Abstract
The general transcription factor IIE (TFIIE) is essential for transcription initiation by RNA polymerase II (RNA pol II) via direct interaction with the basal transcription/DNA repair factor IIH (TFIIH). TFIIH harbors mutations in two rare genetic disorders, the cancer-prone xeroderma pigmentosum (XP) and the cancer-free, multisystem developmental disorder trichothiodystrophy (TTD). The phenotypic complexity resulting from mutations affecting TFIIH has been attributed to the nucleotide excision repair (NER) defect as well as to impaired transcription. Here, we report two unrelated children showing clinical features typical of TTD who harbor different homozygous missense mutations in GTF2E2 (c.448G>C [p.Ala150Pro] and c.559G>T [p.Asp187Tyr]) encoding the beta subunit of transcription factor IIE (TFIIEβ). Repair of ultraviolet-induced DNA damage was normal in the GTF2E2 mutated cells, indicating that TFIIE was not involved in NER. We found decreased protein levels of the two TFIIE subunits (TFIIEα and TFIIEβ) as well as decreased phosphorylation of TFIIEα in cells from both children. Interestingly, decreased phosphorylation of TFIIEα was also seen in TTD cells with mutations in ERCC2, which encodes the XPD subunit of TFIIH, but not in XP cells with ERCC2 mutations. Our findings support the theory that TTD is caused by transcriptional impairments that are distinct from the NER disorder XP.
Introduction
The basal transcription factor IIH (TFIIH) has essential functions in RNA pol I and RNA pol II transcription, as well as nucleotide excision repair (NER) of ultraviolet (UV)-induced DNA lesions. TFIIH is a multi-protein complex comprised of a 7-subunit core (XPB, XPD, p62, p52, p44, p34, and TTDA) and a 3-subunit cyclin activating kinase (CAK) complex formed by CDK7, MAT1, and cyclin H.1 In RNA pol I transcription, the TFIIH ATPase activity of XPB is required for proper transcription of ribosomal DNA.2, 3 In RNA pol II transcription, the helicase activity of TFIIH subunit XPB and the ATPase activity of XPD are involved in promoter opening. The TFIIH kinase activity of CDK7 is required for RNA pol II phosphorylation at the C-terminal domain (CTD). In NER, the helicase activity of XPD and the ATPase activity of XPB open the DNA strand around a UV-induced lesion in order to permit incision and repair of the damaged DNA strand.4
Mutations in ERCC2 (OMIM: 126340) and ERCC3 (OMIM: 133510), encoding the TFIIH subunits XPD and XPB, respectively, cause the rare autosomal-recessive disorders xeroderma pigmentosum (XP) and trichothiodystrophy (TTD) (OMIM: 610651, 278730, 608780).5 All the mutations found in TTD-affected individuals result in reduced steady-state levels of the entire TFIIH complex (Table S1) and impaired functioning in repair and transcription (reviewed in Stefanini et al.6). Although individuals with XP have a 10,000-fold increased risk of skin cancer, individuals with TTD have normal skin cancer risk even though they might present with cutaneous photosensitivity. In addition, individuals with TTD show developmental defects including neurological and hair abnormalities (Figure S1). Most of these individuals with XP or TTD are compound heterozygous and both alleles might contribute to the phenotype.4, 7, 8, 9 This complex phenotypic variability is associated with different mutation(s) in ERCC2 or ERCC3 that, in addition to interfering with the role of TFIIH in repair, differentially affect the multiple functions of TFIIH in transcription, including basal and activated transcription. Features of XP are thought to be associated with ERCC2 and ERCC3 mutations primarily affecting DNA repair,5, 8 whereas ERCC2 and ERCC3 mutations associated with TTD show transcriptional alterations in addition to impaired NER4, 9, 10, 11 (see also references in Compe and Egly4 and Stefanini et al.6).
In addition to the NER-defective form of TTD due to mutations in genes encoding TFIIH subunits (OMIM: 601675), an additional TTD form (OMIM: 234050) characterized by the absence of clinical and cellular photosensitivity, normal response to UV light, and TFIIH steady-state level has been reported. A small proportion of the non-photosensitive TTD case subjects harbor mutations in TTDN1 (trichothiodystrophy non-photosensitive 1 [MPLKIP]12, 13, 14 [OMIM: 609188]) whose product seems to be involved in cell cycle progression15 although its precise function still remains unknown. Recently, an X-linked form of non-photosensitive TTD was found associated with a nonsense mutation in RNF113A (ring finger protein 113A [OMIM: 300953]) in two Australian cousins.16 The molecular function of RNF113A remains unknown.
Here we report two unrelated non-photosensitive children with TTD from different parts of the world who were studied by scientists from Europe and the US. The affected children’s cells were NER proficient and did not have mutations in any of the genes known to be associated with TTD. In searching for TTD-associated genes that could cause the TTD phenotype, the European group employed a targeted approach by studying several basal transcription factors that act in concert with TFIIH whereas the US group applied whole-exome sequencing technology. These different approaches led to the identification of two homozygous missense mutations in the same gene, GTF2E2 (OMIM: 189964), encoding the beta subunit of the basal transcription factor II E (TFIIEβ). This transcription factor is essential for the assembly and stabilization of the RNA pol II pre-initiation complex (PIC) at the transcription start site.17 TFIIE enters the PIC after RNA pol II, recruits TFIIH to the PIC, stimulates RNA pol II CTD phosphorylation by the CDK7 subunit of TFIIH, and regulates the helicase activity of XPB to catalyze the open complex formation, leading to promoter clearance.18, 19, 20 The two individuals with mutations in GTF2E2 studied here have normal NER but reduced cellular levels of both subunits of the TFIIE complex (TFIIEα and TFIIEβ). In addition, the pathological alterations in TFIIEβ resulted in decreased amounts of the overall TFIIE complex and of phosphorylated TFIIEα. Reduced levels of phosphorylated TFIIEα were also seen in TTD cells with ERCC2 (XPD) mutations but not in XP cells with mutations in ERCC2.
