Abstract
Nonsense-mediated decay (NMD) is an important process that is best known for degrading transcripts that contain premature stop codons (PTCs) to mitigate their potentially harmful consequences, although its regulatory role encompasses other classes of transcripts as well. Despite the critical role of NMD at the cellular level, our knowledge about the consequences of deficiency of its components at the organismal level is largely limited to model organisms. In this study, we report two consanguineous families in which a similar pattern of congenital anomalies was found to be most likely caused by homozygous loss-of-function mutations in SMG9, encoding an essential component of the SURF complex that generates phospho-UPF1, the single most important step in NMD. By knocking out Smg9 in mice via CRISPR/Cas9, we were able to recapitulate the major features of the SMG9-related multiple congenital anomaly syndrome we observed in humans. Surprisingly, human cells devoid of SMG9 do not appear to have reduction of PTC-containing transcripts but do display global transcriptional dysregulation. We conclude that SMG9 is required for normal human and murine development, most likely through a transcriptional regulatory role, the precise nature of which remains to be determined.
Keywords: NMD, SMG1, UPF1, brain atrophy, developmental delay, cleft palate, microphthalmia, congenital heart disease
Introduction
The field of transcriptional regulation has grown rapidly in the recent past, and completely novel cellular mechanisms, e.g., control of transcript levels by other RNA species, have emerged 1. Transcripts destined for translation have the potential to encode harmful peptides if the reading frame is altered, a mechanism that underpins the pathogenesis of many human genetic diseases2, 3. Fortunately, the transcriptional regulatory machinery has the capacity to address at least some of these errors. One noteworthy process in this regard is nonsense-mediated decay (NMD). Through NMD, transcripts that contain a premature stop codon (PTC) are degraded such that the cell is protected from a potential dominant-negative effect or toxic gain of function exerted by the truncated protein4, 5, 6.
NMD is a complex process that involves RNA-protein and protein-protein interactions, and it has been observed in all eukaryotes studied to date6. The current model suggests that exon-exon junctions are flagged by a UPF2-UPF3B complex (exon junction complex [EJC])7, 8. If this complex is encountered downstream of a stop codon, this will trigger a series of events. The single most critical step in this process involves the phosphorylation of UPF1 by SMG1, a process that requires the dual action of two ancillary proteins (SMG8 and SMG9) in the SURF (SMG1, UPF1, and the eukaryotic release factors 1 and 3 [eRF1 and eRF3]) complex, which assembles on ribosomes that encounter a PTC9. Upon its phosphorylation, UPF1 releases eRF1 and eRF3, recruits SMG5, SMG6, and SMG7 and halts translation. SMG5–SMG7-mediated exonucleolytic decay and SMG6-mediated endonucleolytic decay lead to degradation of the PTC-containing transcript10, 11, 12, 13, 14, 15. It is worth highlighting that NMD also operates in a partially redundant EJC-independent manner15, 16.
Importantly, NMD also acts on “physiological” transcripts, and at least 10% of human transcripts are subject to NMD regulation17, 18, 19. These transcripts are not limited to those harboring PTCs introduced normally through alternative splicing, but also include transcripts with a multitude of other features, including those in which the normal stop codon is separated from the poly(A)-binding protein by a long 3′ UTR, those with a 3′ UTR that spans an intron, and those with an upstream open reading frame20, 21, 22, 23.
The consequences of deficiency of NMD components in humans in the developmental context are largely unknown, with the exception of UPF3B (MIM: 300298), loss-of-function mutations of which result in intellectual disability (MIM: 300676) 23. In this study, we show that deficiency of another NMD component, SMG9, although compatible with embryonic viability, is associated with a number of major malformations.
Materials and Methods
Human Subjects
Affected individuals were phenotyped via standard clinical evaluation, including appropriate imaging and laboratory tests. All available family members were recruited after they gave informed consent and were enrolled in an institutional review board (IRB)-approved research protocol (KFSHRC RAC no. 2080006). Blood was collected in EDTA tubes for DNA extraction and in Na-heparin tubes for the establishment of lymphoblastoid cell lines.
Autozygome Mapping and Linkage Analysis
We determined the entire set of autozygosity intervals per genome (autozygome) as described before24, 25. In brief, runs of homozygosity (ROH) ≥2 Mb in size, used as surrogates of autozygosity, were determined from genome-wide SNP genotypes obtained on the Axiom SNP Chip platform (Affymetrix) via AutoSNPa26. We performed linkage analysis with the GeneHunter multipoint linkage analysis algorithm by employing a fully penetrant autosomal-recessive model27.
Exome Sequencing
Exome capture was performed with a TruSeq Exome Enrichment kit (Illumina) according to the manufacturer’s protocol. Samples were prepared as an Illumina sequencing library, and in the second step, the sequencing libraries were enriched for the desired target with the Illumina Exome Enrichment protocol. The captured libraries were sequenced with an Illumina HiSeq 2000 Sequencer. The reads were mapped by the Burrows-Wheeler Aligner against the UCSC Genome Browser hg19 reference genome. The SNPs and indels were detected by SAMtools. Variants from whole-exome sequencing were filtered such that only novel or very low frequency (0.1%) coding or splicing homozygous variants within the shared autozygome of the three affected individuals were considered as likely causal variants. The frequency of variants was determined via publicly available variant databases (1000 Genomes, the Exome Variant Server, and the Exome Aggregation Consortium [ExAC] Browser), as well as a database of 734 in-house ethnically-matched exomes.
