | 2.9 | PNN | 2.9 | MERTK | −4.3 | CD164L2 | −3.8 |
| GALNTL5 | 2.9 | SCO1 | 2.9 | HIST1H2AJ | −4.2 | OR6N2 | −3.8 |
| WDR24 | 2.9 | OR2H1 | 2.8 | AK4 | −4.1 | TINAG | −3.7 |
| GABRA3 | 2.9 | OPN4 | 2.8 | LOC100128977 | −4.1 | MUC17 | −3.6 |
| MCCD1 | 2.8 | WDYHV1 | 2.8 | TCEB3 | −4.0 | MYL10 | −3.6 |
| FBXL16 | 2.8 | GTF2E1 | 2.8 | FUT2 | −4.0 | TCEB3 | −3.5 |
| PROCR | 2.8 | PSMB3 | 2.8 | CRHR1 | −3.9 | FNDC9 | −3.5 |
| PLRG1 | 2.7 | NFU1 | 2.7 | IMP5 | −3.9 | GDF3 | −3.5 |
| OR2H1 | 2.7 | CD8A | 2.7 | SLC17A2 | −3.9 | LMAN1L | −3.5 |
| C2orf89 | 2.7 | RXRB | 2.6 | ERBB2IP | −3.9 | LILRA1 | −3.4 |
| FXYD2 | 2.6 | UBE2Q2 | 2.6 | NDST1 | −3.8 | TADA2A | −3.3 |
| GTF2E1 | 2.6 | CRP | 2.6 | HDAC1 | −3.8 | ZNF556 | −3.3 |
| SPINK8 | 2.6 | MARK1 | 2.6 | CD164L2 | −3.8 | SUSD4 | −3.2 |
| USP48 | 2.6 | LURAP1 | 2.5 | SYF2 | −3.8 | SYF2 | −3.2 |
| DONSON | 2.5 | NAIF1 | 2.5 | TAB1 | −3.8 | HIBADH | −3.2 |
| SACM1L | 2.5 | GGCX | 2.5 | PDE6B | −3.7 | UFL1 | −3.2 |
| ZNF230 | 2.5 | SIK2 | 2.5 | COL4A4 | −3.6 | C8orf74 | −3.2 |
| GGT5 | 2.5 | SPAG17 | 2.5 | HSPA1A | −3.6 | GPR107 | −3.2 |
| KRTAP13‐1 | 2.4 | DLG1 | 2.4 | MUSK | −3.5 | GBA | −3.2 |
| LURAP1 | 2.4 | MYH11 | 2.4 | ARHGAP17 | −3.5 | OR5AR1 | −3.2 |
One gene that appears to be of interest is MC4R, melanocortin 4 receptor, which is the highest ranked when the narrow category of variants is used and the third highest using the broad category, implying that there is an excess of rarer, functional variants among SZ subjects. In fact, though, MC4R variants have previously been reported to both increase and decrease the risk for obesity rather than having an effect on schizophrenia (Vaisse et al., 1998; Yeo et al., 1998; Mergen et al., 2001; Miraglia et al., 2002; Heid et al., 2005; Young et al., 2007; Chambers et al., 2008; Loos et al., 2008). To assist in understanding this result more fully, the raw output from the SCOREASSOC program is presented in Table 3. This shows that there is a broadly similar distribution of variants between OB and SZ except that two nonsynonymous variants, at positions 18:58038832 and 18:58039276, are more frequent in SZ subjects. There is also a single, highly weighted stop variant at 18:58039478 in a SZ subject which would also make a contribution to the high MLP for this gene. The two nonsynonymous variants are rs52820871 (chr18.hg19:g.58038832T>G) and rs2229616 (chr18.hg19:g.58039276C>T), which are already well established to be associated with lower BMI (Geller et al. 2004; Heid et al. 2005; Stutzmann et al. 2007; Young et al. 2007; Wang et al. 2010; Evans et al. 2014; Malzahn et al. 2014). The stop variant is rs13447324 (chr18.hg19:g.58039478G>T), Y35*, which is likewise well‐established as a cause of autosomal dominant obesity (Hinney et al., 1999; Sina et al., 1999; Farooqi & O'Rahilly, 2006). Thus, these results for MC4R are consistent with the previously reported effects of the two nonsynonymous variants as being protective against obesity but are anomalous in that the stop variant is observed in an SZ subject and not in any OB subject.