Material and Methods
Affected Individuals, Cell Culture, UV Sensitivity Testing, and Transfection
The affected children were studied under protocols approved by the Institutional Review Boards at the US NIH and Vrije Universiteit Brussel. Written informed consent was obtained from the parents of the children. Normal, XP, and TTD primary human skin fibroblasts from the Human Genetic Mutant Cell Repository and lymphoblastoid cell lines from Fisher BioServices (NCI Frederick Central Repository) or established in Pavia, Italy, were cultured as described.21 The study was performed on primary fibroblasts of the affected individuals TTD28PV and TTD379BE and the parents of TTD379BE and on lymphoblastoid cells of TTD28PV, the parents and healthy sister of TTD28PV, and on a healthy donor, C6PV. Investigations on TFIIEα were extended to primary fibroblasts of six TTD- and six XP-affected individuals with mutations in ERCC2 (XPD) (Table S1). Normal primary fibroblasts from four genetically unrelated healthy donors (C3PV, C377RM, C1609RM, C16354BE) and from the phenotypically normal parents of TTD379BE were analyzed in parallel. For proliferating and non-proliferating cultures, 3 × 105 fibroblasts were seeded in 6-cm Petri dishes and cultured for 3 and 10 days, respectively, before processing. Cell survival after UV irradiation was measured using CellTiter96 Non-Radioactive Cell Proliferation Assay (Promega). After UV host cell reactivation, DNA repair assays were performed by transfection of 1 μg of UVC-irradiated (1,000 or 500 J/m2) or non-irradiated luciferase expression vector into 2.5 × 105 fibroblasts using Lipofectamine 2000 (Life Technologies) according to the vendor’s protocol. Luciferase activity was measured as described.22 Transient expression of recombinant TFIIEαFlag was obtained by transfecting 2 μg of pMP2-TFIIEαFlag plasmid into 1 × 106 C3PV primary fibroblasts by Amaxa Nucleofector technology (Lonza). Cells were processed 48 hr after transfection. Evaluation of unscheduled DNA synthesis (UDS) and recovery of RNA synthesis (RRS) were carried out according to routine procedures.23, 24, 25
DNA Isolation and Sequencing
We isolated DNA from blood of TTD-affected individuals who had been examined at the NIH Clinical Center under protocols approved by the National Cancer Institute Institutional Review Board. Genomic DNA from blood or cell lines was isolated via DNeasy Blood & Tissue Kit (QIAGEN), purified via Genomic DNA Clean & Concentrator (Zymo Research), and used for whole-exome sequencing or Sanger sequencing via GTF2E2 reverse primer GTF2E2-R1 (Table S2) to confirm the mutation detected by whole-exome sequencing.
In TTD28PV primary fibroblasts, the complete coding region of GTF2E2 was analyzed by sequencing of PCR-amplified cDNA, and the mutation was investigated in the relevant genomic DNA region of the family members. Additional TTD-affected subjects were screened for mutations in GTF2E2 and GTF2E1 by directed sequencing of either the cDNA or genomic DNA using PCR and sequencing primers (Table S2).
Whole-Exome Sequencing and Data Analysis
Exonic DNA was captured using Agilent SureSelect Human All Exon V5+UTRs target enrichment kit (Agilent Technologies), which targets 75 megabases (21,522 genes and 359,555 exons). Exome libraries were prepared according to manufacturers’ recommended protocols. Sequencing was performed via an Illumina HiSeq2000 (Illumina), and the sequencing was run as 2 × 101 base pairs with TruSeq V3-HS reagents. The HiSeq Real Time Analysis (RTA 1.12.4.2) was used for processing image files, and the Illumina CASAVA_v1.8.2 was used to demultiplex and convert binary base calls and qualities to fastq format. The TTD379BE sample had 127 million reads and more than 93% of the bases had quality values of Q30 and above. The sequencing reads were trimmed of adapters and of low-quality bases using Trimmomatic and were aligned to human hg19 reference genome (GRCh37/UCSC hg19) using the short read alignment component in Burrows-Wheeler Aligner BWA v.0.7.26 The mean alignment percentage was 98.9% mapped to reference genome, and 68% of the mapped reads were on target within the capture regions. The mean depth of coverage was 106× with more than 99.7% of the target regions covered at least 10×, and 94.54% target regions were covered at least 30× sequencing depth.
The Genome Analysis Toolkit (GATK_v2.5, developed at Broad Institute) was used to perform variant discovery and genotyping. SNPs and indels were called according to the GATK’s Best Practices. In brief, the Mapped BAM files were modified to add read groups, marked for duplicates, and sorted with Picard software. The alignments were locally realigned around candidate indels and alignment base quality scores were recalibrated. After the mapped read pre-processing, the variants were initially called with the Unified Genotyper. The raw SNPs and indels were further processed via the GATK’s variant quality score recalibration (VQSR) workflow. For further analysis, we selected variants that were within the target regions covered by the SureSelect All Exon V5+URT kit. In addition, we used a Mendelian inheritance model and performed trio analysis to assess the Mendelian error rates. The analysis-ready SNPs and INDELs were then annotated for functional significance with Variant Effect Predictor (VEP, Ensembl) and ANNOVAR software. The annotated non-synonymous variants were further filtered based on SIFT27 and PolyPhen228 and classified as “not benign” or “not tolerated,” respectively. We also applied variant annotation and interpretation analyses generated through the use of QIAGEN’s Ingenuity Variant Analysis software for validation and further evaluation of the prioritized variants.
Quantitative Real-Time PCR
Total RNA was extracted using RNAqueous extraction kit (Life Technologies) or the RNeasy Mini Kit (QIAGEN). Transcript levels of GTF2E2 and GAPDH (for normalization) were measured on a Biorad CFX96 or a LightCycler 480 (Roche). Primer sequences are in Table S2.
RNA Interference
GTF2E2 or CDK7 silencing was performed as previously described.9 In brief, 8 × 105 primary fibroblasts were transfected with 360 μmol control (AllStar negative control, QIAGEN Sciences), GTF2E2 or CDK7 siRNA (FlexiTube siRNA, QIAGEN Sciences) using the HiPerfect Transfection Reagent (QIAGEN Sciences) and incubated at 37°C for different time points. Transfected cells were processed for qPCR and immunoblot analysis.
Phosphoprotein Enrichment
Phosphoprotein-enriched fractions were obtained using the Pro-Q Diamond Phosphoprotein Enrichment Kit (Molecular Probes by Life Technologies). In brief, 8 × 105 primary fibroblasts were lysed in 500 μL of supplied lysis buffer and the clarified supernatant was loaded on Pro-Q Diamond columns. Phosphorylated and unphosphorylated protein enriched fractions were eluted in 5 mL and 1.25 mL of elution buffer, respectively, and thereafter concentrated to a volume of 50 μL by using Amicon Ultra-4 Columns (Millipore). Concentrated fractions were diluted in Laemmli buffer and analyzed by immunoblotting.
Phosphatase Treatment
Protein dephosphorylation was carried out using calf intestinal alkaline phosphatase (CIP). Primary fibroblasts were lysed in culture dishes via directly scraping in CIP buffer (100 mM NaCl, 50 mM Tris, 10 mM MgCl2, 1 mM dithiothreitol [pH 7.9]) supplemented with EDTA-free Protease Inhibitor Cocktail (Roche Diagnostic). Cell lysates were incubated at 37°C for different time points after addition of 3 U/μL CIP (Sigma Aldrich). Inhibition of CIP activity was obtained by adding 1× PhosSTOP (Roche Diagnostic) to the cell lysates. After treatment, samples were diluted in Laemmli buffer and analyzed by immunoblotting.
Immunoprecipitation
Immunoprecipitation was performed on HeLa cells or C3PV primary fibroblasts lysed in RIPA buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton, 0.25% Na-deoxycholate, 0.1% SDS) according to standard protocols. Primary antibodies utilized were anti-TFIIEα (2A1), anti-Flag (Sigma), and mouse IgG (Santa Cruz cat# sc-2025; RRID: AB_737182).