Real-Time RT-PCR and Immunoblotting
For RT-PCR and relative qRT-PCR, total RNA from affected-individual and control-individual lymphoblastoid cell lines was extracted with the QIAamp RNA Mini Kit (QIAGEN), and DNase was treated by the RNase-Free DNase Set (QIAGEN), according to the manufacturer’s recommendations. Preparation of the cDNA was carried out with the iScriptTM cDNA synthesis kit and Poly T oligonucleotide primers (Applied Biosystems). Two sets of primers for SMG9 (MIM: 613176) cDNA were designed to specifically amplify the cDNA (SMG9 exons 5–7 and 5–8). Relative qRT-PCR for the expression of SMG9, VIM (MIM: 193060), TNS3 (MIM: 606825), EGR1 (MIM: 128990), UCHL1 (MIM: 191342), SPINT2 (MIM: 605124), RORA (MIM: 600825), and VCAN (MIM: 118661) was performed with SYBR Green and an Applied Biosystems 7500 Fast Real-Time PCR System (Table S3).
For immunoblotting, total protein was extracted from one affected-individual and three control-individual lymphoblastoid cells. Anti-SMG9 antibody was purchased from Sigma Aldrich (SAB2107730). The membrane was blocked with 5% milk powder in PBS with Tween for 1 hr at room temperature (RT) and incubated with the primary antibody (dilution 1/500) overnight at 4°C, followed by stringency washes and treatment with secondary antibody for signal detection.
Global Transcriptional Profiling and Statistical Analysis
Lymphoblastoid cells from one affected individual and three healthy control individuals (two females and one male, age range 2–6 years) were used for global expression profiling, performed in triplicate for each sample, via Affymetrix’s GeneChip Human Genome U133 Plus 2.0 Arrays. Sample handling, cDNA synthesis, cRNA labeling and synthesis, hybridization, washing and scanning of chips, and all related quality controls were performed according to the manufacturer’s instructions. In brief, a total of 200 ng of RNA was reverse transcribed to synthesize the first strand of cDNA with the help of oligo-dT primers containing T7 flanking sequence. After second-strand synthesis, in vitro transcription was carried out with labeled aRNA. After purification and fragmentation of aRNA, the samples were hybridized overnight onto U133 Plus 2.0 Arrays. Finally, after being washed to remove the unbound transcripts, the hybridized microarrays were scanned and the intensity (CEL) files with the acquisition and initial quantification of array images were generated with Expression Console 1.3. Significantly modulated genes were defined as those with absolute fold change (FC) > 2 and adjusted p value < 0.05. We used the Benjamini-Hochberg step-up procedure to control the false discovery rate (FDR) at 5% 28. Functional pathway, gene ontology (GO), and network analyses were performed with Ingenuity Pathways Analysis (IPA) 6.3 (Ingenuity Systems) and DAVID Bioinformatics Resources29. A right-tailed Fisher’s exact test was used to calculate a p value determining the probability that the biological function (or pathway) assigned to that dataset is explained by chance alone. For GO analysis, we used the background list of genes expressed in lymphoblastoid cell lines that was obtained from The Gene Expression Barcode project. Statistical analyses were performed with the MATLAB software packages (Mathworks) and Partek Genomics Suite. P values were calculated via two-sided tests. P values of less than 0.05 were considered statistically significant.
Generation of Smg9−/− Mice
A null mutation was generated in mouse Smg9 via CRISPR/Cas9 mutagenesis. A guide RNA was designed against exon 2 (first coding exon) with the online tool CRISPR Design, and a single guide with minimal off-target potential and sequence TCTACGGGATAGAGCGGCGG was selected. Production of sgRNA and microinjection was performed as previously described30. In brief, an oligonucleotide that includes a T7 promoter and the guide sequence above is annealed to a reverse primer that includes the entire stem-loop sequence and is amplified by PCR. The resulting PCR products were in vitro transcribed and purified, and the quality of the RNA products was confirmed with an Agilent Bioanalyzer 2100.
All animal procedures were conducted according to relevant national and international guidelines (AALAC and IACUC) and have been approved by The Jackson Laboratory animal care and use committee (protocol no. 99066). C57BL/6NJ and CB6F1/J mouse strains were used as embryo donors and pseudopregnant recipient dams, respectively. Cas9 mRNA (100 ng/μl) (TriLink Biotechnologies) and sgRNA (50 ng/μl) were injected into the pronuclei of zygotes. Oviduct transfers into pseudopregnant dams were performed on the same day. Founder mice were screened by PCR and Sanger sequencing, identifying several putative mutants. A single allele was chosen for expansion and analysis, which was confirmed by TOPO TA cloning and sequencing (Life Technologies). Genotyping protocols and mice are available at The Jackson Laboratory mouse strain datasheet.