Table 3.
Output from SCOREASSOC for the analysis of MC4R using the broad category of variants and treating SZ subjects as cases
| OB | SZ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position (hg19, chr18) | AA | AB | BB | MAF | AA | AB | BB | MAF | Weight | Variant effect | VCF annotation |
| 58038462 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 9.99 | DOWNSTREAM | T>C |
| 58038470 | 981 | 1 | 0 | 0.0005 | 1391 | 0 | 0 | 0.0000 | 9.99 | DOWNSTREAM | C>G |
| 58038489 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 9.99 | DOWNSTREAM | G>A |
| 58038514 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 9.99 | DOWNSTREAM | C>T |
| 58038524 | 979 | 3 | 0 | 0.0015 | 1386 | 6 | 0 | 0.0022 | 9.93 | DOWNSTREAM | G>A |
| 58038612 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | C>T:PolyPhen:benign(0.048) |
| 58038826 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | C>T:PolyPhen:possibly_damaging(0.45) |
| 58038829 | 982 | 0 | 0 | 0.0000 | 1390 | 2 | 0 | 0.0007 | 99.85 | NON_SYNONYMOUS_CODING | C>T:PolyPhen:probably_damaging(1) |
| 58038832 | 968 | 14 | 0 | 0.0071 | 1361 | 31 | 0 | 0.0111 | 96.62 | NON_SYNONYMOUS_CODING | T>G:PolyPhen:benign(0.008) |
| 58038989 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 49.96 | SYNONYMOUS_CODING | G>A |
| 58039013 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 49.96 | SYNONYMOUS_CODING | A>G |
| 58039049 | 981 | 1 | 0 | 0.0005 | 1392 | 0 | 0 | 0.0000 | 49.96 | SYNONYMOUS_CODING | C>T |
| 58039203 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | G>A:PolyPhen:probably_damaging(0.997) |
| 58039215 | 982 | 0 | 0 | 0.0000 | 1390 | 2 | 0 | 0.0007 | 99.85 | NON_SYNONYMOUS_CODING | T>C:PolyPhen:probably_damaging(0.99) |
| 58039219 | 981 | 1 | 0 | 0.0005 | 1392 | 0 | 0 | 0.0000 | 99.92 | NON_SYNONYMOUS_CODING | C>A:PolyPhen:probably_damaging(0.996) |
| 58039276 | 964 | 18 | 0 | 0.0092 | 1337 | 55 | 0 | 0.0198 | 94.55 | NON_SYNONYMOUS_CODING | C>T:PolyPhen:benign(0.042) |
| 58039301 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 49.96 | SYNONYMOUS_CODING | G>A |
| 58039402 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | C>T:PolyPhen:probably_damaging(0.985) |
| 58039473 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | T>A>C:PolyPhen:benign(0) |
| 58039478 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 199.85 | STOP_GAINED | G>T |
| 58039552 | 982 | 0 | 0 | 0.0000 | 1391 | 1 | 0 | 0.0004 | 99.92 | NON_SYNONYMOUS_CODING | T>C:PolyPhen:benign(0) |
| 58039642 | 981 | 1 | 0 | 0.0005 | 1391 | 1 | 0 | 0.0004 | 49.92 | 5PRIME_UTR | G>C |
The table shows genotype counts, frequencies, weights and effects for each variant. The weighted scores were calculated for each subject and the means compared. Mean scores OB = 3.4, SZ = 7.0, t(2372 df) = 3.8, p = 0.00015, SLP = 3.8.
Another gene of interest is NLGN2, neuroligin 2, which is ranked fourth when using the broad category of variants. This codes for a postsynaptic protein and a previous study reported novel, functional mutations in NLGN2, each occurring in one or two subjects with schizophrenia (Sun et al., 2011). The SCOREASSOC output shows that the SLP of 3.5 reflects a general excess of rare variants among SZ subjects, mostly not affecting amino acid sequence. This effect is spread across dozens of variants, such that one cannot definitively identify any which might individually be associated with risk of schizophrenia.
