Abstract
Infantile encephalopathies are a group of clinically and biologically heterogeneous disorders for which the genetic basis remains largely unknown. Here, we report a syndromic neonatal encephalopathy characterized by profound developmental disability, severe hypotonia, seizures, diminished respiratory drive requiring mechanical ventilation, brain atrophy, dysgenesis of the corpus callosum, cerebellar vermis hypoplasia, and facial dysmorphism. Biallelic inactivating mutations in TBCK (TBC1-domain-containing kinase) were independently identified by whole-exome sequencing as the cause of this condition in four unrelated families. Matching these families was facilitated by the sharing of phenotypic profiles and WES data in a recently released web-based tool (Geno2MP) that links phenotypic information to rare variants in families with Mendelian traits. TBCK is a putative GTPase-activating protein (GAP) for small GTPases of the Rab family and has been shown to control cell growth and proliferation, actin-cytoskeleton dynamics, and mTOR signaling. Two of the three mutations (c.376C>T [p.Arg126∗] and c.1363A>T [p.Lys455∗]) are predicted to truncate the protein, and loss of the major TBCK isoform was confirmed in primary fibroblasts from one affected individual. The third mutation, c.1532G>A (p.Arg511His), alters a conserved residue within the TBC1 domain. Structural analysis implicated Arg511 as a required residue for Rab-GAP function, and in silico homology modeling predicted impaired GAP function in the corresponding mutant. These results suggest that loss of Rab-GAP activity is the underlying mechanism of disease. In contrast to other disorders caused by dysregulated mTOR signaling associated with focal or global brain overgrowth, impaired TBCK function results in progressive loss of brain volume.
Keywords: exome sequencing, Mendelian disease, infantile encephalopathy, mTOR signaling, GTPase-activating protein
Main Text
Severe infantile encephalopathy is a non-specific clinical condition that can be caused by hypoxemia, hemorrhage, toxins (e.g., hyperbilirubinemia or withdrawal from selective serotonin-reuptake inhibitors or narcotics), and mutations in multiple genes, such as PURA (MIM: 600473), UNC80 (MIM: 612636), or NALCN (MIM: 611549).1, 2, 3, 4, 5, 6, 7, 8, 9 Over the past 5 years, substantial progress has been made toward defining the genetic basis of infantile encephalopathies; however, the genetic cause remains unknown for a large proportion of affected individuals, and these conditions have few distinguishing clinical features, making it challenging to stratify affected individuals for gene-discovery efforts. Whole-exome sequencing (WES) of multiple independent families affected by non-specific infantile encephalopathies is a powerful way to discover shared candidate genes among affected individuals, who upon further clinical evaluation are often found to share phenotypic features delineating a distinctive condition within this otherwise non-specific category. Here, we report on a syndromic infantile encephalopathy characterized by severe developmental disability, brain atrophy, early-onset focal seizures, central respiratory failure, and facial dysmorphism. WES in four families led to the discovery of mutations in TBCK, a gene encoding a putative Rab-specific GTPase-activating protein (GAP), as the underlying molecular cause.
Clinical data and biological samples were obtained from four unrelated families after written informed consent was provided, and studies for each family were approved by the respective institutional review boards of the University of Washington (Seattle), the Genetics Services Advisory Committee (New South Wales), the Università La Sapienza (Rome), and the Children’s Hospital of Philadelphia (Philadelphia). A single affected individual with neonatal-onset encephalopathy, brain atrophy with cerebellar malformation, and later-onset seizures (individual A-II-1 in family A; Figure 1, Table 1, and Table S2) was referred to one of us with the tentative diagnosis of Joubert syndrome (MIM: PS213300) with atypical features. Twenty-seven genes previously implicated in Joubert syndrome were screened,10 but no mutations predicted to be pathogenic were found. Upon re-evaluation, the clinical and MRI features (i.e., early respiratory failure, epilepsy, developmental regression, dysmorphic features, and absence of classic imaging features) were inconsistent with the diagnosis of Joubert syndrome, which suggested that this person instead had a condition involving a hindbrain malformation that had not been previously delineated. Subsequently, WES was performed by the University of Washington Center for Mendelian Genomics (UW-CMG) as described previously.11 Single-nucleotide variants (SNVs) were annotated with the SeattleSeq137 Annotation Server. SNVs categorized as intergenic, coding synonymous, in the UTR, near a gene, or intronic were excluded. Variants flagged as low quality or potential false positives (quality score ≤ 50, long homopolymer run > 4, low quality by depth < 5, or within a cluster of SNPs), as well as those with a mean alternative allele frequency > 0.005 in the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (ESP6500) or in an internal WES database of ∼700 exomes, were also excluded. Allowing for recessive and X-linked inheritance, this filtering left 30 candidate genes (Table S1); however, 27 genes could be excluded because they contained one variant present as homozygous in the Exome Aggregation Consortium (ExAC) Browser or with a CADD score ≤ 15.12 Only one gene, TBCK, harbored a biallelic truncating variant (chr4: 107,183,260 G>A [UCSC Genome Browser build hg19], c.376C>T [p.Arg126∗] [GenBank: NM_001163435.2]), so TBCK was considered the best candidate. No other individuals with TBCK mutations were known at the time, so this candidate gene was internally archived in 2013 and later in Geno2MP,13 a recently released web-based tool that was developed by the UW-CMG and that links phenotypic descriptions to rare-variant genotypes in ∼3,800 exomes from families affected by Mendelian conditions.
