Abstract
Glycosylphosphatidylinositol (GPI) is a glycolipid that anchors >150 various proteins to the cell surface. At least 27 genes are involved in biosynthesis and transport of GPI-anchored proteins (GPI-APs). To date, mutations in 13 of these genes are known to cause inherited GPI deficiencies (IGDs), and all are inherited as recessive traits. IGDs mainly manifest as intellectual disability, epilepsy, coarse facial features, and multiple organ anomalies. These symptoms are caused by the decreased surface expression of GPI-APs or by structural abnormalities of GPI. Here, we present five affected individuals (from two consanguineous families from Egypt and Pakistan and one non-consanguineous family from Japan) who show intellectual disability, hypotonia, and early-onset seizures. We identified pathogenic variants in PIGG, a gene in the GPI pathway. In the consanguineous families, homozygous variants c.928C>T (p.Gln310∗) and c.2261+1G>C were found, whereas the Japanese individual was compound heterozygous for c.2005C>T (p.Arg669Cys) and a 2.4 Mb deletion involving PIGG. PIGG is the enzyme that modifies the second mannose with ethanolamine phosphate, which is removed soon after GPI is attached to the protein. Physiological significance of this transient modification has been unclear. Using B lymphoblasts from affected individuals of the Egyptian and Japanese families, we revealed that PIGG activity was almost completely abolished; however, the GPI-APs had normal surface levels and normal structure, indicating that the pathogenesis of PIGG deficiency is not yet fully understood. The discovery of pathogenic variants in PIGG expands the spectrum of IGDs and further enhances our understanding of this etiopathogenic class of intellectual disability.
Introduction
With an estimated prevalence of approximately 1%,1 intellectual disability is one of the most common disorders of the human population. Different definitions are available, but the general consensus is that the IQ should be below 70 and symptoms should appear during the development period and affect all aspects of adaptive functioning (conceptual, social, and practical).2, 3 Intellectual disability has several etiologies, and its genetic causes, including chromosomal aneuploidies, copy-number variants (CNVs), and monogenic disorders, are estimated to account for 25%–50% of the total cases. The identification of genic variants responsible for monogenic disorders has been revolutionized since the widespread use of high-throughput sequencing.4, 5
Glycosylphosphatidylinositol (GPI) is a glycolipid that anchors more than 150 proteins to the cell surface. GPI-anchored proteins (GPI-APs) have various important roles on the cell surface. At least 27 genes are involved in the biosynthesis and transport of GPI-APs. Among them, those that are involved in biosynthesis of GPI are called PIG (phosphatidylinositol glycan) genes, and those that are involved in the modification of GPI after attachment to proteins are called PGAP (post-GPI attachment to proteins) genes.6
The first reported inherited GPI deficiency (IGD) was PIGM (MIM: 610273) deficiency in individuals suffering from portal thrombosis and seizures without intellectual disability.7, 8 The mutation was located in the promoter, which disrupted the binding of transcriptional factor SP1 and decreased promoter acetylation, leading to decreased expression of PIGM. Many more IGDs have been recently found via whole-exome sequencing (WES). Most IGDs caused by the defect in PIG genes, such as PIGA (MIM: 311770),9, 10, 11, 12, 13 PIGQ (MIM: 605754),14 PIGY (MIM: 610662),15 PIGL (MIM: 605947),16, 17 PIGW (MIM: 610275),18 PIGV (MIM: 610274),19, 20 PIGN (MIM: 606097),21, 22, 23 PIGO (MIM: 614730),24, 25, 26 and PIGT (MIM: 610272),27, 28 are partial deficiencies given that complete loss of function of these genes causes embryonic death.29 These IGDs show decreased amounts of various GPI-APs and can be diagnosed by flow cytometry of granulocytes.25 The major symptoms include intellectual disability, epilepsy, and coarse facial features. Although the symptoms are very broad, other characteristic features sometimes seen include a tented upper lip, brachytelephalangy with hypoplastic nails, hearing loss, and multiple organ anomalies, such as an aganglionic megacolon and kidney or anorectal anomalies.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27
In PGAP1 (MIM: 611655) deficiency, surface levels of GPI-APs are not affected. Similarly, GPI-AP levels in PGAP3 (MIM: 611801) deficiency are variable and sometimes not affected. Therefore, abnormalities in these deficiencies are most likely caused by the abnormal structures of proteins’ membrane anchors. In these cases, the individuals with complete deficiencies are alive and show only severe intellectual disability, often with seizures and hypotonia.30, 31, 32 Hyperphosphatasia is a useful marker for diagnosing defects in PIG genes involved in later steps of the GPI-biosynthesis pathway, such as PIGV and PIGO, because the protein part of alkaline phosphatase (ALP) is secreted from the endoplasmic reticulum (ER) without attachment to the GPI anchor. PGAP2 (MIM: 615817) and PGAP3 deficiencies also show hyperphosphatasia32, 33, 34 because once ALP is anchored to the membrane by GPI, it is released from the cell surface. Thus, the symptoms vary in severity depending upon the degree of the defect and/or position in the pathway of the affected gene.
Here, we report five individuals (from three different families) who suffer from intellectual disability and hypotonia and carry loss-of-function variants in PIGG (phosphatidylinositol glycan anchor biosynthesis, class G). PIGG is the enzyme that attaches ethanolamine phosphate (EtNP) to the second mannose. Different from other PIG genes, PIGG is not essential for GPI-AP biosynthesis. GPI-APs in PIGG-deficient cells are present at normal levels and have normal structures because EtNP on the second mannose is removed by PGAP5 (HGNC-approved gene symbol: MPPE1 [MIM: 11900]) in the ER physiologically.35 However, the discovery of individuals with PIGG deficiency reveals that this step is also important for neurological development.
Subjects and Methods
Clinical Reports of Affected Individuals
This study was approved by the institutional review boards of Osaka University (Japan), Yokohama City University School of Medicine (Japan), Yamagata University (Japan), University of Geneva (Switzerland), and National Research Centre (Egypt). Ethical approval for molecular genetics research studies and data use was obtained from the South Yorkshire Research Ethics Committee (reference no. 11/H1310/1; UK). Informed consent was obtained from all examined persons or their guardians.
The affected individuals belong to three different families identified in three different studies. An overview of the clinical findings, along with a comparison to the phenotype of other IGDs, can be found in Table 1, and detailed clinical descriptions can be found in the Supplemental Note. In brief, the individuals in family EG (V:1 [EG01] and V:5 [EG02], now aged 24 and 14 years, respectively) are the offspring of a consanguineous marriage between first cousins originating from Egypt. They presented with hypotonia, seizures (at 4 months of age) that were not easily controlled by drug therapy, and profound uniform developmental delay or intellectual disability. The individual in family JP (V:6 [JP01]) was born to non-consanguineous healthy Japanese parents (Figure 1). Seizures were first noted at the age of 10 months. She developed severe psychomotor developmental delay with no speech development, autistic features, and growth retardation with a poor appetite. The individuals in family PK (V:9 [PK01] and V:10 [PK02], presently 12 and 10 years old, respectively), are offspring of a mating between first cousins of Pakistani origin (Figure 1). They had severe delay in their motor development. The first individual has had a single episode of seizures, whereas the second individual has not had any epileptic episodes.