Protein Isolation, Immunoblotting, and Immunofluorescence
Whole-cell lysates were prepared for immunoblotting as described previously.22 TFIIH, TFIIE, TBP, TFIIB, and TTDN1 proteins were analyzed by lysing cell pellets of primary fibroblasts or lymphoblasts in urea buffer (62.5 mM Tris [pH 6.8], 4 M urea, 2% SDS, 10% glycerol, 0.005% bromophenol blue, 3% β-mercaptoethanol) as previously described.29, 30 TFIIEα phosphorylation in primary fibroblasts was investigated by direct lysis in culture dishes via scraping in 2× Laemmli buffer (125 mM Tris [pH 6.8], 4% SDS, 20% glycerol, 0.01% bromophenol blue, 6% β-mercaptoethanol) supplemented with PhosSTOP (Roche Diagnostic), and proteins were separated on 4%–20% gradient gels (Protean TGX, BioRad).31
For immunofluorescence analysis, cells were labeled with different sizes of polystyrene beads (normal cells 0.99 μm, affected subject cells 2.17 μm) as described before.32 Cells were grown on microscope cover glass (Thermo Scientific) and UV irradiated (254 nm, 100 J/m2) through a polycarbonate isopore membrane (pore size, 5 μm; diameter, 25 mm; Millipore) as described33 or left unirradiated. Immunofluorescence labeling and imaging was performed as described.22
The following commercial primary antibodies were used for immunoblotting at the indicated dilutions: mouse-anti-γ-tubulin (Sigma-Aldrich cat# T6557; RRID: AB_477584), 1:10,000; mouse-anti XPA (Santa Cruz cat# sc-73272; RRID: AB_1131404), 1:100; mouse-anti-GTF2E1 (Novus Biologicals cat# H00002960-B01; RRID: AB_2279378), 1:100; rabbit-anti-GTF2E2 (Novus Biologicals cat# NBP1-87931; RRID: AB_1102880), 1:150; rabbit-anti-XPB (Santa Cruz cat# sc-293; RRID: AB_2262177), 1:150; rabbit-anti-XPD (Santa Cruz cat# sc-20696; RRID: AB_2100152), 1:150; rabbit-anti XPC (Santa Cruz cat# sc-30156; RRID: AB_2241587), 1:200; rabbit anti-TTDN1 (Abcam cat# ab34309; RRID: AB_870614), 1:500; mouse-anti-Pol II-Ser5 (Covance, cat# MMS-134R; RRID: AB_10119940), 1:500; mouse anti-phosphoserine, anti-phosphothreonine, and anti-phosphotyrosine (525288, Detection kit, Calbiochem), 1:250. Mouse antibodies against the TFIIH subunits CDK7 (2F8), p44 (1H5), p62 (3C9), and XPD (2F6), the TFIIE subunits α (2A1) and β (1C2), TBP (3G3), TFIIB (4A10), and RNA pol II (7C2) were a gift from J.M. Egly and were all diluted 1:2,000.
Statistical Analysis
Statistical analysis was performed with a two-tailed Student’s t test. Fisher F-ratio at a probability level of 0.05 was used to compare variances among the analyzed groups. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
Results
Clinical Descriptions of Affected Children
TTD379BE
TTD379BE (IV-1 in Figure 1C), a 10-year-old Asian boy, was diagnosed with TTD at age 1 year (Table 1 and Figure S1). He was born in India at term with low birth weight (2.4 kg; 10th percentile). As with other individuals with TTD,13, 34, 35 the pregnancy was complicated by mild intrauterine growth retardation. His skin was normal with no collodion membrane and no sun sensitivity but he was noted to have very dry skin and sparse and slow-growing hair. His gross motor development was delayed. Esotropia was surgically corrected at age 5 years. He had cognitive delays noted at age 3 years. He is described as a very happy, overly friendly child.
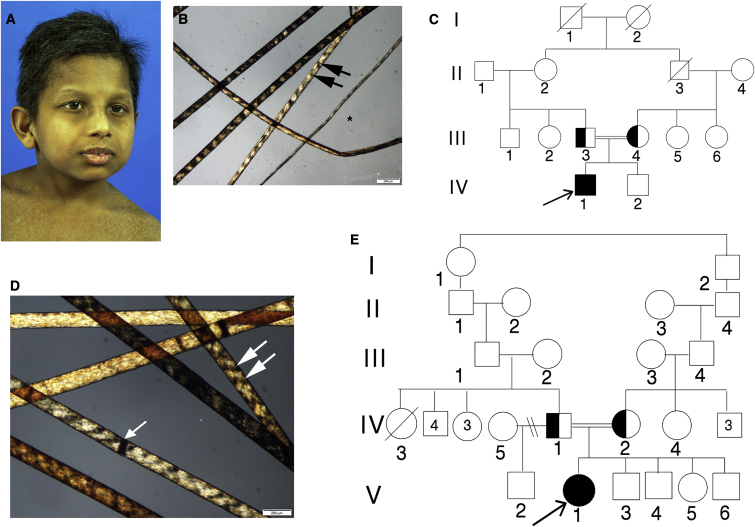
Figure 1.
TTD-Affected Children TTD379BE and TTD28PV
(A) TTD379BE, a 10-year-old boy from India with mild micrognathia, low-set ears, and short hair.
(B) Hair of affected child TTD379BE showing alternating dark and light “tiger tail” banding (arrows) with polarized microscopy typical of TTD.61 Hair shafts vary in diameter (∗).
(C) Pedigree of TTD379BE (IV-1). His clinically normal parents (III-3 and III-4) are first cousins.
(D) Hair of 16-year-old affected girl, TTD28PV, showing tiger tail banding with polarized microscopy (arrows). The hair shafts also have dark transverse bands that indicate sites of hair fractures (trichoschisis) (arrowhead).
(E) Pedigree of TTD28PV (V-1). Her parents (IV-1 and IV-2) are consanguineous.
Table 1.
Clinical Abnormalities in TTD- or XP-Affected Individuals with ERCC2 Mutations or TTD-Affected Individuals with GTF2E2 Mutations
|
TTD (ERCC2) Features |
TTD379BE (GTF2E2) (c.448G>C [p.Ala150Pro]) |
TTD28PV (GTF2E2) (c.559G>T [p.Asp187Tyr]) |
XP (ERCC2) Features |
Normal |
|
|---|---|---|---|---|---|
| repair + TRANSCRIPTION defecta | TRANSCRIPTION defect | TRANSCRIPTION defect | REPAIR + transcription defect | normal | |
| Clinical Features | |||||
| Dry skin | yes | yes | yes | yes | no |
| Ichthyosis | yes | yes | yes | no | no |
| Short, brittle hair | yes | yes | yes | no | no |
| Hair tiger-tail banding | yes | yes | yes | no | no |
| Short stature | yes | yes | yes | no | no |
| Microcephaly | yes | yes | yes | no | no |
| Developmental delay | yes | yes | yes | yes/no | no |
| Happy personality | yes | yes | yes | no | no |
| Acute burning on minimal sun exposure | yes | no | no | yes | no |
| Recurrent infections | yes | no | no | no | no |
| Increased freckle-like pigmentation | no | no | no | yes | no |
| Skin cancer |
no |
no |
no |
yes |
no |
|
Clinical Lab Studies | |||||
| Low RBC mean corpuscular volume | yes | yes | yes | no | no |
| Elevated Hemoglobin A2 | yes | yes | yes | no | no |
| Dilated brain ventricles (CT or MRI) | no | no | no | yes | no |
Terms in all capital letters indicate greater contribution to the phenotype.