To produce homozygous embryos, timed matings were performed, where day “0” was assumed to be the midpoint of the prior light cycle after the appearance of a copulation plug. Dissections were performed at the time indicated. Embryos were photographed, fixed in 4% paraformaldehyde (24 hr for embryonic day 15.5 [E15.5] and 48 hr for E18.5), stabilized in an acrylamide-based hydrogel31, and stained in Lugol’s solution (Sigma catalog no. L6146-1L) for 24 hr (E15.5) or 48 hr (E18.5). Samples were then rinsed with 1× PBS and embedded in 1% agarose for imaging.
Imaging was conducted on a Bruker Skyscan 1172 X-ray scanner (Bruker BioSpin). Images were reconstructed with NRecon software (Bruker BioSpin) and surface rendered with Imaris software (Bitplane).
For the qRT-PCR of Smg9, RNA was purified from E15 embryonic tissue. Isolated RNA was subject to DNA degradation. cDNA was generated with a High Capacity RNA-to-cDNA Kit (Applied Biosystems), followed by RNA degradation. A TaqMan (Applied Biosystems) assay was designed to amplify cDNA for Smg9 transcripts. Primers were designed to span the exon 2 and exon 3 junction with the probe lying directly over the exon 2 and exon 3 junction site to eliminate genomic DNA amplification. Smg9 qPCR was performed with Applied Biosystems TaqMan reagents, and Gapdh was used as the internal control, multiplexed to optimize accuracy. TaqMan assay was amplified via conditions specified by Applied Biosystems. We used a reaction volume of 10 μl containing 40 ng cDNA. Thermocycler conditions included an initial hold at 50° for 2 min, polymerase activation hold at 95° for 20 s, 40 cycles of 95°, a 1 s denaturation, and a 20 s 60° anneal and extension time. The analysis was performed with Viia7 analysis software. Samples were run in triplicate. Wild-type expression was calculated with n = 1. Each biological replicate is represented on the bar graphs.
Results
Clinical Report
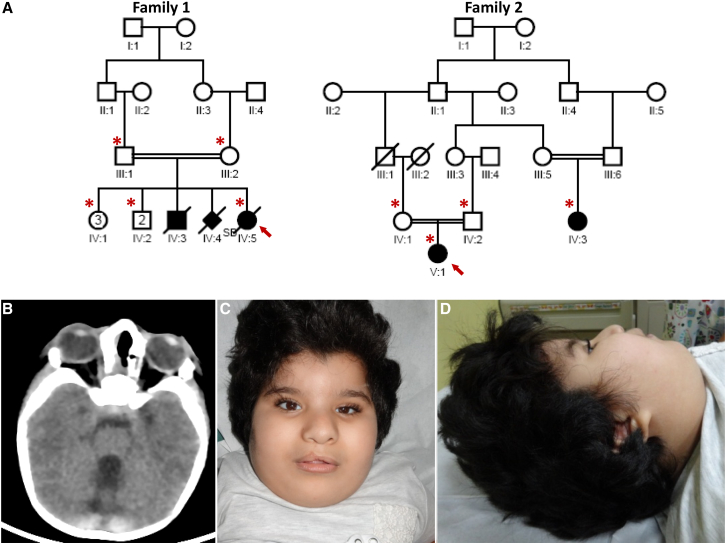
The proband in family 1(IV:5) is a Saudi Arabian female delivered at term by elective cesarean section to a 33-year-old gravida 12, para 8 mother with history of three abortions. The parents are first-degree cousins (Figure 1A). One sibling died at the age of one year and had multiple congenital anomalies similar to the proband’s, as well as a cleft palate (no records available). There is history of a stillborn girl who also had a similar pattern of multiple congenital anomalies (no records available). She was found to have craniofacial dysmorphism, microphthalmia, and major brain and heart malformations and died at 7 weeks of age. Family 2 consists of first-cousin healthy parents of Qatari origin with one child (index, V:1) and a first cousin (IV:3) who was also found to have a similar phenotype in the form of craniofacial dysmorphism, congenital heart disease, and brain malformation (Figure 1A). For clinical details of the three affected individuals, see the Supplemental Note and Table 1.
Figure 1.
Identification of an Autosomal-Recessive Multiple Congenital Anomalies Syndrome
(A) Pedigrees of the two study families showing the consanguineous nature of the parents. The index individual is indicated in each pedigree by an arrow, and asterisks denote individuals whose DNA was available for analysis.
(B) CT scan for the index individual in family 1 (IV:5) showing cerebellar vermis hypoplasia.
(C and D) Facial images of the index individual in family 2 (V:1) showing a right-sided cleft lip (status post repair), a full and everted lower lip, a widow’s peak, hypertelorism, small eyes, posteriorly rotated ears with attached lobules, and a broad nasal bridge.
Table 1.