Table 1.
An Overview of the Affected Individuals’ Symptoms and a Comparison with the Different Aspects of the Already Identified IGDs
|
Clinical Syndrome |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCAHS2 | Ohtahara Syndrome | HPMRS6 | CHIME Syndrome | HPMRS5 | Inherited GPI Deficiency | HPMRS1 | MCAHS1 | HPMRS2 | IGD Identified in This Study | MCAHS3 | MRT42 | HPMRS4 | HPMRS3 | |||
| MIM no. | 300868 | 3008350 | 616809 | 280000 | 616025 | 610293 | 239300 | 614080 | 614749 | NA | 615398 | 611655 | 615716 | 614207 | ||
| Associated gene (MIM no.) | PIGA (311770) | PIGQ (605754) | PIGY (610662) | PIGL (605947) | PIGW (610275) | PIGM (610273) | PIGV (610274) | PIGN (606097) | PIGO (614730) | PIGG (not assigned) | PIGT (610272) | PGAP1 (611655) | PGAP3 (611801) | PGAP2 (615187) | ||
| GPI-biosynthesis stepa | GPI biosynthesis (ER cytoplasmic side) | GPI biosynthesis (ER lumen side) | GPI transamidase component (ER) | removal of inositol-linked acyl chain (ER) | lipid remodeling in Golgi | |||||||||||
| step 1 | step 1 | step 1 | step 2 | step 4 | step 6 | step 7 | step 8 | step 10 | step 11 | step 12 | step 13 | step 16 | step 17 | |||
| No. of families (affected individuals) | 9 (13) | 1 (1) | 2 (4) | 6 (7) | 1 (1) | 2 (3) | 14 (20) | 9 (17) | 4 (6) | 1 (EG01 and EG002) | 1 (JP01) | 1 (PK01 and PK02) | 3 (7) | 5 (7) | 4 (6) | 4 (9) |
| Mode of inheritance (locus) | X-linked (Xp22.2) | AR (16p13.3) | AR (4q22.1) | AR (17p11.2) | AR (17q12) | AR (1q23.2) | AR (1p36.11) | AR (18q21.33) | AR (9p13.3) | AR (4p16.3) | AR (4p16.3) | AR (4p16.3) | AR (20q13.12) | AR (2q33.1) | AR (17q12) | AR (11p15.4) |
| Clinical Features | ||||||||||||||||
| DD or ID | + | + | +/− | + | + | + | + | + | +/− | + | + | + | + | + | + | + |
| Seizures | + (<1 year) | + (<1 year) | +/− | + (<1 year) | + (<1 year) | + | + | + (<1 year) | + | + (4 months) | + (10 months) | +/− | + (<1 year) | +/− | +/− | +/− |
| Hypotonia | + | + | +/− | + | + | − | + | + | + | + | + | + | + | + | + | + |
| Head circumference | ND | ND | − or microcephaly | ND | ND | ND | ND | − or microcephaly | − or microcephaly | ND | ND | ND | − or microcephaly | − or microcephaly | − or microcephaly | ND |
| Facial dysmorphism | +/− | NA | + | + | + | − | + | + | + | + | − | − | + | − | + | +/− |
| Hearing impairment | + | NA | − | + | − | − | +/− | − | +/− | − | − | − | +/− | − | − | +/− |
| Joint contractures | + | + | +/− | ND | ND | ND | ND | ND | ND | hyperlaxity | − | − | ND | ND | ND | ND |
| Skeletal anomalies | ND | ND | short fingers and small feet, proximal limb shortening, hip dysplasia | ND | ND | ND | hypoplastic terminal phalanges | ND | hypoplastic terminal phalanges | − | − | − | osteopenia, scoliosis delayed bone age, short arms | ND | ND | ND |
| Skin anomalies | +/− | − | − | +/− | − | − | − | − | − | − | − | − | − | − | − | − |
| Congenital heart defects | +/− | − | − | +/− | − | − | +/− | +/− | +/− | − | − | − | +/− | − | − | +/− |
| Vesicoureteral reflex or anomalies in urinary tract | +/− | − | +/− | +/− | − | − | +/− | +/− | − | − | − | − | +/− | − | − | − |
| Anorectal anomalies | − | − | − | − | − | − | +/− | +/− | +/− | − | − | − | +/− | − | − | +/− |
| CNS abnormalities in MRI | +/− | + | +/− | +/− | + | − | + | +/− | +/− | thin corpus callosum | − | cerebellar hypoplasia, cerebral atrophy | + | +/− | +/− | +/− |
| Increased serum alkaline phosphatase | +/− (mild) | − | +/− (mild) | +/− | + | − | + | − | + | − | − | NA | − (hypophosphatasia) | − | + | + |
| Decreased GPI-AP | + | NA | + | + | + | + | + | + | + | − | − | − | + | − | +/− | + |
Abbreviations are as follows: +, present; −, absent; +/−, variable; AR, autosomal recessive; MCAHS, multiple congenital anomalies-hypotonia-seizures syndrome; CHIME, colobomas, congenital heart disease, ichthyosiform dermatosis, mental retardation, and ear anomalies; HPMRS, hyperphosphatasia with mental retardation syndrome; MRT42, mental retardation, autosomal recessive 42; DD, development delay; ID, intellectual disability; NA, not applicable; and ND, not determined.
Based on the review by Kinoshita.6
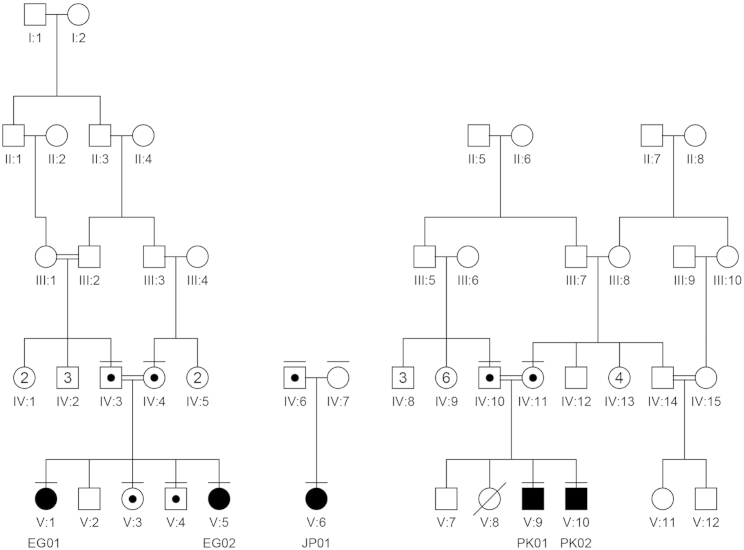
Figure 1.
Family Pedigrees
The affected individuals are noted as EG01, EG02, JP01, PK01, and PK02. The family members for whom DNA was available are noted with a horizontal bar above their respective symbols. Affected individuals are homozygous or compound heterozygous for the causative variants, and family members are verified carriers. In family JP, the second variant is a de novo CNV that encompasses PIGG.