At age 10 years, he appeared younger than his chronologic age (Figure 1A). His height, weight, and head circumference were <3rd percentile. His scalp hair was short, thick, and brittle. His hair showed typical alternating dark and light “tiger tail” banding on polarized microscopy (Figure 1B). He did not have the large number of freckle-like pigmented lesions in sun-exposed sites that are present in individuals with xeroderma pigmentosum (XP)5 (Table 1 and Figure S1). There was thick, coarse ichthyosiform scaling. There was no evidence of lenticular opacities. Neurological evaluation showed abnormal long tract signs and cerebellar dysfunction. His deep tendon reflexes were 1–2+. He had marked cognitive delays including a speech articulation disorder with features of attention deficit disorder. Formal IQ testing was not performed. He had slight sensorineural hearing loss bilaterally. His parents are first cousins (III-3 and III-4 in Figure 1C). Both parents and his brother (IV-2 in Figure 1C) are clinically normal.
In laboratory tests, the MCV was reduced to 64.5 (normal 74.4–86.1) and his hemoglobin electrophoresis showed elevated Hb A2 (Table 1 and Figure S1) as described in other individuals with TTD.36 He had normal bone age and no evidence of osteosclerosis, osteopenia, or hip abnormality as seen in some individuals with TTD.13, 34 However, he had probable craniosynostosis of coronal sutures. CT exam showed a morphologically normal brain with no gross atrophy or calcifications. There was decreased attenuation throughout the white matter, possibly representing a leukoencephalopathy.
TTD28PV
TTD28PV (V-1 in Figure 1E), a 16-year-old Moroccan girl, weighed 3,110 g at 38.5 weeks gestation (Table 1 and Figure S1) with an uncomplicated pregnancy. There was no notion of a collodion membrane and she was not sun sensitive. She had a patent ductus arteriosus at birth that was closed by catheterization. She had brittle hair with tiger-tail banding under polarized microscopy (Figure 1D) and lamellar ichthyosis of her skin. She could sit at 1 year, walk independently at 35 months, and spoke 1 word at 33 months. She suffered from chronic rhinosinusitis but did not need hospitalization for infections or other problems. She wears glasses (−0.5 D). She is very friendly, always laughing with good social interaction.
At 16 years her height and weight were <<3rd percentile. She had bilateral pes cavus with Babinski in extension. She had an IQ of 40 and was in special education classes for children with moderate intellectual disability. She had growth hormone deficiency and has been treated with growth hormone with good response. Hb A2 and Hb F were elevated and MCV was low, indicating microcytosis as seen in other TTD-affected individuals (Figure S1).13, 34 She has mild hearing loss. MRI of the brain at 2 years and at 10 years was normal. X-ray of the pelvis at 21 months showed bilateral coxa valga and delayed bone age (10 years at 13 years of age).
The parents (IV-1 and IV-2 in Figure 1E) are from Morocco and are distantly related although their precise ancestry is not known. They have five other children (V-2 to V-6 in Figure 1E) who are phenotypically normal (Figure 1E). For further details, see Supplemental Note.
Normal UV Damage Repair Ability in TTD379BE and TTD28PV Cells
TTD cells with mutations in ERCC3 (XPB), ERCC2 (XPD), or GTF2H5 (TTDA [OMIM: 608780]) are defective in the removal of UV-induced DNA damage by NER.6, 29, 37, 38 Unlike the other TTD cells (Table 2 and Figure S1), cells from both TTD379BE and TTD28PV have normal post-UV DNA repair as shown by their normal sensitivity to UV irradiation (Figure 2A), host cell reactivation (repair of a UV-irradiated luciferase vector) that was not lower than normal (Figures 2B and S2), and normal removal of 6-4 photoproducts (Figure 2C) and cyclobutane pyrimidine dimers (Figure 2D). We also found normal recruitment of NER proteins (XPC, XPA, XPB, and XPD) to UV-induced localized DNA damage (Figure 2E). In addition, in primary fibroblasts from TTD28PV, we analyzed the efficiency of NER by evaluating unscheduled DNA synthesis (UDS) and recovery of RNA synthesis (RRS) after UV irradiation (Figure S2). After exposure to 10 and 20 J/m2 UV radiation, the level of UDS corresponded to 95% of that in normal cells. As with the normal donor, 24 hr after irradiation with 20 J/m2, the case subject’s cells recovered RNA synthesis to the level in un-irradiated cells. Therefore, both clinical and cellular studies lead to the classification of both TTD28PV and TTD379BE children in the non-photosensitive group of TTD. However, no alterations were found in MPLKIP (TTDN1 [OMIM: 609188]) that accounts for about 10%–20% of the non-photosensitive TTD-affected case subjects.
Table 2.
Laboratory Abnormalities in TTD- or XP-Affected Individuals with ERCC2 Mutations or TTD-Affected Individuals with GTF2E2 Mutations
|
TTD (ERCC2) Features |
TTD379BE (GTF2E2) (c.448G>C [p.Ala150Pro]) |
TTD28PV (GTF2E2) (c.559G>T [p.Asp187Tyr]) |
XP (ERCC2) Features |
Normal |
|
|---|---|---|---|---|---|
| repair + TRANSCRIPTION defecta | TRANSCRIPTION defect | TRANSCRIPTION defect | REPAIR + transcription defect | normal | |
| DNA Repair | |||||
| Reduced post-UV cell survival and host cell reactivation | yes | no | no | yes | no |
| Reduced repair of cyclobutane pyrimidine dimer photoproducts | yes | no | no | yes | no |
| Reduced XPB and XPD proteins |
yes |
no |
no |
yes |
no |
| Transcription | |||||
| Reduced TFIIEβ protein | no | yes | yes | no | no |
| Reduced TFIIEα protein | no | yes | yes | no | no |
| Reduced phosphorylated TFIIEα protein in confluent cells | yes | yes | yes | no | no |
Terms in all capital letters indicate greater contribution to the phenotype.
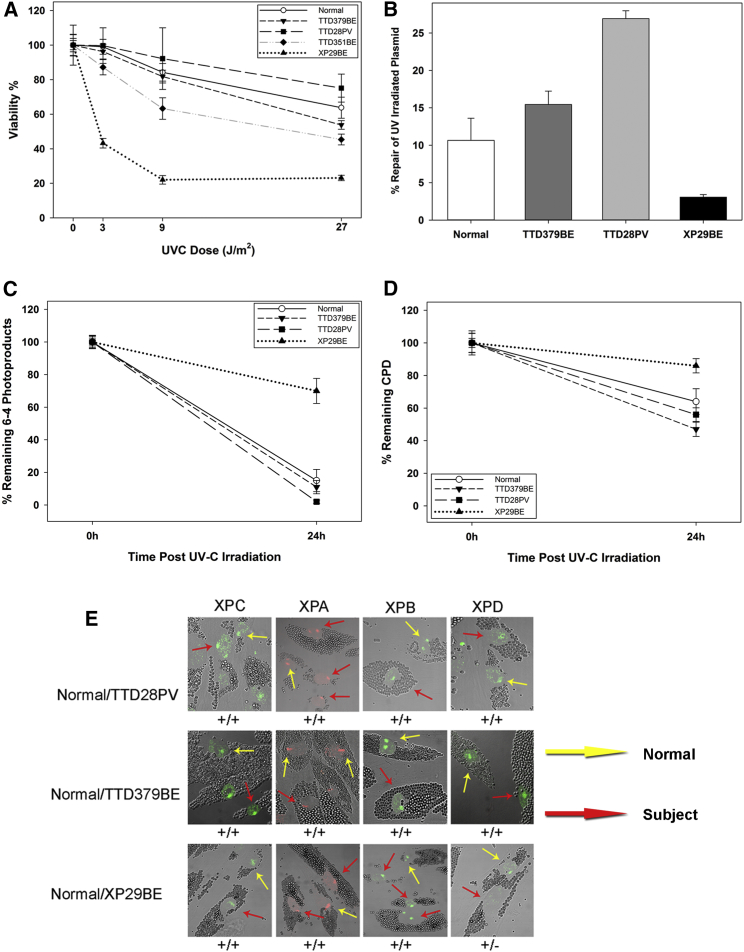
Figure 2.