Findings for genes which may be of interest are summarised in Table 4. One phenomenon to be aware of is that because of LD relationships between variants the same signal can be picked up by multiple genes. This is the case for CRHR1, MAPK, STH and IMP5. A haplotype spanning all these genes is somewhat commoner in OB than SZ subjects. If it represents a real signal then CRHR1 seems to be the gene mostly likely to be responsible. Likewise, both HIST1H2AJ and HIST1H4A are highly ranked. However the variants making the most substantial contributions to the MLP for each gene are only 2 Mb apart and are in strong LD with each other, so there is in fact only one signal.
Table 4.
List of some of the highest and lowest ranked genes with explanatory notes
| Symbol | SLP | Analysis | Gene name | Comments |
|---|---|---|---|---|
| MC4R | 3.8 | SZ, broad | Melanocortin 4 receptor | Increased frequency among SZ subjects of two nonsynonymous variants previously reported to be protective against obesity |
| NLGN2 | 3.5 | SZ, broad | Neuroligin 2 | Codes for postsynaptic protein and regarded as candidate gene for schizophrenia. Overall generally increased numbers of rare variants in SZ subjects with no individual variant strongly associated |
| CRP | 3.4 | SZ, broad | C‐reactive protein, pentraxin‐related | Involved in immunity and inflammation systems. Variants generally commoner in SZ subjects. Nonsynonymous variant at 1:159683814 is present in 17 SZ and 2 OB subjects. This is rs77832441, which has been reported to be associated with reduced CRP levels |
| DONSON | 3.4 | SZ, narrow | Downstream neighbour of SON | Function unknown. Nonsynonymous variants at 21:34950728 seen in 21 SZ against 7 OB subjects and at 21:34955922 in 10 SZ and 0 OB subjects |
| GABRA3 | 2.9 | SZ, broad | GABA A receptor, alpha 3 | Some previous reports of involvement of GABA receptors in schizophrenia. However, the gene is on the X chromosome and the result is likely an artifact due to counting hemizygote males as homozygotes |
| GTF3A | 4.1 | SZ, broad | General transcription factor IIIA | Result is driven by modest excess of many different variants |
| IL36B | 2.9 | SZ, broad | Interleukin 36, beta | Involved in inflammation. There are stop variants at 2:113785602 and 2:113788694 in 7 and 2 SZ subjects and no OB subjects |
| ADCYAP1R1 | 3.2 | SZ, narrow | Adenylate cyclase activating polypeptide 1 (pituitary) receptor | Previous reports of association with schizophrenia and involvement in adipose tissue expandability. Nonsynonymous variants at 7:31104520 and 7:31124376 commoner in SZ than OB subjects |
| ARSA | 2.9 | SZ, narrow | Arylsulfatase A | Mutations in this gene are the known cause of metachromatic leucodystrophy, which can have features similar to schizophrenia. A few very rare nonsynonymous and splice site variants are seen only in SZ cases and the common nonsynonymous variant at 22:51065361 (rs6151415) has MAF 0.085 in SZ and 0.061 in OB subjects |
| DLG1 | 2.4 | SZ, narrow | Discs, large homolog 1 (drosophila) | Involved in synaptogenesis. A number of nonsynonymous variants somewhat commoner in SZ than OB subjects and non‐synonymous variant at 3:196792663 occurs in 10 SZ and no OB subjects |
| SIK2 | 2.5 | SZ, narrow | Salt‐inducible kinase 2 | Known to be involved in lipid homeostasis and adipogenesis. A number of nonsynonymous variants occur only in SZ subjects and nonsynonymous variant at 11:111590605 occurs in 14 SZ and 2 OB subjects |
| SLAIN1 | 3.3 | SZ, narrow | SLAIN motif family, member 1 | Involved in neurodevelopment. Nonsynonymous variant at 13:78320801 occurs in 18 SZ subjects and 1 OB subject |
| UBE2Q2 | 2.6 | SZ, narrow | Ubiquitin‐conjugating enzyme E2Q family member 2 | Differentially expressed in mice with diet‐induced obesity. Excess of several nonsynonymous variants in SZ versus OB subjects |
| ZNF 507 | 3.3 | SZ, narrow | Zinc finger protein 507 | Disruption associated with neurodevelopmental disorders. Several nonsynonymous variants common in SZ subjects and nonsynonymous variant at 19:32844995 occurs in 9 SZ and no OB subjects |