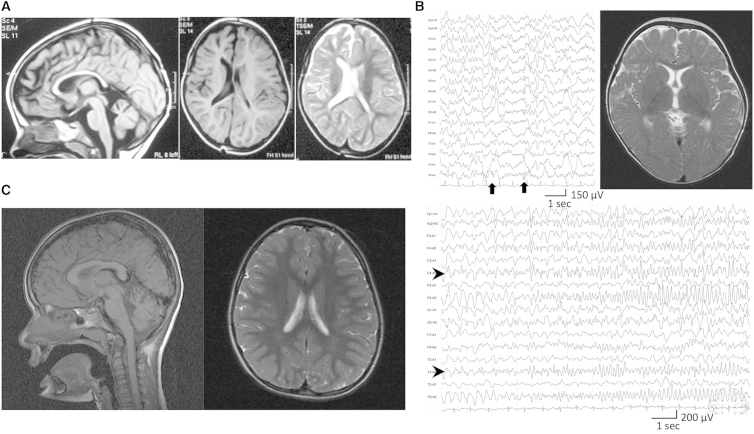
All affected individuals from the three families had either no dysmorphic features (families JP and PK) or no common features (family EG), and the common symptoms were hypotonia, severe developmental delay, and seizures. Electroencephalography (EEG) of the individuals in family EG showed that seizures were temporal and had secondary generalization, whereas EEG of the individual in family JP showed that seizure waves were in the right parietotemporal regions and propagated to the right hemisphere (Figure 2B, lower panel). Brain MRI showed a thin corpus callosum and asymmetry of the lateral ventricles in both individuals in family EG (Figure 2A), normal findings in the individual in family JP (Figure 2B, right upper panel), and cerebellar hypoplasia and mild cerebral atrophy in both individuals in family PK (Figure 2C).
Figure 2.
MRI and EEG of the Affected Individuals
(A) MRI of individual EG02 when she was 2.5 years old. (Left panel) Mid-sagittal T1-weighted MRI shows the thin corpus callosum. (Right panel) Axial plane in T1- and T2-weighted MRI shows asymmetry of the lateral ventricles.
(B) MRI and EEG of individual JP01. (Upper left panel) Interictal EEG during sleep at the age of 3 years shows focal spikes on the right posterior area (C4-P4-O2-T6, arrows). (Lower panel) Ictal EEG at 14 months shows that low-amplitude fast waves arose from right centrotemporal lesions (C4-T4, arrowheads) and then propagated to the entire right hemisphere. According to the ictal discharge, the individual developed eye deviation and head turning to the right after clonic seizures of the left upper extremity. (Upper right panel) T2-weighted brain MRI at the level of basal ganglia shows normal appearance at 11 months.
(C) MRI of individual PK01. (Left panel) Mid-sagittal T1-weighted MRI shows cerebellar hypoplasia. (Right panel) Axial plane in T2-weighted MRI shows mild cerebral atrophy.
Variant Identification
Egyptian Family
Variant identification was performed as previously described.36 In brief, all family members for whom DNA was available (noted horizontal bars, Figure 1) were genotyped with the HumanOmniExpress Bead Chip (Illumina), and the genotypes were used for identifying runs of homozygosity in each individual. Simultaneously in the eldest individual, the exome was captured with SureSelect Human All Exon V3 (Agilent), and the produced libraries were sequenced in an Illumina HiSeq 2000. Variant identification was performed with publicly available algorithms and then with genotyping data, and the variants identified through exome sequencing were combined with CATCH (Consanguinity Analysis through Common Homozygosity)37 for the creation of a final list of candidate variants. All identified variants were confirmed by Sanger sequencing, and family segregation was confirmed.
Japanese Family
Genomic DNA was captured with the SureSelect Human All Exon V5 (Agilent Technologies) and sequenced on an Illumina HiSeq 2000 with 101 bp paired-end reads. Exome-data processing, variant calling, and variant annotation were performed as described previously.38 Copy-number analysis using WES data was performed with eXome Hidden Markov Model v.1.0 (XHMM) as previously described.39, 40 Microdeletion involving PIGG was validated by quantitative real-time PCR (qPCR) on a Rotor-Gene Q thermal-cycling system (QIAGEN) with DNA from the affected individual and parents. PCR was performed in a volume of 15 μl containing 10 ng of genomic DNA, 1× Rotor-Gene SYBR Green PCR Master Mix (QIAGEN), and 1.0 μM of each primer. qPCR was carried out with the relative standard-curve method with four standard samples (30, 10, 3.33, and 1.11 ng DNA). Two primer sets for exons 7 and 12 of PIGG and two reference primer sets for an area on chromosomes 9 and 15 were used. Relative copy numbers of test regions were independently calculated in comparison with those of the two reference regions and were averaged. Primer information is available on request.
Pakistani Family
Target capture was performed with the Agilent SureSelect All Exon V4 Exome Enrichment Kit according to the manufacturer’s standard protocols. Sequencing of 150 bp paired-end reads was performed with an Illumina MiSeq. Reads were aligned to GRCh37 with Novoalign (Novocraft Technologies) and processed with the Genome Analysis Toolkit (GATK) and Picard (see Web Resources) for realignment of short insertions and deletions (indels) and removal of duplicate reads. Depth of coverage of the consensus coding sequence (CCDS) was assessed with the GATK, which showed that >94% of CCDS bases were covered by at least five good-quality reads (minimum Phred-like base quality of 17 and minimum mapping quality of 20). Single-nucleotide variants (SNVs) and indels were called with the UnifiedGenotyper feature of the GATK.41, 42
Generation of LCLs, PI-PLC Treatment, and Analysis by Fluorescence-Activated Cell Sorting
Lymphoblastoid cell lines (LCLs) were generated from the affected individuals’ B-lymphocytes and cultured in RPMI 1640-R2405 (Sigma-Aldrich) supplemented with 10% fetal calf serum. The LCLs were transfected with an empty pMEoriP vector or pMEoriP-humanPIGG-GST. Cells were suspended in 0.8 ml of Opti-MEM and electroporated with 20 μg each of the plasmids at 260 V and 960 mF with a Gene Pulser (Bio-Rad). Permanently transfected cells were obtained by selection with 0.5 μg/ml puromycin. Those cells were treated with or without 10 units/ml of phosphatidylinositol-specific phospholipase C (PI-PLC; Molecular Probe) for 1.5 hr at 37°C. Surface expression of GPI-APs was determined by cell staining with phycoerythrin (PE)-conjugated mouse anti-human CD59 (5H8).