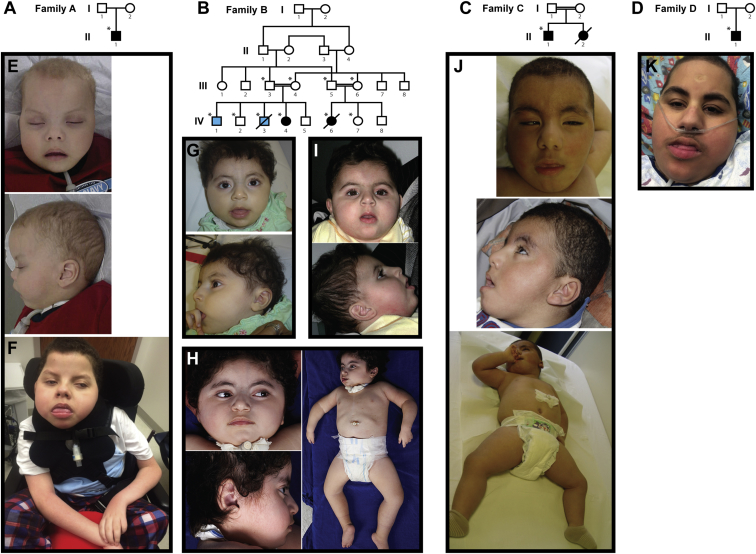
Figure 1.
Pedigrees and Pictures of Individuals with TBCK-Related Encephalopathy
(A–D) Pedigrees for families A–D (A–D, respectively), in whom loss-of-function mutations in TBCK segregate with disease. Solid black fill indicates individuals affected by TBCK-related encephalopathy. Light blue fill in family B indicates individuals affected by a hematologic phenotype without neurodevelopmental features; neither is homozygous for p.Lys455∗. WES was performed on individuals marked with asterisks.
(E–J) Images show the similar facial features and hypotonia in individual A-II-1 at 25 months (E) and 13 years (F), individual B-IV-4 at 17 months (G) and 4 years, 3 months (H), individual B-IV-6 at 18 months (I), individual C-II-1 at 21 months (J), and individual D-II-1 at 14 years. See Table 1 for detailed clinical information on each affected individual and Figure 2 and Table S3 for imaging information.
Table 1.
Clinical Features of Individuals with TBCK-Related Encephalopathy
|
Affected Individuals |
|||||
|---|---|---|---|---|---|
| A-II-1 | B-IV-4 | B-IV-6 | C-II-1 | D-II-1 | |
| Mutation | c.376C>T (p.Arg126∗) | c.1363A>T (p.Lys455∗) | c.1363A>T (p.Lys455∗) | c.1532G>A (p.Arg511His) | c.376C>T (p.Arg126∗) |
| Demographics | |||||
| Age | 14 years | 4 years | 10 years (deceased) | 24 months | 14 years |
| Sex | male | female | female | male | male |
| Ethnicity | Puerto Rican | Lebanese | Lebanese | Egyptian | Puerto Rican |
| Consanguinity | no | first cousin | first cousin | first cousin | no |
| Family history | mother and sister with cleft lip and palate, sister with arrhythmia | two brothers with hematologic disorder | two male cousins with hematologic disorder | similarly affected sister (deceased) | negative |
| Physical and Neurological Features | |||||
| Birth weight | 4.15 kg | 4.2 kg | 3.78 kg | unknown | 2.72 kg |
| Birth OFC | 38.5 cm (75th percentile) | 37 cm (90th percentile) | 35 cm (50th percentile) | unknown | unknown |
| Current OFC | 55 cm (57th percentile) | 53.5 cm (75th percentile) | 53 cm (50th percentile) | 49 cm (50th–70th percentile) | 53 cm (19th percentile) |
| Developmental disability | profound | profound | profound | profound | profound |
| Developmental regression | yes | yes | yes | no | yes |
| Reflexes | decreased or absent | decreased or absent | proximally increased, distally areflexic | decreased or absent | decreased or absent |
| Severe hypotonia | yes | yes | yes | yes | yes |
| Seizures | focal, refractory | focal, controlled on AED | tonic seizures, controlled on AED | no (abnormal EEG) | mixed focal and generalized, controlled on AED |
| Age of onset of seizures | 2.5 years | 25 months | 6 years | NA | 11 months (with fever), 15 months (afebrile) |
| Eyes and vision | cataracts, ptosis, cortical visual impairment | cortical visual impairment | visual impairment | bilateral optical atrophy, severe esotropia | cortical visual impairment, normal ERG |
| Hearing | normal | normal | normal | normal | normal |
| Respiration | chronic respiratory insufficiency, tracheostomy at 4 months | chronic respiratory insufficiency, tracheostomy at 2 years | chronic respiratory insufficiency, tracheostomy at 4 years | hypoventilation | chronic respiratory insufficiency, BiPAP at night, O2 as necessary via NC |
| Feeding | G-tube | G-tube | G-tube | G-tube | G-tube |
| Other features | right preauricular pit, gingival hyperplasia, macroglossia, neurogenic bladder (vesicostomy), subdural hematoma (unclear etiology), metabolic stroke, osteoporosis (three femur fractures), hypertriglyceridemia | mild virilization of external genitalia as a neonate (subsequently normalized), turricephaly, osteoporosis | muscle fasciculations, increased muscle bulk, elevated creatine kinase (800 IU/L) during viral infection (not repeated), osteoporosis | right-sided aortic arch, 11 ribs, turricephaly, hypertrichosis | hypertriglyceridemia, tongue fasciculations, intermittent hyponatremia, osteoporosis |
| Facial Features | |||||
| Bitemporal narrowing | yes | mild | mild | mild | yes |
| Arched eyebrows | yes | yes | yes | no | yes |
| Deep-set eyes | no | yes | yes | no | yes |
| High nasal bridge | yes | yes | yes | yes | yes |
| Anteverted nares | yes | yes | yes | mild | no |
| Exaggerated Cupid’s bow | yes | yes | yes | mild | yes |
| Coarse features | yes | no | no | yes | yes |
| Macroglossia | yes | no | no | no | yes |
| Gingival hyperplasia | yes | yes | unknown | unknown | no |
Abbreviations are as follows: AED, anti-epileptic drug; BiPAP, bilevel positive airway pressure; EEG, electroencephalogram; ERG, electroretinogram; G-tube, gastrostomy tube; NA, not applicable; NC, nasal cannula; O2, oxygen; and OFC, occipital frontal circumference.