Normal DNA Repair in Primary Fibroblasts from TTD379BE and TTD28PV
(A) Post-UV cell survival in normal, TTD379BE, TTD28PV, XP29BE (XP/XP-D), and TTD351BE (TTD/XP-D) cells. Bars indicate SEM.
(B) Post-UV host cell reactivation in normal, TTD379BE, TTD28PV, and XP29BE (XP/XP-D) fibroblasts. Cells were transfected with an UV-C-irradiated (1,000 J/m2) luciferase reporter vector and incubated for 48 hr. Repair of the plasmid is expressed as induced light units of active luciferase compared to a non-irradiated luciferase plasmid. Two experiments each in triplicate were performed. Bars indicate SEM.
(C and D) Repair of 6-4 photoproducts (6-4PP) (C) and cyclobutane pyrimidine dimers (CPD) (D) measured by immunofluorescence in normal, TTD379BE, TTD28PV, and XP29BE (XP/XP-D) fibroblasts. Cells were irradiated with 100 J/m2 UVC through a filter with 5 μm pores to generate localized DNA damage. 100 nuclei were scored. Bars indicate SEM.
(E) Immunofluorescence analysis of NER proteins in normal (incubated with 1 μm beads, yellow arrows) and TTD379BE or TTD28PV (incubated with 2 μm beads, red arrows) cells loaded on the same coverslip and irradiated with 100 J/m2 UVC through a filter with 5 μm pores to generate localized DNA damage. Post-UV localization of XPD protein to the damaged sites is not detected (−) in XP29BE (XP/XP-D) cells but is present (+) at normal levels in TTD28PV and TTD379BE cells.
Reduced Levels of TFIIE Complex in TTD28PV and TTD379BE Cells
The link between TTD and transcription alterations prompted us to investigate the cellular levels of several RNA pol II-basal transcription factors. Normal amounts of TBP, TFIIB, TFIIH complex, as well as TTDN1 were found in TTD379BE and TTD28PV cells (Figure 3A). In contrast, immunoblotting studies revealed drastic reductions in the levels of both α and β subunits of TFIIE complex in primary fibroblasts from TTD28PV and TTD379BE (Figure 3A) and in lymphoblasts of TTD28PV (Figure 3B) compared to TTD28PV’s healthy relatives and genetically unrelated normal donors. Immunofluorescence staining of TTD379BE’s cells confirmed reduced level of TFIIEβ protein compared to normal control cells (Figure 3C). In addition, the reduction of both TFIIEα and TFIIEβ did not affect the nuclear distribution after localized UV irradiation, providing additional evidence that TFIIE is not involved in NER (Figure 3D). Overall, these findings point to the involvement of TFIIE in the pathological phenotype of affected children TTD28PV and TTD379BE.
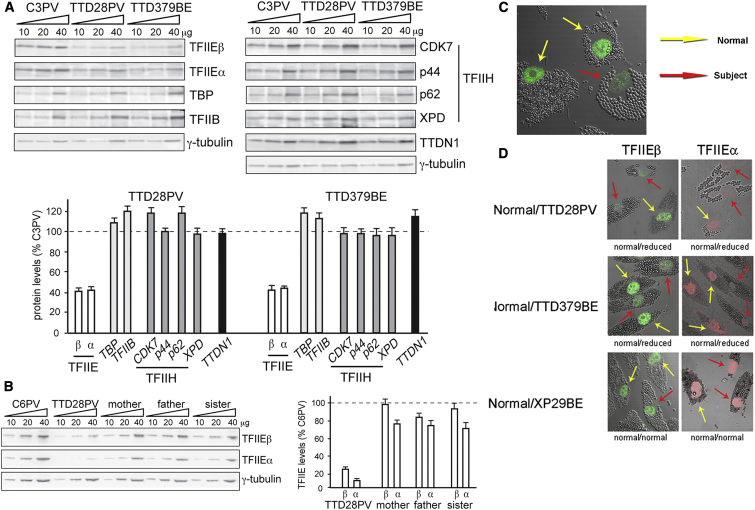
Figure 3.
Reduced Steady-State Levels of TFIIE and Normal Levels of Other General Transcription Factors in Primary Fibroblasts from Affected Individuals TTD379BE and TTD28PV
(A) Immunoblot analysis of whole-cell lysates with antibodies against the α and β subunits of TFIIE, TBP, TFIIB, the CDK7, p44, p62, and XPD subunits of TFIIH, and TTDN1. γ-tubulin was used as loading control. The amount of each protein was first expressed as the mean value of the levels observed in the three increasing concentrations of the cell lysate and normalized to the γ-tubulin content. The protein levels in both TTD cell strains were then expressed as percentages of the corresponding values in the normal (C3PV) cells. The reported values are the means of at least two independent experiments. Bars indicate the SE.
(B) Anti-TFIIEα and anti-TFIIEβ immunoblot analysis of whole-cell lysates from lymphoblastoid cells of TTD28PV and her unaffected mother, father, and sister. The amount of the analyzed proteins was determined as described in (A); the levels of TFIIEα and TFIIEβ are expressed as percentages of the corresponding values in the normal C6PV lymphoblasts. The mean levels of two independent experiments are reported. Bars indicate the SE.
(C) Reduced immunofluorescence of TFIIEβ protein in TTD379BE cells (incubated with 2 μm beads, red arrow) compared to normal cells (incubated with 1 μm beads, yellow arrows).
(D) Immunofluorescence detection of reduced levels of TFIIEβ and TFIIEα in TTD28PV and TTD379BE cells. TTD and normal cells were labeled as in Figure 2C and irradiated with 100 J/m2 UVC through a filter with 5 μm pores to generate localized DNA damage. The TFIIE proteins were reduced in the TTD cells. There was no localization of TFIIE proteins at the site of localized DNA damage in the TTD or normal cells.