Functional Assay of PIGG: Determining PIGG-Dependent Generation of Mature GPI by Analyzing GPI Mannolipids
Affected individuals’ LCLs were labeled with 14C-mannose. A total of 106 cells were incubated for 1 hr in a medium containing 100 μg/ml glucose and 10 μg/ml tunicamycin (Sigma) and then incubated in the same medium containing 0.5 μCi/ml 14C-mannose (American Radiolabeled Chemicals) for 1 hr. Lipids were extracted and partitioned into n-buthanol. Glycolipids were separated on Kiesel gel 60 (Merck) and detected by a BAS-1500 image analyzer (Fujifilm). H8 indicated a complete GPI anchor, and H7 indicated a GPI-anchor intermediate without EtNP at the second mannose. A fourth mannose was added to the third mannose in H7 and H8 for the generation of H7′ and H8′, respectively. Each affected individual’s cell was permanently transfected with PIGG cDNA or an empty vector. The positive control was lipid extracts from radiolabeled PIGK-deficient K562 cells in which the complete GPI anchor (H8) had accumulated.
Western Blotting
Lipofectamine 2000 (Invitrogen) was used for transiently transfecting HEK293 cells with SRα-promoter-driven cDNA encoding wild-type or mutant PIGG tagged with hemagglutinin (HA) at the N terminus (pME HA-hPIGG). Two days later, lysates were applied to SDS-PAGE, and HA-tagged PIGG was assessed by western blotting using anti-HA antibody.
Analysis by Fluorescence-Activated Cell Sorting
Surface levels of GPI-APs were determined by cell staining with Alexa 488-conjugated inactivated aerolysin (fluorescently-labeled inactive toxin aerolysin; Protox Biotech) and the following appropriate antibodies: PE-conjugated mouse anti-decay accelerating factor (DAF; IA10), anti-CD16 (3G8), anti-CD24 (ML5), anti-CD59 (5H8), and anti-CD48 (BJ40) antibodies (Biolegend). Cells were analyzed by flow cytometry (MACSQuant Analyzer 10, Miltenyi Biotec) with Flowjo software (v.9.5.3, Tommy Digital).
Generation of PIGG-Knockout HEK293 Cells
PIGG-knockout cells were generated from HEK293 cells via the CRISPR/Cas9 system. The human-codon-optimized Streptococcus pyogenes Cas9 and chimeric guide RNA expression plasmid pX330 were obtained from Addgene. The seed sequences for the SpCas9 target sites in PIGG exons 1 and 2 were selected, and a pair of annealed oligonucleotides designed according to these sequences were cloned into the BbsI sites of pX330. Lipofectamine 2000 was used for transfecting HEK293 cells with two kinds of pX330 containing the target sites. The candidate PIGG-knockout clones were obtained by limiting dilution and further selected by PCR and direct sequencing.
Results
Exome Sequencing and Genotyping of Family EG
After exome sequencing, 42,995,036 reads were on target in that they covered 89.12% of the coding exons as defined in RefSeq at 8×. Of the 20,119 high-quality exonic variants identified, 10,187 were synonymous and 9,021 were missense. After genotyping, we verified the correct family structure and identified that the parents share 17.9% of their genomes.
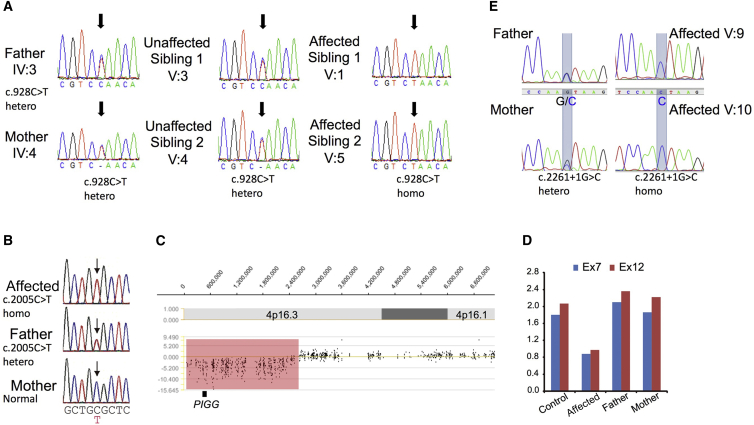
These data were combined through CATCH,37 and after analysis of the results, three variants remained for further evaluation (Table S1) in genes PIGG (c.928C>T [p.Gln310∗] [GenBank: NM_001127178.2]), COQ5 (MIM: 616359; c.319G>A [p.Gly107Arg] [GenBank: NM_032314.3]), and RGS12 (MIM: 602512; c.188A>T [p.Gln63Leu] [GenBank: NM_002926.3]). All variants were confirmed by Sanger sequencing, and family segregation confirmed that the affected individuals, as well as the unaffected siblings, are heterozygous for the variants (Figure 3A). Variants in PIGG and COQ5 are not present in publically available databases, whereas the variant in RGS12 has a cumulative frequency of 4.95e−5 in the Exome Aggregation Consortium (ExAC) Browser.43 The variant in PIGG is nonsense, whereas the other two are missense.
Figure 3.
Identified Variants
(A) Sequencing chromatograms of the identified variant in family EG. The parents and unaffected siblings are heterozygous, whereas the affected individuals are homozygous for the c.928C>T mutation (noted by the black arrow).
(B) Sequencing chromatograms of the identified variant in family JP. The father has the heterozygous c.2005C>T variant, but the mother does not (noted by the black arrow).
(C) XHMM analysis using WES data of individual JP01. A microdeletion involving PIGG (chr4: 60,226–2,452,836) is indicated.
(D) qPCR analysis of exons 7 and 12 of the PIGG genome from individual JP01 and her parents shows a heterozygous PIGG deletion in the affected individual, but not in her mother, suggesting that the deletion occurred de novo on the maternal chromosome.
(E) Sequencing chromatograms of the identified variant in family PK are noted by the blue transparent boxes. The parents are heterozygous, and the affected individuals are homozygous for the c.2261+1G>C mutation.
PIGG was considered the most prominent candidate because the variant identified causes a clear loss of function. A recent inquiry of the ExAC Browser verified that no homozygous loss-of-function variants have been reported and that the most common loss-of-function variant, c.1515G>A (p.Trp505∗) (GenBank: NM_001127178.2), has a minor allele frequency (MAF) of 0.0012 in the European population. This translates to a frequency of less than 1 in 600,000 individuals, compatible with a rare autosomal-recessive disorder.
Exome Sequencing of Family JP
We have been conducting WES analysis of inherited intractable disorders, including infantile epilepsy, as a Japan national project since 2011. Searching WES data of 437 individuals with infantile epilepsy, we identified a PIGG variant (c.2005C>T [p.Arg669Cys] [GenBank: NM_001127178.2]) in the affected individual in family JP, and it seemed homozygous (no reads for the reference allele and 57 reads for the variant allele). The c.2005C>T variant was found in 4 of 575 in-house control exomes and has a MAF of 0.06% in the East Asian population according to the ExAC Browser. Web-based prediction tools suggested that the c.2005C>T variant is pathogenic (Table S1). Sanger sequencing using DNA of the affected individual and her parents revealed that the father has the heterozygous c.2005C>T variant but that the mother does not (Figure 3B). Because the affected individual showed only the variant T allele, one or two alleles with the c.2005C>T variant from her father should exist, and the maternal allele might be missing in the affected individual. In fact, a microdeletion involving PIGG (chr4: 60,226–2,452,836) was indicated by XHMM analysis using WES data (Figure 3C). qPCR analysis confirmed the heterozygous PIGG deletion in the affected individual, but not in her mother, suggesting that the microdeletion should have occurred de novo on the maternal chromosome (Figure 3D).