In late 2014, two affected cousins in a second family were ascertained on the basis of an apparently novel neurodegenerative phenotype including cerebellar hypoplasia (family B; Figure 1, Table 1, and Table S2). Samples from the extended family underwent high-density genotyping and WES by the UW-CMG as described previously.11 Linkage analysis conducted under a recessive model, assuming a causal allele frequency of 0.0001 and full penetrance (f = 0,0,1.0), yielded two peaks (chr4: 88,837,705–109,066,366 and chr7: 81,735,015–112,087,033 [hg19]) with a maximum parametric LOD score of 2.48. WES data were annotated with the SeattleSeq138 Annotation Server. As with the analysis for family A, variants unlikely to impact protein-coding sequence, variants flagged by the Genome Analysis Toolkit (GATK) as low quality, and variants with an alternative allele frequency > 0.002 in any population in ESP6500, 1000 Genomes (phase 3 release), or the ExAC Browser (v.1.0) were excluded. Copy-number-variant calls were generated from exome data with CoNIFER.14 After the linkage and exome data were overlapped, only three genes, VPS50 (MIM: 616465; chr7: 92,953,034 G>A [hg19], c.1877G>A [p.Arg626Gln] [GenBank: NM_001257998.1]), LRCH4 (chr7: 100,173,865 C>T [hg19], c.1634G>A [p.Arg545His] [GenBank: NM_002319.3], rs370008127), and TBCK (chr4: 107,156,512 T>A [hg19], c.1363A>T [p.Lys455∗] [GenBank: NM_001163435.2], rs376699648), remained as candidates. All three genes were submitted to GeneMatcher and Geno2MP. No matches were found for VPS50 or LRCH4 in either database, but Geno2MP yielded a single individual (individual A-II-1; Figure 1 and Table 1) who had cerebellar malformation and was homozygous for a nonsense variant in TBCK. Sharing of the same candidate gene and an overlapping phenotype suggested that the putative loss-of-function mutations in TBCK are causal.
In parallel, a third family was ascertained on the basis of profound developmental disability associated with brain atrophy before 2 years of age in a son and death of a 12-month-old daughter exhibiting similar features (family C; Figure 1, Table 1, and Table S2). WES was performed on a sample from the affected son (individual C-II-1). Variants were called with an in-house pipeline15, 16, 17 and filtered against public and in-house databases; only clinically associated variants and variants with a alternative allele frequency < 0.001 (dbSNP142), < 0.002 (ExAC), or < 0.01 (in-house database with ∼500 exomes) or an unknown alternative allele frequency were retained. The SnpEff toolbox (v.4.1) was used for predicting the impact of variants, which were filtered until only functionally relevant variants (i.e., missense variants, nonsense variants, coding indel variants, and intronic variants located from −5 to +5 with respect to an exon-intron junction) remained. Functional annotation of variants was performed with SnpEff v.4.1 and dbNSFP v.2.8. Parental first-cousin consanguinity and the death of a female sibling with similar clinical features (i.e., suggestive facies, severe developmental delay, generalized hypotonia, and congenital heart malformation) strongly suggested autosomal-recessive inheritance, and on the basis of the hypothesis of homozygosity by descent, 16 candidate genes were identified. These candidates were stratified through a mixed filtering and prioritization strategy designed to retain genes with variants predicted to be damaging (CADD_phred > 15) and rank them on the basis of their biological relevance to the developmental processes altered in the disorder (GeneDistiller) (see Table S1).18 Among the eight retained genes, TBCK (chr4: 107,154,202 C>T [hg19], c.1532G>A [p.Arg511His] [GenBank: NM_001163435.2]) received the highest score from GeneDistiller and was considered the best candidate. At this point, an inquiry was made about whether TBCK was a candidate gene for any phenotypes being studied at the UW-CMG.
Finally, another individual of Puerto Rican descent and with severe hypotonia, chronic respiratory insufficiency requiring nighttime BiPAP (bilevel positive airway pressure), brain atrophy, and similar facial features (individual D-II-1; Figures 1 and 2) was ascertained in parallel. Clinical exome sequencing identified the same homozygous c.376C>T (p.Arg126∗) variant present in individual A-II-1, supporting the existence of a founder mutation in the Puerto Rican population. The phenotypic features in individual D-II-1 were quite similar to those of the other affected individuals (Table 1 and Tables S2 and S3).