Clinical Details of the Three Affected Individuals Recruited in this Study
| Category | Subcategory | Features | |
|---|---|---|---|
| Family 1, IV:5 | family history | – | parents are 1st degree cousins, one sibling who had died at the age of one year with a similar disease |
| head and neck | head | prominent forehead and occiput | |
| ears | low set malformed ears | ||
| eyes | microphthalmia | ||
| nose | depressed nasal bridge and anteverted nares | ||
| mouth | high arched palate | ||
| skeletal | hands | clenched hands with camptodactyly | |
| neurologic | CNS | Dandy-Walker malformation, cerebellar vermis hypoplasia, hypoplastic corpus callosum | |
| cardiovascular | heart | interrupted aortic arch, hypoplastic tricuspid and aortic valves, large muscular VSD | |
| Family 2, V:1 | family history | – | parents are 1st degree cousins, one similarly affected cousin |
| head and neck | head | narrow forehead | |
| ears | posteriorly rotated ears with attached lobules | ||
| eyes | hypertelorism and poor vision | ||
| nose | broad nasal bridge | ||
| mouth | full and everted lower lip, right-sided cleft lip | ||
| skeletal | hands | syndactyly between 2nd and 3rd toes | |
| neurologic | – | Dandy-Walker malformation, decreased myelination, brain atrophy | |
| cardiovascular | heart | large VSD | |
| Family 2, IV:3 | head and neck | head | NA |
| skeletal | hands | NA | |
| neurologic | – | truncal hypotonia with peripheral hypertonia, brisk deep tendon reflexes, adductor spasm in both lower limbs | |
| cardiovascular | heart | VSD |
Abbreviations are as follows: NA, not available; VSD, ventricular septal defect.
A Multiple Congenital Anomaly Syndrome Is Linked to Loss-of-Function Mutations in SMG9
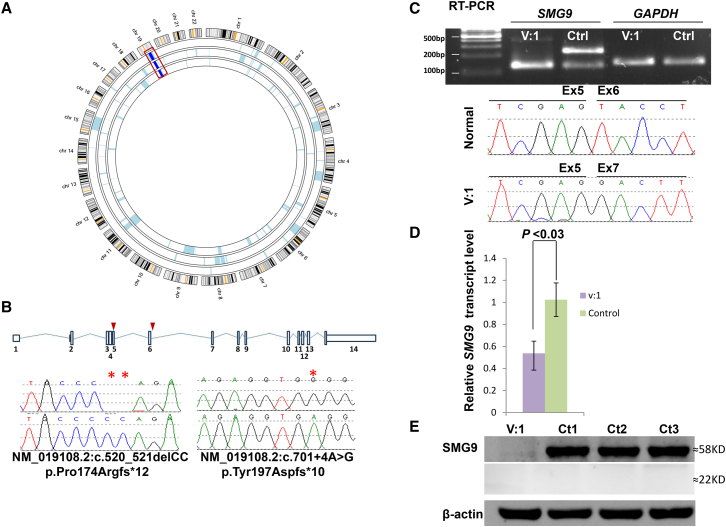
The phenotypic overlap (especially the Dandy-Walker malformation and congenital heart disease) between the affected members of both families, their consanguineous nature, and different geographic origin in Arabia prompted us to consider the possibility that the disease is caused by homozygosity for two distinct founder haplotypes (one in family 1 and another in family 2) that harbor pathogenic variants in the same gene. Indeed, the autozygome of the three affected individuals overlapped on a single locus in which the two families displayed two different haplotypes, and this was further confirmed by linkage analysis (Figure 2A and Figure S1). To identify the likely causal variant within this critical locus, we performed exome sequencing on each of the two probands and achieved 80× mean depth of the target region (target was 52 Mb), and 97% of target regions were covered with at least 10× depth. We only considered homozygous coding and splicing novel variants within the critical locus (Figure S2). Only one variant per exome remained (c.520_521delCC and c.701+4A>G) according to these filters, both affecting the same gene, SMG9 (GenBank: NM_019108.2). Sanger sequencing confirmed homozygosity for these mutations in the probands and heterozygosity in the parents (Figure 2B). The indel in family 1 predicts a frameshift and premature truncation, p.Pro174Argfs∗12. Splice Site Prediction by Neural Network indicates that the intronic variant in family 2 reduces the score of the consensus donor site of exon 6 from 0.98 to 0.65. However, RT-PCR on lymphoblastoid cells from the index individual in family 2 revealed complete skipping of exon 6. The resulting aberrant transcript that predicts frameshift and premature truncation (p.Tyr197Aspfs∗10) was the only recovered transcript in the affected individual, unlike control individuals, who possessed this band at a very low abundance in comparison to the normal exon-6-containing transcript (Figure 2C). Western blot analysis revealed complete lack of SMG9, and the predicted truncated isoform could not be recovered, indicating NMD and/or instability (Figure 2E and Figure S3). Indeed, qRT-PCR showed 50% reduction of the SMG9 transcript, as compared to control individuals (p < 0.03) (Figure 2D). Taken together, our results are consistent with SMG9 deficiency as the likely cause of the multiple congenital anomaly syndrome we observed in the two probands.
Figure 2.
A Multiple Congenital Anomalies Syndrome Is Linked to SMG9 Mutations
(A) AgileMultiIdeogram showing a single homozygous region, on chromosome 19, shared between the three affected individuals from the two families as indicated by the dark blue bars boxed in red.
(B) Upper panel: schematic representation of SMG9 transcript (red triangles indicate the sites of the two mutations). Lower panel: sequence chromatograms of two variants in SMG9 identified in the family (control tracing is shown for comparison and the location of the mutation is denoted by red stars).
(C) RT-PCR gel image and sequence chromatogram show the enhancement of aberrant SMG9 transcript in lymphoblasts derived from affected individual (Aff) as compared with lymphoblasts from a healthy individual of similar age (control), and the absence of the normal SMG9 transcript.