Exome Sequencing and Genotyping of Family PK
Custom Perl scripts were used for removing variants present in dbSNP132 or with a MAF ≥ 0.1% and for annotating functional consequences. We called variants in the autozygous region only with a minimum Phred-like genotype quality of 30 and selected for indels within coding regions, non-synonymous SNVs, and splice-site variants. Variants present in the 1000 Genomes dataset (November 2011), the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server, and another 2500 ethnically matched in-house exomes were also removed. This left a single variant, c.2261+1G>C (GenBank: NM_001127178.1), which was confirmed as present in the affected individuals and segregated with the phenotype in the family according to Sanger sequencing (Figure 3E).
Functional Analysis of the PIGG Mutants
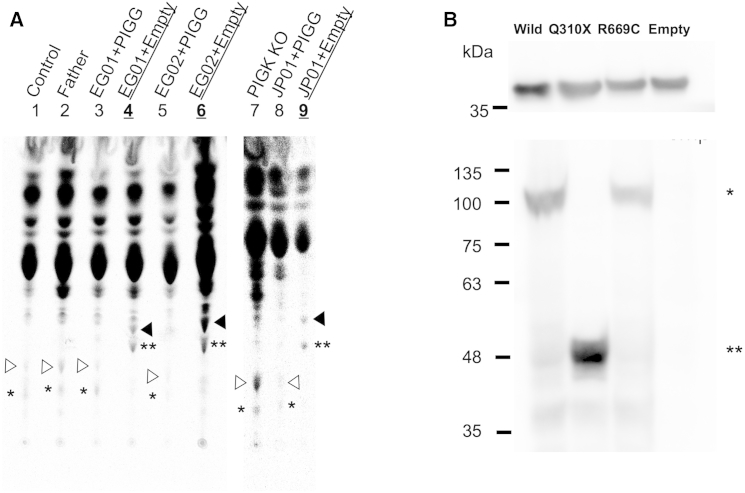
PIGG is an EtNP transferase that mediates conversion of GPI precursor H7 to mature GPI precursor H8 (Figure S2). H8 is normally attached to proteins for the generation of GPI-APs. H7 is also competent for protein attachment, and in the absence of H8, H7 is alternatively used as an anchor. A fourth mannose can be added to H7 and H8 for the generation of H7′ and H8′, respectively. Culturing cells with radioactive mannose and analyzing the metabolically radiolabeled GPI precursors by thin-layer chromatography and phosphoimaging can be used for determining generation of H7, H7′, H8, and H8′ in cells. To determine whether PIGG mutations found in families EG and JP affect functional activity of PIGG, we cultured individuals’ LCLs in a medium containing 14C-mannose and analyzed the metabolically radiolabeled GPI precursors (Figure 4A). H7 and H7′ were accumulated in LCLs from all three affected individuals (black arrowheads [H7] and double asterisks [H7′] in lanes 4, 6, and 9 for individuals EG01, EG02, and JP01, respectively), suggesting that mutant PIGG could not add EtNP to H7 to generate H8. Such accumulation of H7 and H7′ was not seen in LCLs from a healthy individual or the father of affected individuals EG01 and EG02 (lanes 1 and 2). Importantly, these accumulation profiles of affected individuals’ LCLs disappeared after transfection of normal PIGG cDNA (lanes 3, 5, and 8). H8 and H8′ (white arrowheads [H8] and single asterisk [H8′] in lanes 1 and 7, respectively; positive control) were not or were only weakly seen in affected individuals’ LCLs, whereas they were restored by transfection of PIGG cDNA (compare lanes 3 and 4, lanes 5 and 6, and lanes 8 and 9). These results indicate that the PIGG mutations in both families disrupt PIGG functional activity almost completely (Figure 4A).
Figure 4.
Functional Assays
(A) Analysis of mannolipids accumulated in LCLs from the affected individuals. Lanes are as follows: 1, LCLs from a normal individual; 2, LCLs from the father of individuals EG01 and EG02 (IV:3); 3, LCLs (transfected with PIGG cDNA) from individual EG01; 4, LCLs (transfected with an empty vector) from individual EG01; 5, LCLs (transfected with PIGG cDNA) from individual EG02; 6, LCLs (transfected with an empty vector) from individual EG02; 7, K562 PIGK-deficient cells; 8, LCLs (transfected with PIGG cDNA) from individual JP01; and 9, LCLs (transfected with an empty vector) from individual JP01. White arrowheads represent H8, a complete GPI intermediate, and black arrowheads represent H7, a GPI intermediate without ethanolaminephosphate at the second mannose. Asterisks indicate H7′ (∗) and H8′ (∗∗), H7 and H8, respectively, with the fourth mannose.47
(B) Amounts of each mutant PIGG. (Upper panel) Western blot of GAPDH for loading controls. (Lower panel) Western blot of wild-type (∗) or mutant (∗∗) HA-tagged PIGG.
It was predicted that the mutation in family EG would cause the generation of truncated PIGG because of the p.Gln310∗ change, whereas the p.Arg669Cys variant in family JP would not affect the apparent molecular size of PIGG. For analysis of mutant PIGG proteins, amounts of N-terminally HA-tagged wild-type, p.Gln310∗, and p.Arg669Cys PIGG were detected in HEK293 cells by anti-HA antibody after western blotting. p.Gln310∗ PIGG was present at a high level as a truncated protein of 48 kDa, an expected size (Figure 4B). p.Arg669Cys PIGG was present at a level comparable to that of a 100 kDa wild-type PIGG, indicating that the Arg-to-Cys substitution disrupts PIGG functional activity without affecting its stability.
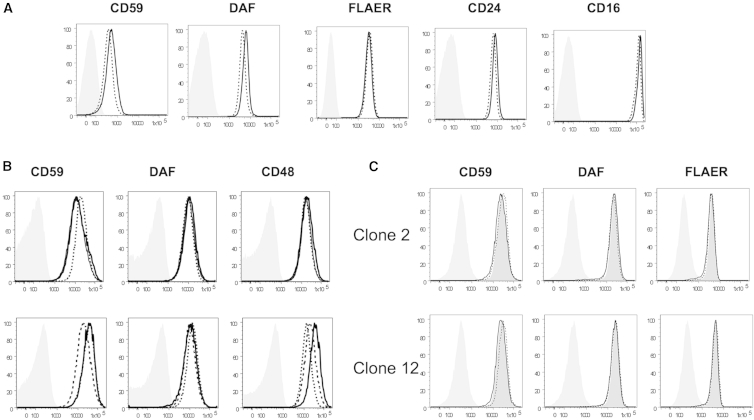
To investigate whether PIGG deficiency affects surface levels of GPI-APs, we used flow cytometry to analyze the granulocytes (Figure 5A) and LCLs from individuals EG01 and EG02 (Figure 5B, upper panels) and the LCLs from JP01 (Figure 4B, lower panels). Surface levels of various GPI-APs were similar on the cells of both affected individuals and healthy individuals, suggesting that they are not affected by loss-of-function mutations in PIGG. Making PIGG-knockout HEK293 cells with the CRISPR/Cas9 system further confirmed that complete loss of PIGG does not affect cell-surface levels of GPI-APs (Figure 5C).