| CRHR1 | ‐3.9 | OB, broad | Corticotropin‐releasing hormone receptor 1 | Previously implicated in physiological pathways including obesity and response to stress. A haplotype of several noncoding variants is somewhat commoner in OB than SZ subjects. This haplotype extends through MAPK, STH and IMP5, accounting for their MLPs |
| HIST1H2AJ | −4.2 | OB, broad | Histone cluster 1, H2aj | Downstream variant at 6:27782031 has MAF 0.14 in OB and 0.11 in SZ subjects |
| HIST1H4A | −4.7 | OB, broad | Histone cluster 1, H4a | 3' UTR variant at 6:26022244 has MAF 0.14 in OB and 0.096 in SZ subjects. However, this variant is in LD with the one at 6:27782031 so these signals are not independent |
| MAPT | ‐4.7 | OB, broad | Microtubule‐associated protein tau | A haplotype of several common variants is slightly commoner among OB subjects |
| MUSK | −3.6 | OB, broad | Interacts with NSF. Splice site variant at 9:113449377 has MAF 0.07 in OB and 0.05 in SZ subjects | |
| NSF | −5.5 | OB, broad | N‐ethylmaleimide‐sensitive factor | Interacts with MUSK. Splice site variant at 17:44788310 has MAF 0.28 in OB and 0.23 in SZ subjects |
| SNORD115 | −4.6 | OB, broad | Small nucleolar RNA, C/D box 115‐15 | In Prader–Willi region and regulates alternative splicing of CRHR1. Several variants have higher MAF in OB than SZ subjects |
| GDF3 | −3.5 | OB, narrow | Growth differentiation factor 3 | Implicated in regulation of adiposity and energy expenditure. Nonsynonymous variant at 12:7842587 has frequency 0.040 in OB and 0.022 in SZ subjects |
| HIBADH | −3.2 | OB, narrow | 3‐hydroxyisobutyrate dehydrogenase | Differentially expressed in T2DM. SLP is driven by splice site or nonsynonymous variants of which 7 are singletons occurring in OB subjects and the other, at 7:27570942, occurs in 8 OB and 3 SZ subjects |
A final point worth making is that for some genes the SLP is driven by just one or two variants with a very marked difference in allele frequencies between cohorts whereas for others there are large numbers of very rare variants, with a tendency for these to occur more commonly in one cohort or the other. For example, in SLAIN1 there is a nonsynonymous variant in 18 SZ subjects and only 1 OB subject. Likewise, in IL36B there are stop variants at two loci which between them are seen in nine SZ subjects and no OB subject, in DLG1 there is a nonsynonymous variant in 10 SZ subjects and no OB subject and in CRP there is a nonsynonymous variant previously claimed to be associated with CRP levels which is present in 17 SZ and two OB subjects. However, in other genes, such as NLGN2, GTF3A and HIBADH a number of very rare variants collectively account for the SLP.
Discussion
The anticonservative nature of the test when using the OB cohort as cases was viewed as slightly puzzling, given that it had previously been shown to behave well when applied to simulated data (Curtis, 2012). The fact that the results treating the SZ cohort as cases were not markedly anticonservative make it seem less likely that this phenomenon might be due to population effects such as stratification or linkage disequlibrium (LD) between variants. An alternative explanation is that the variance of the scores is underestimated when the excess occurs in OB subjects and this might, for example, result from subjects being related to each other or being the offspring of consanguineous matings. However, attempts to model relatedness by duplicating some subjects failed to reproduce the observed Q:Q plots. Likewise, treating homozygotes as heterozygotes, in an attempt to nullify the effects of consanguinity, failed to produce a closer fit to expected values. The weighted burden method does not adjust for population stratification and it assumes that the cohorts are ethnically well matched and that subjects are unrelated. It is not clear how violations of these assumptions might impact on the results obtained.