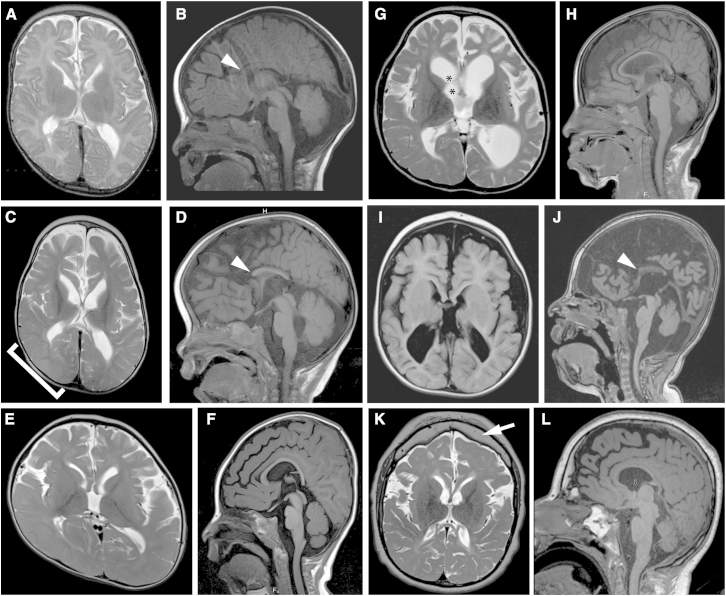
Figure 2.
Brain-Imaging Features in Individuals with TBCK-Related Encephalopathy
(A–D) Axial T2-weighted (A and C) and sagittal T1-weighted images (B and D) show progressive loss of gray- and white-matter volume (most severe in the frontal lobes), demonstrated by increasing ventriculomegaly and extra-axial spaces (cortical and cerebellar) in individual A-II-1 between 22 days of age (A and B) and 29 months of age (C and D). Also present are swelling of the right parieto-occipital lobe, presumably due to a metabolic stroke (bracket in C), a diffusely thin corpus callosum with absent rostrum (arrowheads in B and D), and mild cerebellar vermis hypoplasia with a relatively spared brainstem.
(E and F) Axial T2-weighted (E) and sagittal T1-weighted (F) images show ventriculomegaly, prominent extra-axial spaces, a diffusely thin but complete corpus callosum, and mild cerebellar vermis hypoplasia in individual B-IV-4 at 18 months of age. Right plagiocephaly is also present.
(G and H) Axial T2-weighted (G) and sagittal T1-weighted (H) images show marked ventriculomegaly, prominent extra-axial spaces, a diffusely thin but complete corpus callosum, and mild cerebellar vermis hypoplasia in individual B-IV-6 at 6 years of age. Synechiae are also present in the right frontal horn (asterisks in G).
(I and J) Axial T1-weighted (I) and sagittal T1-weighted (J) images show ventriculomegaly, prominent extra-axial spaces, a diffusely thin corpus callosum with an absent rostrum and anterior body (arrowhead in J), and mild cerebellar vermis hypoplasia in individual C-II-1 at 21 months of age.
(K and L) Axial T2-weighted (K) and sagittal T1-weighted (L) images show mild ventriculomegaly, prominent extra-axial spaces, a diffusely thin corpus callosum, mild cerebellar vermis hypoplasia with a relatively preserved brainstem, and a thick frontal bone (arrow in K) in individual D-II-1 at 14 years of age.
The variants in each family were either rare or not present in reference population databases. Specifically, the maximum alternative allele frequency in any population from ESP6500, 1000 Genomes phase 3, or ExAC Browser v.0.3 was 0.5208% for p.Arg126∗ (families A and D) and 0.0107% for p.Lys455∗ (family B) in ExAC Latinos, whereas p.Arg511His (family C) was not present in any database. Each variant had a high CADD score:12 37.0 for p.Arg126∗, 42.0 for p.Lys455∗, and 35.0 for p.Arg511His.
All affected individuals displayed profound developmental disability (because of which they made little progress beyond infancy) and a relatively homogeneous phenotype (Table 1 and Figure 1). Additionally, individuals A-II-1, B-IV-4, and B-IV-6 exhibited developmental regression (loss of visual fixation and following). All four individuals also required gastrostomy-tube feedings and had decreased respiratory drive, and the three older children required tracheostomies for chronic mechanical ventilation. Individuals A-II-1, B-IV-4, and B-IV-6 also developed focal seizures between 2 and 6 years of age, and individual C-II-1 had an abnormal electroencephalogram before 2 years of age without clinical seizures. Individuals A-II-1, B-IV-4, and C-II-1 had decreased to absent reflexes, whereas individual B-IV-6 developed hyperreflexia. Shared facial features included bitemporal narrowing, arched eyebrows, deep-set eyes, a high nasal bridge, anteverted nares, and an exaggerated “Cupid’s bow” of the upper lip (Figures 1D–1H). Individuals A-II-1 and B-IV-6 developed osteoporosis, which could be either a non-specific effect of chronic, severe disability or a more specific feature of TBCK-related disease. At 3 years of age, individual A-II-1 had an acute neurological decompensation with restricted diffusion of the right parieto-occipital cortex, indicative of a metabolic stroke, from which he recovered to his baseline neurological status. In all four individuals, brain imaging revealed increased ventricular and extra-axial spaces and diffusely decreased white-matter volume, and atrophy was confirmed by serial MRI in individuals A-II-1 and B-IV-4 (Figure 2 and Table S3). Despite the loss of brain volume, microcephaly was not observed. In addition, all individuals showed dysgenesis of the corpus callosum (thinning, partial agenesis, or both), increased T2-weighted FLAIR signal in the periventricular white matter, and cerebellar vermis hypoplasia.