Figure 5.
Flow Cytometric Analysis of Affected Individuals’ Blood and Cell Lines
(A) Surface level of GPI-anchored proteins on the granulocytes from individual JP01. Thick lines represent individual JP01, dotted lines represent a healthy control individual, and gray shadows represent the isotype control.
(B) Surface level of GPI-anchored proteins on the LCLs from individuals EG01 and EG02 (upper panel) and individual JP01 (lower panel). In the upper panels, thick and thin lines represent individuals EG01 and EG02, respectively, and dotted lines represent their father. In the lower panels, thick lines represent affected individual JP01, and dotted and dashed lines represent healthy individuals. Gray shadows represent the isotype control.
(C) Surface level of GPI-anchored proteins on HEK293 cells. PIGG-knockout HEK293 cells were permanently transfected with PIGG cDNA (dotted lines) or an empty vector (thick lines). Dark-gray shadows represent wild-type HEK293 cells, and light-gray shadows represent the isotype control.
Recently, individuals with PGAP1 mutations were reported.30 They had intellectual disability, developmental delay, encephalopathy, cerebral visual impairment, or hereditary spastic paraplegia. The complete-loss-of-function mutations in PGAP1 did not affect the cell-surface levels of GPI-APs but caused structural abnormality of lipid moiety in GPI. Because PGAP1 removes the inositol-linked acyl chain of GPI-APs at the next step after protein attachment to the GPI anchor in the ER (Figure S2), it is possible that PGAP1 might not act on GPI-APs lacking EtNP on the second mannose in cells with mutant PIGG. Additionally, given that PIGG mutations caused neurological phenotypes without causing reduction in the steady-state cell-surface levels of GPI-APs (similarly to PGAP1 deficiencies), we investigated the possibility of an abnormal GPI structure. LCLs from the individuals were treated with PI-PLC, a bacterial enzyme that cleaves GPI between phosphate and glycerol and releases proteins. PI-PLC does not cleave GPI if the inositol residue is acylated. CD59 levels of PIGG-defective cells and PIGG-transfected PIGG-defective cells were similarly reduced by PI-PLC treatment, indicating that elimination of the acyl chain from the inositol normally occurs in LCLs from three affected individuals (Figure S1). Therefore, the structure of GPI in GPI-APs on cells with mutant PIGG would be normal.
Discussion
Various symptoms of IGDs are caused by reduced surface levels of GPI-APs or by abnormal GPI-AP structure. Here, we report PIGG deficiency, one IGD in which cells express normal surface levels of GPI-APs with a normal structure. Nevertheless, individuals from families EG and JP suffer from intellectual disability, hypotonia, and seizures, and individuals from family PK show severe intellectual disability accompanied by ataxia. In individuals from family EG, hypotonia has been remarkable since birth and remains a characteristic symptom of the disorder. Later, it was seen as hyporeflexia and a delay in achieving walking. The affected individual in family JP showed more severe developmental delay, and she could not walk or speak. In both affected individuals from families EG and JP, seizures appeared early in life, and developmental delay was also documented early in life and evolved to uniform intellectual disability. The affected individuals described here did not present with coarse faces, dysmorphic features, or other accompanying symptoms frequently seen in IGDs. Intellectual disability with seizures and severe hypotonia appear to be common characteristics of PIGG deficiency.
Through in vitro testing of affected individuals’ LCLs, we showed that the identified variants completely abolished the function of PIGG, whereas the surface level of GPI-AP was normal. We obtained a similar result with PIGG-knockout HEK293 cells. Apparently H7 was used as an alternative GPI anchor. The EtNP transferred by PIGG to the second mannose is a transient side branch that is removed by phosphodiesterase PGAP5 in the second step after GPI is attached to the protein and that is not present in mature GPI-APs35 (Figure S2). Therefore, the lack of this transient EtNP side branch does not affect steady-state levels of cell-surface GPI-APs on B-lymphoblastoid cells and HEK293 cells (Figures 5B and 5C).
During GPI biosynthesis, EtNP is added to all three mannose residues. PIGN attaches EtNP to the first mannose at the 2-position. PIGG and PIGO attach EtNP at the 6-positions of the second and third mannoses, respectively, and share a common essential cofactor, PIGF. The EtNP attached to the third mannose, so-called “bridging” EtNP, links GPI to proteins. PIGN, PIGO, and PIGG constitute a family of EtNP transferases. In PIGN-defective cells, the cell-surface GPI-APs lack the EtNP side branch on the first mannose, and the surface levels are also affected. PIGN deficiency is characterized by intellectual disability, seizures, and multiple abnormalities. In PIGO-defective cells, the cell-surface GPI-AP levels are decreased because of a lack of or decrease in the “bridging” EtNP. PIGO deficiency causes Mabry syndrome, characterized by hyperphosphatasia, intellectual disability, seizures, and brachytelephalangy. Different from these EtNP transferase deficiencies, PIGG deficiency does not cause decreased surface expression of GPI-APs. We investigated the possibility of structural abnormality in GPI given that PGAP1 does not remove the inositol-linked acyl chain in the absence of EtNP on the second mannose and would thus cause proteins with inositol-acylated GPI to be present on the cell surface. However, this was proven not to be the case (Figure S1). In PIGG-deficient cells, therefore, GPI-APs would be normally remodeled in their fatty acid and transported to the microdomain of the plasma membrane (Figure S2). Consistent with this, PIGG-deficient individuals did not show hyperphosphatasia, which is commonly seen in PGAP3 deficiency, the fatty-acid-remodeling defect.
Another potential defect in PIGG-defective cells is that transport of GPI-APs from the ER to the cell surface is delayed. Recognition and removal of EtNP linked to the second mannose by PGAP5 at the ER exit site is important for efficient packaging of GPI-APs into COPII-coated transport vesicles and subsequent transport to the Golgi apparatus. We investigated this possibility by using PIGG-knockout HEK293 cells expressing doxycycline-inducible GPI-anchored reporter protein, but we did not detect transport delay (data not shown). Although significant delay in GPI-AP transport was not detected in HEK293 cells, we still think that neuronal cells, in which many important GPI-APs need to be quickly transported to distant sites from the cell body, might require PIGG for sufficient protein attachment of GPI or for efficient GPI-AP transport from the ER to the plasma membrane. Unfortunately, the available cellular systems were not able to resolve this issue. A defect in these processes might be relevant to neurological defects seen in individuals with PIGG deficiency.
Although almost nothing is known about the biological function of PIGG in mammalian cells, important functions were reported for GP17, the yeast ortholog of PIGG. Yeast GP17 plays an important role in cell-wall biogenesis, cell separation, and daughter cell growth.44, 45, 46 Extensive additional experiments through the study of animal models are required for further delineating PIGG’s function in the human CNS.