The finding that no genes produce results withstanding correction for multiple testing is in line with that of the previous schizophrenia exome study, which used a somewhat larger sample size. It is becoming apparent that next generation sequencing studies applied to complex diseases in samples numbering the low thousands are hypothesis‐generating and are unlikely to produce results which conclusively implicate individual variants or genes. Nevertheless, some findings do appear to be of interest and worthy of attempts at follow‐up, although there is a question of how much to focus attention on genes which seem to be plausible candidates without running the risk of overlooking novel findings which might point to previously unsuspected mechanisms of pathogenesis. For example, neuronal and inflammatory genes are thought to be involved in the susceptibility to schizophrenia and in this light the results for NLGN2, ARSA, DLG1, SLAIN1, ZNF507, CRP and IL36B seem interesting. Likewise, given previous findings related to obesity, the results for SIK2, CRHR1, SNORD115, GDF3 and HIBADH may be of note, especially in view of the fact that SNORD115 has been reported to be involved in the alternative splicing of CRHR1 (Kishore et al., 2010).
Following up suggestive results in larger samples may not be straightforward. For variants which are not extremely rare this might simply involve carrying out genotyping in additional, larger case‐control cohorts. However, some genes are highlighted on the basis of an excess of many different variants, each occurring in only one or two subjects. Validating these findings might require sequencing the gene in large numbers of subjects. It has been suggested that an alternative approach to following up an extremely rare variant is to carry out family studies of the subjects possessing the variant (Curtis, 2011). Thus, if there are affected relatives who also have the variant one gains confidence that it has an effect whereas an affected relative not sharing the variant casts doubt on its relevance.
The results illustrate a problem with the analytic approach which was originally proposed, which was to test for an excess of rare and/or functional variants in cases compared with controls. In fact, three genes possibly involved in susceptibility to obesity, MC4R, ADCYAP1R1 and SIK2, produced highly ranked positive SLPs indicating that such an excess was occurring in SZ rather than OB subjects. Thus, had the method been applied as intended in a case‐control study with OB chosen as the case phenotype then these findings might have been overlooked. It was argued previously that there were biological and statistical reasons to expect that, when dealing with a fairly rare disease with a deleterious effect on fitness, rare variants identified in a case‐control study would be more likely to increase risk than reduce it (Curtis, 2012). Nevertheless, the example of MC4R makes it clear that one cannot rely on this. In the light of this, it seems that one should attend to both strongly positive and negative SLPs in order to detect either an increase or decrease in variants among cases. Of course, this would not at all address the issue of different variants within the same gene having effects in different directions, in which case a test such as SKAT would be needed alongside the weighted burden test.
Approaches such as this might benefit from an improvement in the ability to predict the likely consequences of a change in DNA sequence. The weighting scheme used was crude and fairly arbitrary. One could easily imagine introducing other considerations, such as utilising SIFT and PolyPhen scores or information on regulatory function (Ng & Henikoff, 2003; Adzhubei et al., 2010). If effects could be predicted accurately then weights could be assigned on a more rational basis. On the other hand, it can be argued that the whole point of performing empirical studies is that one does not know which variants contribute to risk until one sees the extent to which they are associated with a disease phenotype. The effects of varying the weights given to different types of variant were not explored systematically. One might expect that varying the weights would have some impact on the SLPs obtained and their ranks but, as is often the case, the advantages of carrying out exploratory analyses in order to find a more appropriate model may be outweighed by the difficulties in interpreting results obtained from testing multiple scenarios.
The weighted burden test provides a quick, simple and intuitive test summarising the extent to which a gene harbours more functional, rare variants in cases than controls. It can be used to rank genes and highlight genes of interest and one can then look at the results for individual genes and variants in more detail. The method allows the user to test all variants simultaneously in a single analysis, implementing a crude model of variant effects which may have some face validity. On the other hand, it requires that weights be specified in advance and makes no attempt to fit them to the observed data. The method is implemented only for dichotomous phenotypes although it might be possible to extend it to be applied to quantitative measures. In the light of the results obtained, it seems sensible to implement a two‐tailed version of the approach, in that one should test for an excess of variants either in cases or in controls. However, this would not be helpful if some variants within a gene acted to increase risk and others to decrease it and in this situation one would not expect the method to be successful. Thus it should not be used in isolation. Hopefully, it will be possible to refine such approaches further once there is greater knowledge regarding the nature of genetic variation which influences risk of non‐Mendelian disease.
Conflict of Interest
The authors declare they have no conflict of interest.
Supporting information
Disclaimer: Supplementary materials have been peer‐reviewed but not copyedited.