(D) Relative quantitative real-time PCR showing 50% reduction in SMG9 expression in the affected individual in comparison to expression in the control individual. Result is the average for three independent experiments and three control individuals (p < 0.03 on t test).
(E) Immunoblotting with antibody against the N terminal of SMG9 (SAB2107730) showing no detectable band from cells derived from the index individual (Aff) (target mass KDa, 57.7, 54.8) as compared with those derived from the three normal control individuals (Ct1, Ct2, Ct3), as well as the absence of the truncated SMG9 isoform at 22 KDa.
No Evidence of Widespread Perturbation of NMD in SMG9 Deficiency
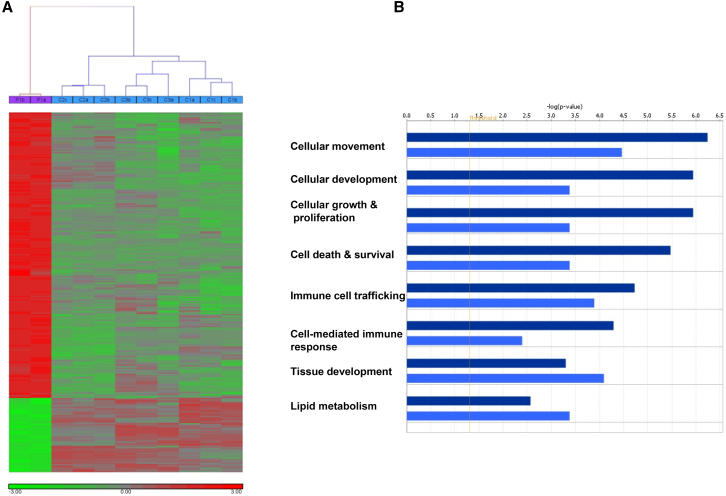
SMG9 has been found to be necessary for SMG1-mediated phosphorylation of UPF1, the single most important step in NMD, so we were surprised by the finding that the index individual in family 2 (V:1), who appears to have complete deficiency of SMG9, shows evidence of NMD of the PTC-containing SMG9 transcript (Figure 2D). Therefore, we set out to test the entire transcriptional profile of the affected individual and compare it to that of control individuals for whom exome sequencing data were available. We first looked for evidence in control individuals that transcripts with PTC are significantly lower than other transcripts in general and found that, despite extreme variability, such was the case (p < 0.01). We then repeated the same test on the affected individual and found that the same pattern was observed (Figure S4 and Table S1). Comparing the expression of 27 common PTC-containing genes in control individuals and in the affected individual revealed no significant difference (p value > 0.05) (Figure S5 and Table S2). However, as shown in Figure 3, the global transcriptional profile of the affected individual and control individuals revealed a pattern of gene dysregulation that clearly distinguished individuals as either affected or controls (Figure 3A). We observed a prevalent upregulation of gene expression as a result of SMG9 deficiency. Indeed, with a stringent statistical threshold (FDR of 5% and FC > 2), 300 probes were differentially expressed (DE), of which 238 probes (171 genes) were upregulated and 62 probes (54 genes) were downregulated, when comparing expression in the affected individual versus that in the control individuals (Figure 3A and Table S1). Validation was carried out on selected genes taken from the most dysregulated ones by quantitative real-time PCR, and the results were in agreement with the microarray (Figure S6). The GO and functional analyses of upregulated DE genes showed an over-representation of genes involved in cellular movement, development, growth and proliferation, immune response, and cell death. On the other hand, lipid metabolism and the tissue development process were significantly enriched categories among the downregulated genes (Figure 3B). Although we cannot exclude the possibility of an effect by SMG9 deficiency on NMD for specific transcripts, our data suggest that there is no widespread impairment of NMD although the transcriptional profile was dysregulated.
Figure 3.
Global Transcriptional Changes Associated with SMG9 Deficiency
(A) Heat-map of genes that were significantly dysregulated in a SMG9-deficient individual in comparison to three control individuals. The hierarchical clustering clearly distinguished individuals as either affected or controls. The expression level of each gene across the samples is scaled to [-3, 3] interval. These mapped expression levels are depicted with a color scale as shown at the bottom of the figure, such that highly expressed genes are indicated in red, intermediate in black, and weakly expressed in green.
(B) GO and functional analysis of DE genes (up- or downregulated). x axis indicates the significance (−log p value) of the functional association that is dependent on the number of genes in a class as well as biological relevance. Dark bars represent upregulated genes, and light bars represent downregulated genes. The threshold line represents a p value of 0.05.