PIGG is the 14th of the 27 known genes of the GPI-biosynthesis pathway to be associated with human disorders, suggesting that pathogenic variants in the other genes participating in GPI biosynthesis remain to be discovered in affected individuals with similar phenotypes. The complete repertoire of genes involved in GPI-biosynthesis disorders will enhance our understanding of the pathophysiology of this form of ID and could result in treatment options for these disorders.
Acknowledgments
We thank the affected individuals and their families for their participation in the study. This study was funded by grants from the Gebert Rüf Stiftung foundation to S.E.A., the von Meissner foundation to P.M., the Japan Agency for Medical Research and Development and Ministry of Health, Labour, and Welfare to Y.M., M.K., and N.M., the Japan Ministry of Education, Culture, Sports, Science, and Technology to N.M., the Japan Society of the Promotion of Science to N.M., Y.M., and H.S., and the Takeda Science Foundation to Y.M., H.S., and N.M. This work was also supported by a Sir Jules Thorn Award for Biomedical Research (JTA/09 to C.A.J. and E.S.). We also thank Dr. Abou Rami, Dr. Yusuke Maeda, and Dr. Christelle Borel for constructive discussions and Mrs. Emilie Falconnet, Pascale Ribaux, Anne Vannier, and Kana Miyanagi for their technical assistance.
Published: March 17, 2016
Footnotes
Supplemental Data include a Supplemental Note, two figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.007.
Contributor Information
Stylianos E. Antonarakis, Email: stylianos.antonarakis@unige.ch.
Yoshiko Murakami, Email: yoshiko@biken.osaka-u.ac.jp.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org/
Clinical Genome Database, http://research.nhgri.nih.gov/CGD/
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
ELAND alignment algorithm, http://www.Illumina.com
ExAC Browser, http://exac.broadinstitute.org/
GeneReviews, http://www.ncbi.nlm.nih.gov/books/NBK1116/
Human Splicing Finder, http://www.umd.be/HSF/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
TMHMM Server, http://www.cbs.dtu.dk/services/TMHMM/
UCSC Genome Browser, www.genome.ucsc.edu
Supplemental Data
References
- 1.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association (2013). DSM-5 Intellectual Disability Fact Sheet, http://www.dsm5.org/documents/intellectual%20disability%20fact%20sheet.pdf.
- 3.World Health Organization Division of Mental Health and Prevention of Substance Abuse (1996). ICD-10 Guide for Mental Retardation, http://www.who.int/mental_health/media/en/69.pdf.
- 4.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 5.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinoshita T. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2014;90:130–143. doi: 10.2183/pjab.90.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almeida A.M., Murakami Y., Layton D.M., Hillmen P., Sellick G.S., Maeda Y., Richards S., Patterson S., Kotsianidis I., Mollica L. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat. Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- 8.Almeida A.M., Murakami Y., Baker A., Maeda Y., Roberts I.A., Kinoshita T., Layton D.M., Karadimitris A. Targeted therapy for inherited GPI deficiency. N. Engl. J. Med. 2007;356:1641–1647. doi: 10.1056/NEJMoa063369. [DOI] [PubMed] [Google Scholar]
- 9.Johnston J.J., Gropman A.L., Sapp J.C., Teer J.K., Martin J.M., Liu C.F., Yuan X., Ye Z., Cheng L., Brodsky R.A., Biesecker L.G. The phenotype of a germline mutation in PIGA: the gene somatically mutated in paroxysmal nocturnal hemoglobinuria. Am. J. Hum. Genet. 2012;90:295–300. doi: 10.1016/j.ajhg.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato M., Saitsu H., Murakami Y., Kikuchi K., Watanabe S., Iai M., Miya K., Matsuura R., Takayama R., Ohba C. PIGA mutations cause early-onset epileptic encephalopathies and distinctive features. Neurology. 2014;82:1587–1596. doi: 10.1212/WNL.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 11.Swoboda K.J., Margraf R.L., Carey J.C., Zhou H., Newcomb T.M., Coonrod E., Durtschi J., Mallempati K., Kumanovics A., Katz B.E. A novel germline PIGA mutation in Ferro-Cerebro-Cutaneous syndrome: a neurodegenerative X-linked epileptic encephalopathy with systemic iron-overload. Am. J. Med. Genet. A. 2014;164A:17–28. doi: 10.1002/ajmg.a.36189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belet S., Fieremans N., Yuan X., Van Esch H., Verbeeck J., Ye Z., Cheng L., Brodsky B.R., Hu H., Kalscheuer V.M. Early frameshift mutation in PIGA identified in a large XLID family without neonatal lethality. Hum. Mutat. 2014;35:350–355. doi: 10.1002/humu.22498. [DOI] [PubMed] [Google Scholar]
- 13.Tarailo-Graovac M., Sinclair G., Stockler-Ipsiroglu S., Van Allen M., Rozmus J., Shyr C., Biancheri R., Oh T., Sayson B., Lafek M. The genotypic and phenotypic spectrum of PIGA deficiency. Orphanet J. Rare Dis. 2015;10:23. doi: 10.1186/s13023-015-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin H.C., Kim G.E., Pagnamenta A.T., Murakami Y., Carvill G.L., Meyer E., Copley R.R., Rimmer A., Barcia G., Fleming M.R., WGS500 Consortium Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum. Mol. Genet. 2014;23:3200–3211. doi: 10.1093/hmg/ddu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilkovski B., Pagnamenta A.T., O’Grady G.L., Kinoshita T., Howard M.F., Lek M., Thomas B., Turner A., Christodoulou J., Sillence D. Mutations in PIGY: expanding the phenotype of inherited glycosylphosphatidylinositol deficiencies. Hum. Mol. Genet. 2015;24:6146–6159. doi: 10.1093/hmg/ddv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng B.G., Hackmann K., Jones M.A., Eroshkin A.M., He P., Wiliams R., Bhide S., Cantagrel V., Gleeson J.G., Paller A.S. Mutations in the glycosylphosphatidylinositol gene PIGL cause CHIME syndrome. Am. J. Hum. Genet. 2012;90:685–688. doi: 10.1016/j.ajhg.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara I., Murakami Y., Niihori T., Kanno J., Hakoda A., Sakamoto O., Okamoto N., Funayama R., Nagashima T., Nakayama K. Mutations in PIGL in a patient with Mabry syndrome. Am. J. Med. Genet. A. 2015;167A:777–785. doi: 10.1002/ajmg.a.36987. [DOI] [PubMed] [Google Scholar]
- 18.Chiyonobu T., Inoue N., Morimoto M., Kinoshita T., Murakami Y. Glycosylphosphatidylinositol (GPI) anchor deficiency caused by mutations in PIGW is associated with West syndrome and hyperphosphatasia with mental retardation syndrome. J. Med. Genet. 2014;51:203–207. doi: 10.1136/jmedgenet-2013-102156. [DOI] [PubMed] [Google Scholar]