Table S1 SLPs for all genes obtained from weighted burden test using broad and narrow sets of variants.
Acknowledgement
Thanks to Sadaf Farooqi for helpful comments on MC4R variants. This study makes use of data generated by the UK10K Consortium, derived from samples from UK10K_NEURO_Iop_Collier, UK10K_NEURO_UKSCZ, UK10K_NEURO_ABERDEEN, UK10K_EURO_NEDINBURGH, UK10K_NEURO_EDINBURGH, UK10K_NEURO_UCL and UK10K_OBESITY_SCOOP. A full list of the investigators who contributed to the generation of the data is available online (http://www.UK10K.org). Funding for UK10K was provided by the Wellcome Trust under award WT091310.
References
- Adzhubei, I. A. , Schmidt, S. , Peshkin, L. , Ramensky, V. E. , Gerasimova, A. , Bork, P. , Kondrashov, A. S. & Sunyaev, S. R. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, J. C. , Elliott, P. , Zabaneh, D. , Zhang, W. , Li, Y. , Froguel, P. , Balding, D. , Scott, J. & Kooner, J. S. (2008) Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 40, 716–718. [DOI] [PubMed] [Google Scholar]
- Curtis, D. (2011) Assessing the contribution family data can make to case‐control studies of rare variants. Ann Hum Genet 75, 630–638. [DOI] [PubMed] [Google Scholar]
- Curtis, D. (2012) A rapid method for combined analysis of common and rare variants at the level of a region, gene, or pathway. Adv Appl Bioinform Chem 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, D. , Vine, A.E. , Mcquillin, A. , Bass, N. J. , Pereira, A. , Kandaswamy, R. , Lawrence, J. , Anjorin, A. , Choudhury, K. & Datta, S. R. (2011) Case‐case genome wide association analysis reveals markers differentially associated with schizophrenia and bipolar disorder and implicates calcium channel genes. Psychiatr Genet 21, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D. S. , Calton, M. A. , Kim, M. J. , Kwok, P.‐Y. , Miljkovic, I. , Harris, T. , Koster, A. , Liu, Y. , Tranah, G. J. , & Ahituv, N. (2014) Genetic association study of adiposity and melanocortin‐4 receptor (MC4R) common variants: Replication and functional characterization of non‐coding regions. PLoS One 9, e96805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi, I. S. & O'rahilly, S. (2006) Genetics of obesity in humans. Endocr Rev 27, 710–718. [DOI] [PubMed] [Google Scholar]
- Geller, F. , Reichwald, K. , Dempfle, A. , Illig, T. , Vollmert, C. , Herpertz, S. , Siffert, W. , Platzer, M. , Hess, C. , & Gudermann, T. (2004) Melanocortin‐4 receptor gene variant I103 is negatively associated with obesity. Am J Hum Genet 74, 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid, I. , Vollmert, C. , Hinney, A. , Döring, A. , Geller, F. , Löwel, H. , Wichmann, H. , Illig, T. , Hebebrand, J. , & Kronenberg, F. (2005) Association of the 103I MC4R allele with decreased body mass in 7937 participants of two population based surveys. J Med Genet 42, e21‐e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney, A. , Schmidt, A. , Nottebom, K. , Heibult, O. , Becker, I. , Ziegler, A. , Gerber, G. , Sina, M. , Gorg, T. , & Mayer, H. (1999) Several mutations in the melanocortin‐4 receptor gene including a nonsense and a frameshift mutation associated with dominantly inherited obesity in humans. J Clin Endocrinol Metab 84, 1483–1486. [DOI] [PubMed] [Google Scholar]
- Kishore, S. , Khanna, A. , Zhang, Z. , Hui, J. , Balwierz, P.J. , Stefan, M. , Beach, C. , Nicholls, R. D. , Zavolan, M. , & Stamm, S. (2010) The snoRNA MBII‐52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos, R. J. , Lindgren, C. M. , Li, S. , Wheeler, E. , Zhao, J. H. , Prokopenko, I. , Inouye, M. , Freathy, R. M. , Attwood, A. P. , & Beckmann, J.S. (2008) Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 40, 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, B. E. & Browning, S. R. (2009) A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genetics 5, e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn, D. , Müller‐Nurasyid, M. , Heid, I. M. , Wichmann, H.‐E. , & Bickeböller, H. (2014) Controversial association results for INSIG2 on body mass index may be explained by interactions with age and with MC4R. Eur J Hum Genet, 22, 1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergen, M. , Mergen, H. , Ozata, M. , Oner, R. , & Oner, C. (2001) Rapid communication: A novel melanocortin 4 receptor (MC4R) gene mutation associated with morbid obesity. J Clin Endocrinol Metab 86, 3448‐3448. [DOI] [PubMed] [Google Scholar]