TBCK encodes a protein with a TBC (Tre-2, Bub2, and Cdc16) domain flanked by an N-terminal kinase-like domain and a rhodanese homology domain at the C terminus, but its function has not been extensively characterized. Because of a lack of key catalytic residues, the kinase domain is presumably inactive.19 Multiple TBCK mRNAs have been reported, and two major isoforms differ in the presence or absence of the portion that encodes the N-terminal kinase-like domain (Figure 3A).20 For characterizing the impact of the truncating mutations, the amount of TBCK was evaluated by western blot with the use of skin fibroblasts obtained from individual A-II-1, who is homozygous for c.376C>T (p.Arg126∗) (Figure 3B). As expected, two major bands at ∼101 and 71 kDa, representing the long (full-length) protein and the shorter isoform without the N-terminal kinase-like domain,20 respectively, were observed in two control fibroblast lines, and the full-length isoform was more abundant. In contrast, the full-length protein was nearly absent in the fibroblasts from individual A-II-1, and the 71 kDa isoform was also reduced. Although the transcript for the 71 kDa isoform is not directly affected by the mutation, the reduced protein amount indicates that the mutation perturbs the levels of both major TBCK isoforms in fibroblasts.
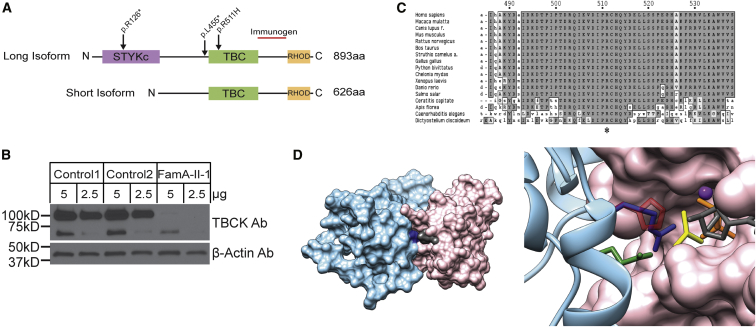
Figure 3.
Biochemical and Structural Characterization of TBCK Mutations
(A) TBCK encodes two isoforms containing a TBC1 domain (TBC) flanked by a rhodanese domain (RHOD) at the C terminus. The long isoform also contains a pseudokinase domain (STYKc) at the N terminus. The location of the identified variants is reported.
(B) Western blot shows that TBCK amounts were dramatically lower in a cell line from individual A-II-1 than in two control cell lines. The TBCK monoclonal antibody (1/250, Sigma HPA039951) recognizes a C-terminal fragment of the protein (depicted as a red line in A). After stripping, β-actin (1/5,000, Sigma A5441) was used as a loading control. The predicted sizes of the TBCK long and short isoforms are 101 and 71 kDa, respectively.
(C) Conservation of the catalytic arginine “finger” (Arg511 in TBCK) in TBCK orthologs.
(D) Homology model of the TBCK TBC1 domain complexed with the Rab33 GTPase. In the overall structure of the complex (left), both proteins are shown in a surface representation; Rab is colored pink, GDP is gray, the TBC1 domain is light blue, and Arg511 is dark blue. In the enlarged view of the active site of the GTPase (right), GDP is reported in gray sticks, and the phosphates are colored orange. AlF3, which in the crystal structure mimics the transition state for GTP hydrolysis, is shown in yellow, and the Mg atom is shown as a purple sphere. The TBC1 domain is reported in ribbon representation, and the side chains of key residues are shown as sticks. The two catalytic “fingers” Arg511 (blue) and Gln546 (green) are shown together with the disease-associated His511, shown in semi-transparent red.
The c.1532G>A (p.Arg511His) mutation in individual C-II-1 is predicted to alter a highly conserved arginine residue located in the TBC1 domain (Figure 3C). This structural unit of approximately 200 amino acids is characteristic of most GAPs that regulate the Rab family of small GTPases.21, 22 TBC domains negatively control Rab function by promoting GTP hydrolysis via stabilization of the transition state of the reaction. Specifically, the TBC domain interacts with the substrate and catalyzes the reaction by using a so-called “dual-finger” mechanism, in which a key Arg residue projects into the Rab active site.23, 24, 25 Because no crystallographic structure of TBCK was available, a homology model of the TBC1 domain of TBCK (residues 467–648) in complex with a Rab protein was constructed for exploring the structural impact of the disease-causing amino acid substitution. This model was generated with DeepView and the SwissModel server26 on the basis of the crystallographic structure of the Gyp1 TBC domain (36% sequence identity with the TBCK TBC domain) complexed with Rab33 bound to GDP and AlF3 (PDB: 2G77).23 TBCK was originally incorrectly classified as an unconventional TBC protein lacking the “arginine finger”;21 however, our modeling data support more recent bioinformatics analyses21 indicating that Arg511 does in fact represent an arginine finger essential for GAP function. The p.Arg511His substitution was introduced in silico with UCSF Chimera,27 and even though the Arg-to-His substitution introduces a side chain that could maintain the overall positive charge under appropriate conditions,28 the introduced residue is significantly bulkier and shorter than Arg and is unable to project into the GTPase active site. In the model, the minimum distance between Arg511 and the GDP phosphate is 3 Å, whereas this distance increases to 7 Å in the mutant. These considerations strongly point to impaired GAP activity as the predicted mechanism of disease associated with the p.Arg511His substitution.