Mutations in GTF2E2 Are Associated with a Non-photosensitive Form of TTD
In TTD379BE, whole-exome sequencing detected a homozygous, non-synonymous missense mutation in GTF2E2 (c.448G>C [p.Ala150Pro]; GenBank: NM_002095.4), which encodes the 34 kDa TFIIEβ protein. Sanger sequencing (Figure 4A) and restriction fragment length polymorphism testing (Figure S3) confirmed the mutation and identified heterozygous mutations in both parents but not in the unaffected brother. In TTD28PV, targeted Sanger sequencing of PCR-amplified cDNA of both TFIIE subunits was performed. No mutations were detected in GTF2E1 whereas GTF2E2 analysis revealed the presence of a homozygous, non-synonymous missense mutation (c.559G>T [p.Asp187Tyr]) (Figure 4B). Sequencing of the relevant genomic DNA region demonstrated that TTD28PV’s parents were heterozygous (Figure 4B). The healthy siblings were either heterozygous (sister) or did not carry the variant (brother) (Figure 4B). The c.448G>C and c.559G>T changes were not reported in dbSNP, and the affected amino acids were conserved across species (Figure 4C). The two variants obtained high pathogenicity scores in three independent prediction algorithms—PolyPhen2,28 SIFT,27 and MutationTaster39 (Table S3)—strongly suggesting a disease-causing effect. The altered amino acids are 14 Å apart on different surfaces of the wing helix 2 (WH2) region of the TFIIEβ protein (Figure 4D). The p.Ala150Pro change would be expected to destabilize the long alpha helix and cause it to bend or locally unfold (W. Yang, personal communication).
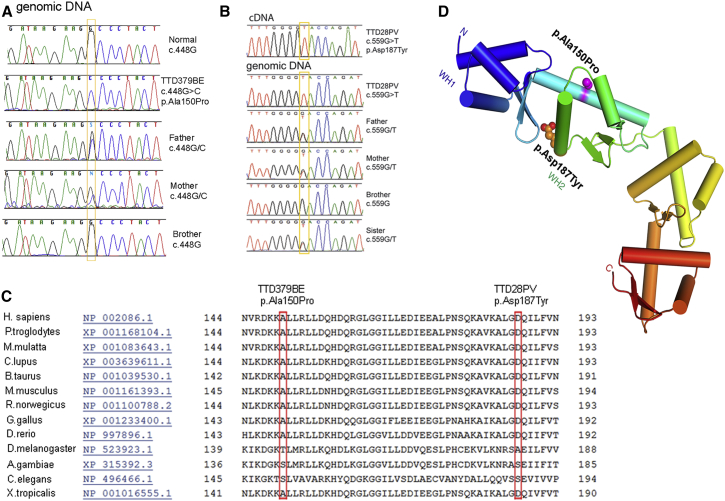
Figure 4.
Identification of Missense Mutations in GTF2E2 in Affected Children TTD379BE and TTD28PV
(A) Electropherograms showing the change identified in GTF2E2 in TTD379BE and his unaffected father and mother but not in the unaffected brother.
(B) Electropherograms showing the change identified in GTF2E2 (GenBank: NM_002095.4, NC_000008.10) in TTD28PV and unaffected father, mother, and sister but not in the unaffected brother. For cDNA numbering, +1 corresponds to the A of the ATG translation initiation codon in the reference sequence.
(C) Conservation of amino acids Ala150 and Asp187 in multiple protein sequence alignment of TFIIEβ regions in 13 species (from HomoloGene: 37573).
(D) Structural location of amino acid changes in TFIIEβ protein resulting from GTF2E2 homozygous missense mutations in TTD-affected individuals. Each winged-helix (WH) domain consists of three helices (cylinders) and a beta-hairpin (two strands). The protein is colored from N to C terminus from blue (WH1 domain), blue-green (WH2 domain), to red (C terminus). The altered amino acids (p.Ala150Pro and p.Asp187Tyr) are 14 Å apart on different surfaces of wing helix 2 (WH2) region of the TFIIEβ protein. The p.Ala150Pro change will destabilize the long alpha helix and cause it to bend or locally unfold (W. Yang, personal communication).
After identification of the mutations in GTF2E2 in the affected children TTD379BE and TTD28PV, we performed Sanger sequencing of GTF2E2 and GTF2E1 in 41 additional individuals with clinically suspected TTD where NER defects or TTDN1 mutations had not been identified (15 individuals from 13 families tested in Italy, 6 individuals from 4 families tested at the NIH, and 20 individuals tested in the UK). No mutations were identified in either GTF2E2 or GTF2E1, suggesting that additional genes are involved in TTD. In our combined cohort of 125 TTD-affected case subjects (48 from Italy, 36 from NIH, and 41 from UK), mutations in GTF2E2 accounted for about 2% of the cases.
GTF2E2 Mutations Affect the Stability of TFIIE Complex
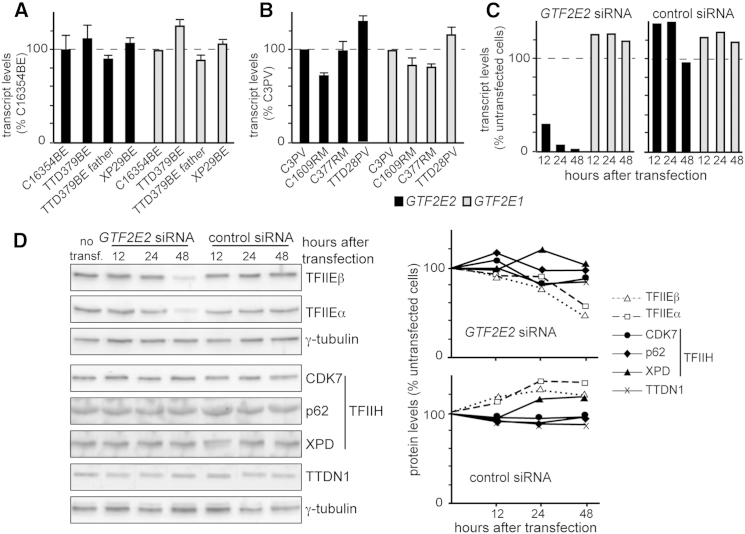
In TTD28PV and TTD379BE, the transcript levels of GTF2E2 and GTF2E1 were in the normal range (Figures 5A and 5B). This result indicates that the mutations in GTF2E2 did not affect the transcription of the gene but impacted TFIIEβ stability because the level of TFIIEβ protein was reduced (Figure 3). This, in turn, affects the stability of the entire TFIIE complex as shown by the equally reduced level of both TFIIE subunits in TTD28PV and TTD379BE cells (Figure 3A).
Figure 5.
GTF2E2 Mutations Do Not Affect the Cellular Amount of GTF2E2 and GTF2E1 Transcripts but Interfere with the Stability of TFIIE Complex
(A) GTF2E2 (black columns) and GTF2E1 (gray columns) transcript levels in primary fibroblasts from the healthy donor C16354BE, affected individual TTD379BE, his father, and affected individual XP29BE (XP/XP-D). Transcript levels were normalized to GAPDH levels and then expressed as percentages of the corresponding value in the healthy donor analyzed in parallel. The reported values are the means of two independent experiments, each done in triplicate. Bars indicate the SE.
(B) GTF2E2 (black columns) and GTF2E1 (gray columns) transcript levels in primary fibroblasts from healthy donors C3PV, C1609RM, and C377RM and affected individual TTD28PV. Transcript levels were expressed as in (A). The reported values are the means of two independent experiments, each done in triplicate. Bars indicate the SE.
(C) GTF2E2 and GTF2E1 transcript levels in C3PV primary fibroblasts 12, 24, and 48 hr after transfection with GTF2E2 or control siRNA. GTF2E2 (black columns) and GTF2E1 (gray columns) transcript levels were normalized to GAPDH and expressed as percentages of the corresponding value in untransfected C3PV cells.