- Miraglia, D. G. E. , Cirillo, G. , Nigro, V. , Santoro, N. , D'urso, L. , Raimondo, P. , Cozzolino, D. , Scafato, D. , & Perrone, L. (2002) Low frequency of melanocortin‐4 receptor (MC4R) mutations in a Mediterranean population with early‐onset obesity. Int J Obes Relat Metab Disord 26, 647–651. [DOI] [PubMed] [Google Scholar]
- Ng, P. C. & Henikoff, S. (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. M. , Moran, J. L. , Fromer, M. , Ruderfer, D. , Solovieff, N. , Roussos, P. , O'dushlaine, C. , Chambert, K. , Bergen, S. E. , & Kähler, A. (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sina, M. , Hinney, A. , Ziegler, A. , Neupert, T. , Mayer, H. , Siegfried, W. , Blum, W. F. , Remschmidt, H. , & Hebebrand, J. (1999) Phenotypes in three pedigrees with autosomal dominant obesity caused by haploinsufficiency mutations in the melanocortin‐4 receptor gene. Am J Hum Genet 65, 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann, F. , Vatin, V. , Cauchi, S. , Morandi, A. , Jouret, B. , Landt, O. , Tounian, P. , Levy‐Marchal, C. , Buzzetti, R. , & Pinelli, L. (2007) Non‐synonymous polymorphisms in melanocortin‐4 receptor protect against obesity: The two facets of a Janus obesity gene. Hum Mol Genet 16, 1837–1844. [DOI] [PubMed] [Google Scholar]
- Sun, C. , Cheng, M.‐C. , Qin, R. , Liao, D.‐L. , Chen, T.‐T. , Koong, F.‐J. , Chen, G. , & Chen, C.‐H. (2011) Identification and functional characterization of rare mutations of the neuroligin‐2 gene (NLGN2) associated with schizophrenia. Hum Mol Genet 20, 3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UK10K Consortium . (2015) The UK10K project identifies rare variants in health and disease. Nature. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse, C. , Clement, K. , Guy‐Grand, B. , & Froguel, P. (1998) A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet 20, 113–114. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Ma, J. , Zhang, S. , Hinney, A. , Hebebrand, J. , Wang, Y. , & Wang, H. J. (2010) Association of the MC4R V103I polymorphism with obesity: A Chinese case–control study and meta‐analysis in 55,195 individuals. Obesity 18, 573–579. [DOI] [PubMed] [Google Scholar]
- Wheeler, E. , Huang, N. , Bochukova, E. G. , Keogh, J. M. , Lindsay, S. , Garg, S. , Henning, E. , Blackburn, H. , Loos, R. J. , & Wareham, N. J. (2013) Genome‐wide SNP and CNV analysis identifies common and low‐frequency variants associated with severe early‐onset obesity. Nat Genet 45, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. C. , Lee, S. , Cai, T. , Li, Y. , Boehnke, M. , & Lin, X. (2011) Rare‐variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G. S. , Farooqi, I. S. , Aminian, S. , Halsall, D. J. , Stanhope, R. G. , & O'rahilly, S. (1998) A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20, 111–112. [DOI] [PubMed] [Google Scholar]
- Young, E. H. , Wareham, N. J. , Farooqi, S. , Hinney, A. , Hebebrand, J. , Scherag, A. , O'rahilly, S. , Barroso, I. , & Sandhu, M. S. (2007) The V103I polymorphism of the MC4R gene and obesity: Population based studies and meta‐analysis of 29 563 individuals. Int J Obes 31, 1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclaimer: Supplementary materials have been peer‐reviewed but not copyedited.
Table S1 SLPs for all genes obtained from weighted burden test using broad and narrow sets of variants.