TBCK was recently documented to play a role in the control of cell proliferation, cell size, and actin-cytoskeleton dynamics.19, 20 Althought the lack of several residues with critical function in mediating the activity of the kinase domain supports the view that TBCK is catalytically inactive,29, 30 the conservation of the key catalytic residues of the TBC1 domain, as well as the identification of a disease-causing mutation specifically targeting one of the two key residues of this domain (Arg511), points to the relevance of Rab-GAP activity for TBCK function, even though its physiological targets have not been identified.19 Indeed, the “Arg finger” is conserved in virtually all TBC domains with GAP activity,21, 22, 25 and it is largely accepted that unconventional TBC domains lacking this residue do not stimulate GTP hydrolysis in Rab proteins but rather have different functions.21, 22, 25 Of note, one of the human unconventional TBC proteins (TBC1D26) and its Chlamydomonas reinhardtii ortholog have a histidine residue in place of the “catalytic” arginine, and no GAP activity was detected in the algal protein.31 Mutation of the “catalytic” Arg in TBC-domain-containing proteins is commonly used as a tool for investigating their function and identifying their substrate Rabs,22, 32, 33, 34, 35 given that substitution of this amino acid greatly reduces the catalytic efficiency of these GAPs.14
TBCK’s role in controlling cell proliferation and growth and actin-cytoskeleton organization has been reported to be mediated by modulation of the mTOR (mammalian target of rapamycin) signaling network and transcriptional regulation of components of the mTOR complex.19 The mTOR pathway is a growth-regulating network, which in an activated state promotes angiogenesis, cell growth, and cell proliferation. A number of developmental brain disorders, collectively termed “TORopathies,” have been shown to result from dysregulated mTOR signaling.36, 37 These disorders are characterized by disorganized cortical lamination, seizures, and cytomegaly.38 Tuberous sclerosis complex (TSC [MIM: PS191100]) is caused by heterozygous loss-of-function mutations in TSC1 (MIM: 605284) or TSC2 (MIM: 191092), encoding proteins that negatively regulate the mTOR pathway. Similarly, in addition to playing a well-documented role in cancer, germline or postzygotic activating de novo mutations in genes encoding components of the PI3K-AKT3-mTOR pathway cause the overlapping hemimegalencephaly phenotypes megalencephaly-capillary malformation-polymicrogyria syndrome (MIM: 602501; usually caused by somatic mutations in PIK3CA [MIM: 171834]) and megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome (MIM: PS603387; usually caused by germline mutations in PIK3R2 [MIM: 603157], CCND2 [MIM: 123833], and AKT3 [MIM: 611223]). More recently, germline gain-of-function mutations in MTOR (MIM: 601231) have been described in two families affected by features overlapping the megalencephaly and RASopathy spectra of disorders.36, 39 These TORopathies are all caused by activation of the mTOR pathway and are generally characterized by brain overgrowth at a global (hemimegalencephaly spectrum) or focal (TSC) level. In contrast, loss of TBCK function is associated with loss of brain volume. In combination with available biochemical and functional data supporting a positive modulatory role for TBCK on mTOR signaling,19 our findings suggest that loss of TBCK function affects this signaling network in a way that opposes what is observed in other mTOR-related disorders and, importantly, that over-inhibition of the mTOR pathway might lead to this distinct and severe phenotype. Some of the affected individuals were found to have moderate mitochondrial dysfunction that did not meet Walker diagnostic criteria40 for a primary respiratory chain disorder and was not associated with increased brain lactate in the three individuals evaluated by magnetic resonance spectroscopy (Table S3). Given that the mTOR pathway is known to positively affect mitochondrial biogenesis41 and negatively regulate autophagy and mitophagy (reviewed in Bockaert and Marin42) and that several individuals showed decreased mitochondrial enzyme levels, we hypothesize that the progression of the disease might be due in part to decreased mitochondrial biogenesis and/or increased mitophagy.
In summary, we have established that biallelic mutations in TBCK cause a severe neurodevelopmental disorder whose major features include profound developmental delay or cognitive deficit, brain atrophy without microcephaly, dysgenesis of the corpus callosum, abnormal white-matter signal, cerebellar vermis hypoplasia, seizures, diminished respiratory function, and distinctive facies. Further strengthening this view is the recent report of a similar individual exhibiting severe developmental disability, poor feeding, abnormal eye movements, epilepsy, facial dysmorphism, severe hypotonia, and diffuse brain atrophy associated with a homozygous canonical splice-site mutation (c.1897+1G>A [GenBank: NM_001163435.1]) in TBCK.43 Structural and molecular-modeling analyses predict a key role for the substituted residue in mediating the Rab-GAP activity of the protein, suggesting that loss of TBCK GAP function is sufficient to cause disease. Of note, many encephalopathies are caused by de novo dominant mutations and therefore have low risk of recurrence, so distinguishing TBCK-related encephalopathy is essential for accurate recurrence-risk counseling and reproductive planning. Recognition of this disorder will also make it possible to delineate the natural history of TBCK-related disease and provide more precise prognostic information essential for guiding decisions about invasive treatments such as tracheostomy and gastrostomy in neonates and young children.