- 19.Krawitz P.M., Schweiger M.R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 20.Horn D., Wieczorek D., Metcalfe K., Barić I., Paležac L., Cuk M., Petković Ramadža D., Krüger U., Demuth S., Heinritz W. Delineation of PIGV mutation spectrum and associated phenotypes in hyperphosphatasia with mental retardation syndrome. Eur. J. Hum. Genet. 2014;22:762–767. doi: 10.1038/ejhg.2013.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maydan G., Noyman I., Har-Zahav A., Neriah Z.B., Pasmanik-Chor M., Yeheskel A., Albin-Kaplanski A., Maya I., Magal N., Birk E. Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J. Med. Genet. 2011;48:383–389. doi: 10.1136/jmg.2010.087114. [DOI] [PubMed] [Google Scholar]
- 22.Ohba C., Okamoto N., Murakami Y., Suzuki Y., Tsurusaki Y., Nakashima M., Miyake N., Tanaka F., Kinoshita T., Matsumoto N., Saitsu H. PIGN mutations cause congenital anomalies, developmental delay, hypotonia, epilepsy, and progressive cerebellar atrophy. Neurogenetics. 2014;15:85–92. doi: 10.1007/s10048-013-0384-7. [DOI] [PubMed] [Google Scholar]
- 23.Brady P.D., Moerman P., De Catte L., Deprest J., Devriendt K., Vermeesch J.R. Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic congenital diaphragmatic hernia. Eur. J. Med. Genet. 2014;57:487–493. doi: 10.1016/j.ejmg.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Krawitz P.M., Murakami Y., Hecht J., Krüger U., Holder S.E., Mortier G.R., Delle Chiaie B., De Baere E., Thompson M.D., Roscioli T. Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 2012;91:146–151. doi: 10.1016/j.ajhg.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuki I., Takahashi Y., Okazaki S., Kawawaki H., Ehara E., Inoue N., Kinoshita T., Murakami Y. Vitamin B6-responsive epilepsy due to inherited GPI deficiency. Neurology. 2013;81:1467–1469. doi: 10.1212/WNL.0b013e3182a8411a. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura K., Osaka H., Murakami Y., Anzai R., Nishiyama K., Kodera H., Nakashima M., Tsurusaki Y., Miyake N., Kinoshita T. PIGO mutations in intractable epilepsy and severe developmental delay with mild elevation of alkaline phosphatase levels. Epilepsia. 2014;55:e13–e17. doi: 10.1111/epi.12508. [DOI] [PubMed] [Google Scholar]
- 27.Kvarnung M., Nilsson D., Lindstrand A., Korenke G.C., Chiang S.C., Blennow E., Bergmann M., Stödberg T., Mäkitie O., Anderlid B.M. A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT. J. Med. Genet. 2013;50:521–528. doi: 10.1136/jmedgenet-2013-101654. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima M., Kashii H., Murakami Y., Kato M., Tsurusaki Y., Miyake N., Kubota M., Kinoshita T., Saitsu H., Matsumoto N. Novel compound heterozygous PIGT mutations caused multiple congenital anomalies-hypotonia-seizures syndrome 3. Neurogenetics. 2014;15:193–200. doi: 10.1007/s10048-014-0408-y. [DOI] [PubMed] [Google Scholar]
- 29.Nozaki M., Ohishi K., Yamada N., Kinoshita T., Nagy A., Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab. Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 30.Murakami Y., Tawamie H., Maeda Y., Büttner C., Buchert R., Radwan F., Schaffer S., Sticht H., Aigner M., Reis A. Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet. 2014;10:e1004320. doi: 10.1371/journal.pgen.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch D.G., Boonstra F.N., Kinoshita T., Jhangiani S., de Ligt J., Cremers F.P., Lupski J.R., Murakami Y., de Vries B.B. Cerebral visual impairment and intellectual disability caused by PGAP1 variants. Eur. J. Hum. Genet. 2015;23:1689–1693. doi: 10.1038/ejhg.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard M.F., Murakami Y., Pagnamenta A.T., Daumer-Haas C., Fischer B., Hecht J., Keays D.A., Knight S.J., Kölsch U., Krüger U. Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 2014;94:278–287. doi: 10.1016/j.ajhg.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krawitz P.M., Murakami Y., Rieß A., Hietala M., Krüger U., Zhu N., Kinoshita T., Mundlos S., Hecht J., Robinson P.N., Horn D. PGAP2 mutations, affecting the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation syndrome. Am. J. Hum. Genet. 2013;92:584–589. doi: 10.1016/j.ajhg.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen L., Tawamie H., Murakami Y., Mang Y., ur Rehman S., Buchert R., Schaffer S., Muhammad S., Bak M., Nöthen M.M. Hypomorphic mutations in PGAP2, encoding a GPI-anchor-remodeling protein, cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2013;92:575–583. doi: 10.1016/j.ajhg.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., Kinoshita T. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 2009;139:352–365. doi: 10.1016/j.cell.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 36.Makrythanasis P., Nelis M., Santoni F.A., Guipponi M., Vannier A., Béna F., Gimelli S., Stathaki E., Temtamy S., Mégarbané A. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat. 2014;35:1203–1210. doi: 10.1002/humu.22617. [DOI] [PubMed] [Google Scholar]
- 37.Santoni F.A., Makrythanasis P., Antonarakis S.E. CATCHing putative causative variants in consanguineous families. BMC Bioinformatics. 2015;16:310. doi: 10.1186/s12859-015-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitsu H., Nishimura T., Muramatsu K., Kodera H., Kumada S., Sugai K., Kasai-Yoshida E., Sawaura N., Nishida H., Hoshino A. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013;45:445–449, e1. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 39.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M., Handsaker R.E., McCarroll S.A., O’Donovan M.C., Owen M.J. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyatake S., Koshimizu E., Fujita A., Fukai R., Imagawa E., Ohba C., Kuki I., Nukui M., Araki A., Makita Y. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an ‘exome-first’ approach. J. Hum. Genet. 2015;60:175–182. doi: 10.1038/jhg.2014.124. [DOI] [PubMed] [Google Scholar]
- 41.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ExAC (2015). Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv, http://biorxiv.org/content/early/2015/10/30/030338. [DOI] [PMC free article] [PubMed]
- 44.Benachour A., Sipos G., Flury I., Reggiori F., Canivenc-Gansel E., Vionnet C., Conzelmann A., Benghezal M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 1999;274:15251–15261. doi: 10.1074/jbc.274.21.15251. [DOI] [PubMed] [Google Scholar]
- 45.Fujita M., Yoko-o T., Okamoto M., Jigami Y. GPI7 involved in glycosylphosphatidylinositol biosynthesis is essential for yeast cell separation. J. Biol. Chem. 2004;279:51869–51879. doi: 10.1074/jbc.M405232200. [DOI] [PubMed] [Google Scholar]
- 46.Richard M., De Groot P., Courtin O., Poulain D., Klis F., Gaillardin C. GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans. Microbiology. 2002;148:2125–2133. doi: 10.1099/00221287-148-7-2125. [DOI] [PubMed] [Google Scholar]
- 47.Hong Y., Maeda Y., Watanabe R., Inoue N., Ohishi K., Kinoshita T. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 2000;275:20911–20919. doi: 10.1074/jbc.M001913200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.