(D) Immunoblot analysis of whole-cell lysates from C3PV primary fibroblasts 12, 24, and 48 hr after transfection with GTF2E2 or control siRNA. Antibodies against the α and β subunits of TFIIE, the CDK7, p62, and XPD subunits of TFIIH, TTDN1, and γ-tubulin (loading control) were used. The levels of the analyzed proteins were normalized to γ-tubulin and expressed as percentages of the corresponding values in untransfected C3PV cells.
The quantitative relationship between the α and β subunits of TFIIE was investigated by silencing of GTF2E2 (via RNAi) in normal skin fibroblasts. We observed that GTF2E2 silencing resulted in reduced mRNA and protein levels of TFIIEβ (Figures 5C and 5D). The reduced TFIIEβ amount did not affect the mRNA level of GTF2E1 but instead caused a striking reduction (about 50%) of TFIIEα protein, suggesting that a stable complex between both TFIIE subunits was required to maintain normal levels of TFIIEα and TFIIEβ. No reduction was observed in the cellular concentration of TFIIH subunits or of TTDN1 with GTF2E2 RNAi confirming that quantitative alterations of TFIIEβ specifically interferes with the stability of the whole TFIIE complex without affecting the cellular concentration of the other factors involved in TTD pathogenesis.
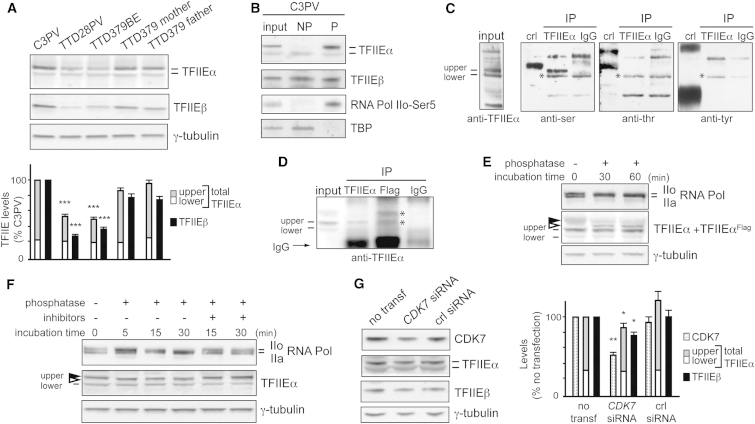
GTF2E2 Mutations Interfere with the Phosphorylation of TFIIEα
Two distinct bands can be resolved for TFIIEα by immunoblotting of primary fibroblast lysates prepared by directly scraping cells in Laemmli buffer (Figure 6A). In normal fibroblasts, the upper band of the doublet represents 75% (SE ±1.7) of the total TFIIEα amount. In GTF2E2 mutant cells from affected children TTD28PV and TTD379BE, no significant variations were observed in the level of the lower band whereas the upper band was drastically reduced (29% compared to normal, p < 0.0005), thus accounting for the reduced amount of total TFIIEα (50% compared to normal, p < 0.0005).
Figure 6.
Reduced Amount of Phosphorylated TFIIEα in Primary Fibroblasts of Affected Individuals TTD379BE and TTD28PV and in Normal C3PV Fibroblasts after CDK7 Silencing
(A) Immunoblot analysis of lysates obtained by directly scraping normal (C3PV and TTD379BE parents) and TTD (TTD28PV and TTD379BE) cells in Laemmli buffer. The levels of the upper (gray) and lower (white) forms of TFIIEα and of TFIIEβ (black) were normalized to γ-tubulin and expressed as percentage of the corresponding values in C3PV cells. The reported values are the means of at least two independent experiments. Bars indicate the SE. The statistically significant reduction of TFIIEα (∗∗∗p < 0.0005; Student’s t test) refers to the upper band, no significant difference was observed for the lower band.
(B) Immunoblot analysis of TFIIEα and TFIIEβ in unphosphorylated (NP) and phosphorylated (P) protein-enriched fractions of cell lysates from normal C3PV fibroblasts. TBP and RNA pol IIo-Ser5 were used as positive controls of the NP and P fractions, respectively.
(C) Immunoprecipitation of TFIIEα or IgG from HeLa total cell lysates and identification of the phosphorylated residue. The TFIIEα or IgG immunoprecipitates (IP) were analyzed by immunoblotting using phosphoaminoacid-specific antibodies. The positive controls (crl) are made of proteins phosphorylated on serine (ser), threonine (thr), or tyrosine (tyr), provided by the kit. Asterisks (∗) indicate non-specific bands.
(D) Immunoprecipitation of TFIIEα, TFIIEαFlag, or IgG from cell lysates of C3PV normal fibroblasts transfected with the pMP2-TFIIEαFlag DNA plasmid expressing the TFIIEαFlag recombinant protein. The immunoprecipitates (IP) were analyzed by immunoblotting using anti-TFIIEα antibodies. Asterisks (∗) indicate the upper and lower bands of TFIIEαFlag.
(E) Immunoblot analysis of TFIIEα after phosphatase treatment of the cell lysate from C3PV fibroblasts transfected with the pMP2-TFIIEαFlag DNA plasmid. The lysate was incubated at 37°C with 3 U/μL of calf intestinal phosphatase for the indicated time points. Arrowheads indicate the phosphorylated (black) and dephosphorylated (white) upper form of TFIIEαFlag. As positive control, the phosphorylation status of RNA Pol IIo over the unphosphorylated RNA pol IIa was investigated in parallel. γ-tubulin is the loading control.
(F) Immunoblot analysis of TFIIEα in the cell lysate from normal C3PV fibroblasts after phosphatase treatment. The lysate was incubated at 37°C with 3 U/μL of calf intestinal phosphatase in the presence or absence of phosphatase inhibitors (PhosSTOP 1×) for the indicated time points. Arrowheads indicate the phosphorylated (black) and dephosphorylated (white) upper form of TFIIEα. As positive control, the phosphorylation status of RNA Pol IIo over the unphosphorylated RNA pol IIa was investigated in parallel. γ-tubulin is the loading control.
(G) Immunoblot analysis of whole-cell lysates from C3PV fibroblasts 72 hr after transfection with CDK7 siRNA or control siRNA. The levels of CDK7 and the upper and lower forms of TFIIEα and TFIIEβ were normalized to the γ-tubulin content and expressed as percentage of the corresponding value in untransfected C3PV cells. The reported values are the mean of two independent experiments. Bars indicate the SE. The statistically significant reduction of TFIIEα (∗p < 0.05; ∗∗p < 0.005; Student’s t test) refers to the upper band; no significant difference is observed for the lower band.
Because it was previously demonstrated by in vitro kinase assays that TFIIEα can be phosphorylated by TFIIH kinase activity,20 we investigated whether the different electrophoretic mobility of the two TFIIEα bands is the result of different phosphorylation states. By a phosphoprotein-enrichment assay, we observed that the upper band of TFIIEα was exclusively present in the phospho-protein-enriched fraction of normal C3PV fibroblasts whereas the lower band was more abundant in the unphosphorylated fraction (Figure 6B). We next immunoprecipitated the TFIIEα protein from a total cell lysate and immunoblotted with antibodies specific for phosphorylated serine, threonine, or tyrosine residues. As shown in Figure 6C, the upper band was recognized only by the anti-serine antibody, thus demonstrating that TFIIEα is serine phosphorylated. Unfortunately, we were not able to assess the phosphorylation status of the lower band in these experiments, because it ran with the same mobility as a non-specific band (indicated by asterisk in Figure 6C).