More than 3 years passed between the identification of TBCK as the strongest candidate gene in family A and the identification of TBCK mutations in families B, C, and D. Importantly, even though two families were sequenced through the same center (UW-CMG), their shared phenotype was not initially recognized, in part because the clinicians rightfully focused on different aspects of each affected individual’s condition. The identification of a shared candidate gene through a search of Geno2MP prompted comparison of the clinical findings and led to delineation of an overlapping phenotype. It is anticipated that matchmaking platforms in development, such as Matchmaker Exchange,44 will help to speed up this process, but only if investigators and clinicians worldwide all participate in such data-sharing efforts. Historically, ascertaining persons with a highly similar phenotype and then conducting subsequent gene discovery within that group has been a successful method for understanding new Mendelian phenotypes; however, this approach requires the Mendelian phenotype to be common enough that a single investigator could be expected to encounter multiple affected individuals. The small number of individuals with biallelic mutations in TBCK identified thus far, and the international assemblage of investigators who identified these individuals, underscores the fact that many, if not most, Mendelian phenotypes now being studied will be more rapidly and successfully tackled by widespread sharing of phenotypes, genotypes, and candidate genes.13, 44, 45
Acknowledgments
We thank the families for their participation and support. Our work was supported in part by grants from the NIH National Human Genome Research Institute and National Heart, Lung, and Blood Institute (1U54HG006493 to M.B., D.N., and J.S. and 1RC2HG005608 to M.B., D.N., and J.S.), the NIH National Institute of Neurological Diseases and Stroke (K12NS049453-09 to X.O.G.), the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development (U54HD083091 [Genetics Core and Sub-project 6849] to D.D.), Telethon-Italy (GGP13107 to M.T.), and the Ospedale Pediatrico Bambino Gesù (GeneRare and Ricerca Corrente 2016 to M.T.). The authors would like to thank the University of Washington Center for Mendelian Genomics and all contributors to Geno2MP for the use of data included in Geno2MP. The authors would also like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Published: March 31, 2016
Footnotes
Supplemental Data include three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.01.016.
Contributor Information
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
Dan Doherty, Email: ddoher@uw.edu.
Web Resources
The URLs for data presented herein are as follows:
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org
GeneDistiller, http://www.genedistiller.org/
Geno2MP, http://geno2mp.gs.washington.edu
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Lalani S.R., Zhang J., Schaaf C.P., Brown C.W., Magoulas P., Tsai A.C.-H., El-Gharbawy A., Wierenga K.J., Bartholomew D., Fong C.-T. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am. J. Hum. Genet. 2014;95:579–583. doi: 10.1016/j.ajhg.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt D., Leventer R.J., Simons C., Taft R., Swoboda K.J., Gawne-Cain M., Magee A.C., Turnpenny P.D., Baralle D., DDD study Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J. Med. Genet. 2014;51:806–813. doi: 10.1136/jmedgenet-2014-102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez Y., Kadir R., Volodarsky M., Noyman I., Flusser H., Shorer Z., Gradstein L., Birnbaum R.Y., Birk O.S. UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J. Med. Genet. 2015 doi: 10.1136/jmedgenet-2015-103352. [DOI] [PubMed] [Google Scholar]
- 4.Stray-Pedersen A., Cobben J.-M., Prescott T.E., Lee S., Cang C., Aranda K., Ahmed S., Alders M., Gerstner T., Aslaksen K., Care4Rare Canada Consortium. Baylor-Hopkins Center for Mendelian Genomics Biallelic Mutations in UNC80 Cause Persistent Hypotonia, Encephalopathy, Growth Retardation, and Severe Intellectual Disability. Am. J. Hum. Genet. 2016;98:202–209. doi: 10.1016/j.ajhg.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamseldin H.E., Faqeih E., Alasmari A., Zaki M.S., Gleeson J.G., Alkuraya F.S. Mutations in UNC80, Encoding Part of the UNC79-UNC80-NALCN Channel Complex, Cause Autosomal-Recessive Severe Infantile Encephalopathy. Am. J. Hum. Genet. 2016;98:210–215. doi: 10.1016/j.ajhg.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong J.X., McMillin M.J., Shively K.M., Beck A.E., Marvin C.T., Armenteros J.R., Buckingham K.J., Nkinsi N.T., Boyle E.A., Berry M.N., University of Washington Center for Mendelian Genomics De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am. J. Hum. Genet. 2015;96:462–473. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köroğlu Ç., Seven M., Tolun A. Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J. Med. Genet. 2013;50:515–520. doi: 10.1136/jmedgenet-2013-101634. [DOI] [PubMed] [Google Scholar]
- 8.Al-Sayed M.D., Al-Zaidan H., Albakheet A., Hakami H., Kenana R., Al-Yafee Y., Al-Dosary M., Qari A., Al-Sheddi T., Al-Muheiza M. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am. J. Hum. Genet. 2013;93:721–726. doi: 10.1016/j.ajhg.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan S., Wu Y. Wolters Kluwer; 2015. Etiology and pathogenesis of neonatal encephalopathy. UpToDate.http://www.uptodate.com/contents/etiology-and-pathogenesis-of-neonatal-encephalopathy [Google Scholar]