Smg9−/− Mice Have Reduced Viability and Display Major Brain, Eye, and Cardiovascular Malformations
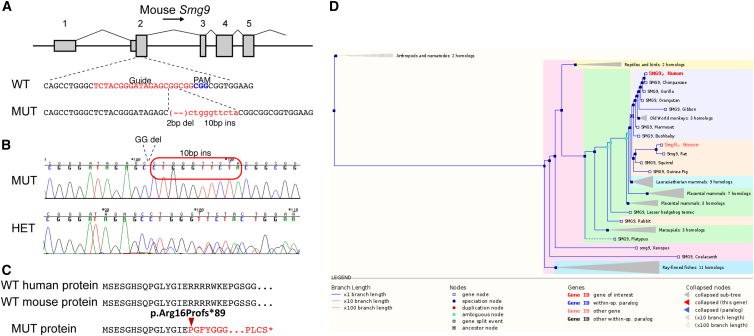
To determine its role in a mammalian model, we attempted to generate a null allele of Smg9 in the mouse by using CRISPR/Cas9. We targeted exon 2 (first coding exon) of Smg9 by using a single guide sequence designed to minimize off-target potential. From eight founders, a single mutation containing a combination of a 2 bp deletion and 10 bp insertion (Figures 4A and 4B) was chosen for expansion and analysis. This mutation is predicted to result in a frameshift and a premature stop at amino acid position 104 (Figure 4C). qRT-PCR on E15.5 embryos showed that Smg9 is reduced by 60%–80%, indicating that the mouse line is at least a severe hypomorph (Figure S7).
Figure 4.
Generation of an Smg9 Null Allele
(A) Schematic of mouse Smg9, showing the position of the CRISPR guide position in exon 2 (first coding exon; exon numbers indicated above the gene model). The mutant allele chosen for analysis contains a 2 bp deletion and simultaneous 10 bp insertion, resulting in a frameshift mutation beginning at 7:24403443 (GRCm38/mm10).
(B) Sequence chromatogram of a TOPO-TA clone of the mutant allele (top) and a heterozygous mutant (bottom), confirming the allele structure. Red box indicates inserted sequence.
(C) Structure of the predicted protein product showing the frameshift at residue 16 and the predicted premature termination at position 104. The aligned human protein is shown above the mouse sequence.
(D) SMG9 phylogenetic tree and conservation across species.
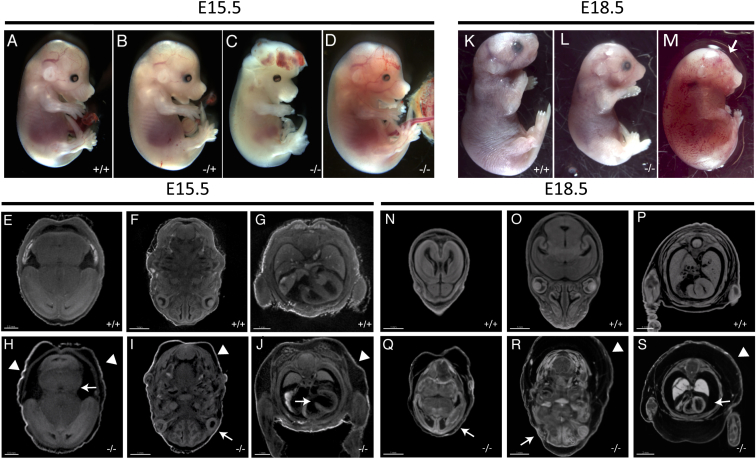
Intercrosses of heterozygous animals yielded no homozygous pups, suggesting embryonic lethality. To identify and characterize embryonic phenotypes of Smg9−/−, we performed timed matings and harvested embryos at E14.5, E15.5, and E18.5 (Table 2). As shown in Figure 5 and Table 2, phenotypes were identified at all stages, and the exact presentation varied significantly between specimens. For example, homozygous embryos examined at E15.5 showed a range of phenotypes including edema, hemorrhage, and exencephaly (Figures 5C and 5D) and in one case, hind-limb preaxial polydactyly (Figure 5D, arrow). These phenotypes are variable and incompletely penetrant, but the majority of homozygotes display some clear phenotypic abnormality, unlike wild-type and heterozygous embryos (Figures 5A and 5B). To examine these phenotypes in more detail, we performed iodine-contrast microCT (computed tomography), which provides a high-resolution 3D dataset with sufficient contrast to observe internal structures. Mutant embryos were clearly smaller and developmentally delayed. Several other features were noted, including edema, observed during gross inspection (Figures 5H–5J, arrowheads). We also noted reduced size in the mid- and hindbrain relative to their stage of development (Figure 5H), microphthalmia (Figure 5I), thin myocardium, and a very prominent atrioventricular septal defect (AVSD) (Figure 5J, arrow). At E18.5, the two homozygotes identified displayed variable levels of edema (Figures 5L and 5M), with evidence of hemorrhage. One specimen was dead (Figure 5M) and showed thin myocardium in sections of microCT volumes (Figure 5S, arrow) and anophthamia (Figures 5M and 5R, arrow) of one eye, consistent with the phenotypes observed at E15.5. Thus, our findings support a role for Smg9 in brain, heart, and eye development.
Table 2.
Summary of Genotypes and Phenotypes Observed in Smg9 Mouse Embryos
| Stage (n) |
Genotype |
Mutant Phenotypes |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| +/+ | −/+ | −/− | Developmental Delay | Brain (uCT) | Edema | Heart (uCT) | Eye (uCT) | Limb | |
| E14.5 (4) | 1 | 1 | 2 | 2/2 | 2/2 | 2/2 | NA | NA | 1/4 |
| E15.5 (38) | 13 | 17 | 8 | 8/8 | 7/7 | 7/8 | 7/7 | 7/7 | 1/8 |
| E18.5 (13) | 6 | 5 | 2 | 2/2 | 2/2 | 1/2 | 1/2 | 1/2 | 0/2 |
NA, not applicable.