- 10.Bachmann-Gagescu R., Dempsey J.C., Phelps I.G., O’Roak B.J., Knutzen D.M., Rue T.C., Ishak G.E., Isabella C.R., Gorden N., Adkins J., University of Washington Center for Mendelian Genomics Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J. Med. Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong J.X., Burrage L.C., Beck A.E., Marvin C.T., McMillin M.J., Shively K.M., Harrell T.M., Buckingham K.J., Bacino C.A., Jain M., University of Washington Center for Mendelian Genomics Autosomal-Dominant Multiple Pterygium Syndrome Is Caused by Mutations in MYH3. Am. J. Hum. Genet. 2015;96:841–849. doi: 10.1016/j.ajhg.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations Impairing GSK3-Mediated MAF Phosphorylation Cause Cataract, Deafness, Intellectual Disability, Seizures, and a Down Syndrome-like Facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordeddu V., Redeker B., Stellacci E., Jongejan A., Fragale A., Bradley T.E.J., Anselmi M., Ciolfi A., Cecchetti S., Muto V. Mutations in ZBTB20 cause Primrose syndrome. Nat. Genet. 2014;46:815–817. doi: 10.1038/ng.3035. [DOI] [PubMed] [Google Scholar]
- 17.Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 18.Seelow D., Schwarz J.M., Schuelke M. GeneDistiller--distilling candidate genes from linkage intervals. PLoS ONE. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Yan X., Zhou T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS ONE. 2013;8:e71349. doi: 10.1371/journal.pone.0071349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Li Q., Li Y., Lin J., Yang D., Zhu G., Wang L., He D., Lu G., Zeng C. A long type of TBCK is a novel cytoplasmic and mitotic apparatus-associated protein likely suppressing cell proliferation. J. Genet. Genomics. 2014;41:69–72. doi: 10.1016/j.jgg.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Frasa M.A.M., Koessmeier K.T., Ahmadian M.R., Braga V.M.M. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- 23.Pan X., Eathiraj S., Munson M., Lambright D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 24.Gavriljuk K., Gazdag E.-M., Itzen A., Kötting C., Goody R.S., Gerwert K. Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc. Natl. Acad. Sci. USA. 2012;109:21348–21353. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 26.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Creighton T.E. W.H. Freeman & Company; 1993. Proteins. [Google Scholar]
- 29.Boudeau J., Miranda-Saavedra D., Barton G.J., Alessi D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Scheeff E.D., Eswaran J., Bunkoczi G., Knapp S., Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhogaraju S., Lorentzen E. Crystal structure of a Chlamydomonas reinhardtii flagellar RabGAP TBC-domain at 1.8 Å resolution. Proteins. 2014;82:2282–2287. doi: 10.1002/prot.24597. [DOI] [PubMed] [Google Scholar]
- 32.Liegel R.P., Handley M.T., Ronchetti A., Brown S., Langemeyer L., Linford A., Chang B., Morris-Rosendahl D.J., Carpanini S., Posmyk R. Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. Am. J. Hum. Genet. 2013;93:1001–1014. doi: 10.1016/j.ajhg.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chotard L., Mishra A.K., Sylvain M.-A., Tuck S., Lambright D.G., Rocheleau C.E. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21:2285–2296. doi: 10.1091/mbc.E09-11-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chadt A., Leicht K., Deshmukh A., Jiang L.Q., Scherneck S., Bernhardt U., Dreja T., Vogel H., Schmolz K., Kluge R. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat. Genet. 2008;40:1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- 35.Haas A.K., Fuchs E., Kopajtich R., Barr F.A. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 36.Smith L.D., Saunders C.J., Dinwiddie D.L., Atherton A.M., Miller N.A., Soden S.E., Farrow E.G., Abdelmoity A.T.G., Kingsmore S.F. Exome Sequencing Reveals De Novo Germline Mutation of the Mammalian Target of Rapamycin (MTOR) in a Patient with Megalencephaly and Intractable Seizures. Journal of Genomes and Exomes. 2013;2:63–72. [Google Scholar]
- 37.Crino P.B. mTOR: A pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011;17:734–742. doi: 10.1016/j.molmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Crino P.B. Focal brain malformations: seizures, signaling, sequencing. Epilepsia. 2009;50(Suppl 9):3–8. doi: 10.1111/j.1528-1167.2009.02289.x. [DOI] [PubMed] [Google Scholar]
- 39.Baynam G., Overkov A., Davis M., Mina K., Schofield L., Allcock R., Laing N., Cook M., Dawkins H., Goldblatt J. A germline MTOR mutation in Aboriginal Australian siblings with intellectual disability, dysmorphism, macrocephaly, and small thoraces. Am. J. Med. Genet. A. 2015;167:1659–1667. doi: 10.1002/ajmg.a.37070. [DOI] [PubMed] [Google Scholar]
- 40.Walker U.A., Collins S., Byrne E. Respiratory chain encephalomyopathies: a diagnostic classification. Eur. Neurol. 1996;36:260–267. doi: 10.1159/000117269. [DOI] [PubMed] [Google Scholar]
- 41.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 42.Bockaert J., Marin P. mTOR in Brain Physiology and Pathologies. Physiol. Rev. 2015;95:1157–1187. doi: 10.1152/physrev.00038.2014. [DOI] [PubMed] [Google Scholar]
- 43.Alazami A.M., Patel N., Shamseldin H.E., Anazi S., Al-Dosari M.S., Alzahrani F., Hijazi H., Alshammari M., Aldahmesh M.A., Salih M.A. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krawitz P., Buske O., Zhu N., Brudno M., Robinson P.N. The genomic birthday paradox: how much is enough? Hum. Mutat. 2015;36:989–997. doi: 10.1002/humu.22848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.