Figure 5.
Multiple Developmental Abnormalities in Smg9−/− Mouse Embryos
(A–D) Mutant embryos at E15.5 display multiple developmental defects, including exencephaly (C) and sporadic hemorrhage and edema (D), versus wild-type (A) or heterozygous (B) littermates.
(E–J) Volume slices of iodine-contrast microCT images of the mutant in (D) show multiple defects, including abnormal brain development (arrow, H), microphthalmia (arrow, I), and an atrioventricular septal defect and thin myocardium (arrow, J). Clear edema in the mutant images is indicated by the arrowheads (H–J). Control E15.5 sections from similar planes are shown in (E)–(G).
(K–S) Some embryos survived or were recovered dead at E18.5 (L and M versus K). As with earlier stages, both mutants were developmentally delayed and showed a variable degree of edema. One mutant had anophthalmia of one eye (arrow in M and R). MicroCT images of the mutant in (M) show edema (arrowheads in R and S), a severely malformed brain (Q), and abnormally thin myocardium in (S). Control sections through similar planes are shown in (N)–(P).
Scale bars for (E)–(J) are 0.5 mm (E and H) and 1.0 mm (F, G, I, and J) and for (N)–(S) are 1.0 mm (N, O, Q, and R) and 2.0 mm (P and S).
Discussion
The widespread role of NMD in transcriptional regulation suggests that defects in NMD can have pathological consequences at the organismal level. Interrogating a developmental role of NMD is challenging because many of its components have additional physiological roles. Moreover, NMD, although conserved across eukaryotes as a mechanism, differs in its details between different species such that the results observed in one species are not necessarily applicable to others6, 32. Nonetheless, reduced embryonic viability as a converging phenotype among animal models deficient for at least some components of NMD appears to suggest that NMD does contribute to normal development. For example, knockdown of SMGL-1 and SMGL-2 is embryonically lethal in C. elegans33. In fruitflies, depletion of UPF1 and UPF2 is also embryonically lethal34. Zebrafish morphants for upf1, upf2, smg5, or smg6 have high rates of embryonic lethality35. Finally, mice knocked out for Smg1, Upf1, or Upf2 display early embryonic lethality as well 36, 37, 38.
SMG9 was first identified as a component of NMD in 2009 when it was found, along with SMG8, to co-purify with SMG1, forming a stable SMG1-SMG8-SMG9 complex11. Further characterization of this protein revealed that it is part of the SURF complex and is necessary for the phosphorylation of UFP1 by SMG1. Deletion mapping identified that the presence of both N and C termini is needed for the proper binding to SMG139. Although the consequences of SMG9 deficiency on the NMD machinery are clearly demonstrated, virtually nothing is known about its developmental relevance. As mentioned above, deficiency of various NMD components has been found to result in abnormal development in model organisms, and UPF3B deficiency in humans is linked to intellectual disability40. However, SMG9 deficiency states have not been established in humans or other model organisms. Our study, therefore, fills an important gap in our knowledge of the developmental role of this component of NMD.
The identification of a single disease locus and two independent, homozygous, apparently loss-of-function mutations therein involving the same gene is highly suggestive of a causal relationship between SMG9 mutations and the multiple congenital anomaly syndrome we describe. This is further corroborated by the recapitulation of key developmental defects in the knockout mice we generated via CRISPR/Cas9 mutagenesis in this study. Interestingly, the predominant involvement of the brain is reminiscent of the zebrafish morphants that are knocked down for various NMD components in which the hindbrain is particularly affected 35. However, the mechanism through which SMG9 deficiency results in abnormal embryogenesis remains unclear.
Perturbation of NMD was a prime candidate mechanism in view of the established role of NMD in protecting the cells from the toxic effect of PTC-containing proteins. However, our finding that the SMG9 mutant transcript itself is subject to NMD in the affected individual’s cells suggested to us that this might not be the case. Indeed, subsequent global transcriptional profiling also failed to show evidence of impaired NMD as a result of SMG9 deficiency. Although different transcripts are known to have different sensitivity to NMD, so we cannot exclude the possibility that NMD is defective for certain PTC-containing transcripts, our data suggest that NMD was generally functional. Given that NMD affects at least 10% of human transcripts, many of which lack PTC, it is possible that the generalized transcriptional dysregulation we observed is still reflective of an abnormality in NMD that we could not measure. Interestingly, it has been suggested that SMG9 might play other physiological roles independent of its role in NMD because it was identified in homodimers that are not part of the SURF complex, and this could also be relevant to the transcriptional dysregulation of affected individuals’ cells39.
In conclusion, this study shows that SMG9 mutations most likely cause a distinctive multiple congenital anomaly syndrome in humans and that its deficiency in mice also leads to abnormal embryogenesis and an overlapping developmental profile. Despite the established role of SMG9 in NMD, we show that PTC-containing transcripts do undergo efficient degradation in the context of SMG9 deficiency, although the latter state is still associated with widespread transcriptional dysregulation. Future studies will be needed to further delineate SMG9-related phenotypes in humans and the underlying mechanism, which might be related to a non-NMD role played by this protein.
Acknowledgments
We thank the families in this study for their enthusiastic participation. We thank the Sequencing Core Facility at the King Faisal Specialist Hospital and Research Center for their technical help. This study was supported by King Abdulaziz City for Science and Technology (KACST) grant 13-BIO1113-20 (F.S.A.), NIH grant OD011185 (S.A.M.), KACST grant 11-BIO2072-20 (D.C.), and the King Salman Center for Disability Research (F.S.A.).
Published: March 24, 2016
Footnotes
Supplemental Data include a Supplemental Note, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.010.
Contributor Information
Stephen A. Murray, Email: steve.murray@jax.org.
Fowzan S. Alkuraya, Email: falkuraya@kfshrc.edu.sa.
Web Resources
The URLs for data presented herein are as follows:
Berkeley Drosophila Genome Project NNSplice 0.9, http://www.fruitfly.org/seq_tools/splice.html
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
CRISPR Design, http://crispr.mit.edu
ExAC Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
SAMtools, http://samtools.sourceforge.net/
The Gene Expression Barcode project, http://barcode.luhs.org
The Jackson Laboratory mouse strain datasheet, https://www.jax.org/strain/027253
UCSC Genome Browser hg19, http://genome.ucsc.edu
Supplemental Data
References
- 1.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;7:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 2.Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuvanagiri M., Schlitter A.M., Hentze M.W., Kulozik A.E. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 4.Stalder L., Mühlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y.-F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 6.Isken O., Maquat L.E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 7.Hwang J., Maquat L.E. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behm-Ansmant I., Izaurralde E. Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- 9.Lykke-Andersen J., Shu M.-D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi T., Yamashita A., Kashima I., Schell T., Anders K.R., Grimson A., Hachiya T., Hentze M.W., Anderson P., Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 11.Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita A., Izumi N., Kashima I., Ohnishi T., Saari B., Katsuhata Y., Muramatsu R., Morita T., Iwamatsu A., Hachiya T. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clerici M., Mourão A., Gutsche I., Gehring N.H., Hentze M.W., Kulozik A., Kadlec J., Sattler M., Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada-Katsuhata Y., Yamashita A., Kutsuzawa K., Izumi N., Hirahara F., Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metze S., Herzog V.A., Ruepp M.-D., Mühlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA. 2013;19:1432–1448. doi: 10.1261/rna.038893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 18.Wittmann J., Hol E.M., Jäck H.-M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan W.K., Huang L., Gudikote J.P., Chang Y.F., Imam J.S., MacLean J.A., 2nd, Wilkinson M.F. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 21.Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov P.V., Gehring N.H., Kunz J.B., Hentze M.W., Kulozik A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebbapragada I., Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Alkuraya F.S. Autozygome decoded. Genet. Med. 2010;12:765–771. doi: 10.1097/GIM.0b013e3181fbfcc4. [DOI] [PubMed] [Google Scholar]
- 25.Alkuraya F.S. Current Protocols in Human Genetics. Wiley; 2012. Discovery of rare homozygous mutations from studies of consanguineous pedigrees. Unit 6.12. [DOI] [PubMed] [Google Scholar]
- 26.Carr I.M., Flintoff K.J., Taylor G.R., Markham A.F., Bonthron D.T. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 27.Lindner T.H., Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 1995;57:289–300. [Google Scholar]
- 29.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 30.Guimier A., Gabriel G.C., Bajolle F., Tsang M., Liu H., Noll A., Schwartz M., El Malti R., Smith L.D., Klena N.T. MMP21 is mutated in human heterotaxy and is required for normal left-right asymmetry in vertebrates. Nat. Genet. 2015;47:1260–1263. doi: 10.1038/ng.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong M.D., Spring S., Henkelman R.M. Structural stabilization of tissue for embryo phenotyping using micro-CT with iodine staining. PLoS ONE. 2013;8:e84321. doi: 10.1371/journal.pone.0084321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conti E., Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Longman D., Plasterk R.H., Johnstone I.L., Cáceres J.F. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avery P., Vicente-Crespo M., Francis D., Nashchekina O., Alonso C.R., Palacios I.M. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17:624–638. doi: 10.1261/rna.2404211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittkopp N., Huntzinger E., Weiler C., Saulière J., Schmidt S., Sonawane M., Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medghalchi S.M., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 37.McIlwain D.R., Pan Q., Reilly P.T., Elia A.J., McCracken S., Wakeham A.C., Itie-Youten A., Blencowe B.J., Mak T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoren L.A., Nørgaard G.A., Weischenfeldt J., Waage J., Jakobsen J.S., Damgaard I., Bergström F.C., Blom A.M., Borup R., Bisgaard H.C., Porse B.T. UPF2 is a critical regulator of liver development, function and regeneration. PLoS ONE. 2010;5:e11650. doi: 10.1371/journal.pone.0011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernández I.S., Yamashita A., Arias-Palomo E., Bamba Y., Bartolomé R.A., Canales M.A., Teixidó J., Ohno S., Llorca O. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res. 2011;39:347–358. doi: 10.1093/nar/gkq749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarpey P.S., Raymond F.L., Nguyen L.S., Rodriguez J., Hackett A., Vandeleur L., Smith R., Shoubridge C., Edkins S., Stevens C. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.