To better clarify the relationship between the upper and lower bands of TFIIEα, we transiently transfected C3PV fibroblasts with a DNA plasmid encoding the recombinant TFIIEα protein tagged with Flag epitope (TFIIEαFlag). In the whole-cell extract of transfected cells (Figure 6D, input), the TFIIEα antibody recognized three bands: two corresponding to the endogenous lower and upper band, respectively, and the third one with a molecular weight (MW) higher than that of the endogenous upper band. After immunoprecipitation with anti-TFIIEα antibodies, a clear signal was detected at the MW corresponding to the endogenous TFIIEα upper form. Immunoprecipitation with anti-Flag antibodies followed by anti-TFIIEα immunoblot revealed two protein bands, one corresponding to the form with the highest MW and one with the same size as the endogenous TFIIEα upper form. Thus, the Flag epitope imposed an electrophoretic up-shift to both the upper and lower bands, the latter with an electrophoretic mobility similar to the endogenous TFIIEα upper form.
Next, we incubated the whole extract of TFIIEαFlag-expressing fibroblasts with calf intestinal alkaline phosphatase for 30 and 60 min and the efficiency of the phosphatase treatment was demonstrated by the shift from the IIo to the IIa form of RNA pol (Figure 6E). A reduction in the amount of the TFIIEαFlag upper form was paralleled by the appearance of an extra band with a slightly lower MW (black and white arrowheads, respectively), confirming the phosphorylation of the upper band. However, the mobility of the dephosphorylated form of the upper band of the FLAG-tagged protein was still slower than that of the lower band of FLAG-tagged protein seen in Figure 6D, suggesting that the dephosphorylated TFIIEαFlag form has an additional post-translational modification.
To verify whether this holds true also for the endogenous TFIIEα, we incubated the whole lysate of normal C3PV fibroblasts with calf intestinal alkaline phosphatase for increasing time periods in the absence or presence of phosphatase inhibitors. Already at the shortest incubation time (5 min), the size of the upper band was slightly reduced (shift from the black to the white arrowheads) and persisted over time (Figures 6F and S4) but did not approach that of the lower band. The presence of the phosphatase inhibitors prevented the TFIIEα dephosphorylation, as demonstrated by the lack of any mobility downshift of the TFIIEα upper form.
To evaluate the contribution of the CDK7 serine/threonine kinase to the in vivo phosphorylation of TFIIEα, the CDK7 expression in normal C3PV fibroblasts was investigated via CDK7 RNAi. A substantial decrease of the CDK7 mRNA level (to 6%–8% of normal) as well as a 50%–60% reduction of the CDK7 protein amount was observed after silencing. This was followed by a decrease in the amount of the p44, p62, and XPD subunits of TFIIH complex but not of the transcription factor TBP (Figure S5). This result demonstrates that reduction in CDK7 can influence the cellular concentrations of other proteins in the TFIIH complex. In addition, CDK7 siRNA altered the phosphorylation status of TFIIEα as shown by the small but reproducible and statistically significant decrease (p < 0.05) in the amount of the phosphorylated TFIIEα (upper band) 72–96 hr after siRNA transfection (Figures 6G and S5). Because we were unable to reduce the CDK7 protein concentration to less than 50%, we cannot make a definitive conclusion, but our findings are consistent with the in vitro data demonstrating that TFIIEα is a substrate of the CDK7 kinase activity of TFIIH. Moreover, a statistically significant reduction (p < 0.05) in the amount of the TFIIEβ subunit was also observed in CDK7-silenced fibroblasts, suggesting a possible interaction of TFIIEβ with the phosphorylated form of TFIIEα.
Alterations in the Phosphorylation Status of TFIIEα Are Common in TTD Cells
Cells from photosensitive individuals with TTD typically have a reduced content of the entire TFIIH complex resulting from mutations in genes encoding the XPD, XPB, or TTDA protein subunits of TFIIH (Table S1).6, 29, 30, 40
Because the kinase activity of TFIIH phosphorylates TFIIEα, we asked whether the TFIIH alterations found in the photosensitive TTD-affected case subjects affect TFIIE. We performed immunoblot analysis of fibroblast lysates from six TTD-affected individuals with alterations in the XPD subunit of TFIIH, the most common form of TTD. We found normal cellular amounts of total TFIIEα (100%–117% of that in C3PV) in non-proliferating and proliferating cells (Figures 7 and S6) in agreement with our previous observations.29, 30 Similar to the healthy donors, the amount of the upper form of TFIIEα ranged from 70% to 80% of the total TFIIEα in proliferating (sub-confluent) TTD fibroblasts (Figure S6). In contrast, in non-proliferating (confluent) TTD cells, the level of the upper band was reduced to 40%–60% of the total TFIIEα with a parallel increase in the level of the lower band (Figure 7). In addition, a slight but significant decrease (p < 0.0005) in the cellular content of TFIIEβ subunit was observed, thus resembling the situation previously observed in CDK7-silenced cells (Figure 6G). In contrast, normal levels of the upper and lower bands of TFIIEα were observed in cells from six XP-affected individuals with mutations in ERCC2 (XPD) in proliferating and non-proliferating cells (Figures 7 and S6). Overall, these findings demonstrate that in non-proliferating, confluent cells, XPD alterations associated with the photosensitive TTD phenotype, but not those resulting in XP, impair the phosphorylation of TFIIEα. Because these differences in TFIIEα phosphorylation are present in cells from individuals with TTD with mutations in ERCC2 (Figure 7) and in GTF2E2 (Figure 6A), alterations in the phosphorylation status of TFIIEα appear to be a specific feature of TTD cells.
Figure 7.
TFIIEα Phosphorylation Is Impaired in Non-proliferating TTD/XP-D Primary Fibroblasts
Immunoblot analysis of lysates obtained by directly scraping four normal (C3PV, C1609RM, C377RM, and C16354BE), six TTD/XP-D (TTD8PV, TTD11PV, TTD12PV, TTD22PV, TTD23PV, and TTD24PV), and six XP/XP-D (XP16PV, XP29BE, XP17BE, XP34BE, XP35BE, and XP17PV) fibroblast strains in Laemmli buffer. The levels of the upper (gray) and lower (white) forms of TFIIEα and of TFIIEβ (black) were normalized to γ-tubulin and expressed as percentage of the corresponding values in C3PV cells. The reported values are the means of at least two independent experiments. Bars indicate the SE (∗∗∗p < 0.0005; ns, not statistically significant; Student’s t test).
Discussion
Several general transcription factors assemble into a pre-initiation complex (PIC) to ensure accurate RNA pol II loading at the transcription start site. Among them, the basal transcription factor IIE (TFIIE) is essential for PIC assembly and stabilization.17 Direct interaction of TFIIE (mainly through its TFIIEα subunit) to TFIIH is required for both PIC formation and the transition from initiation to elongation.41 TFIIEα interacts with XPB, p52, and p62 of TFIIH as well as with other transcription factors TFIIB, TFIIFβ, and TBP.42, 43 On a supercoiled DNA template, TFIIE can melt the promoter independently of TFIIH, but on linearized template both TFIIE and TFIIH are necessary for the transition activity from the initiation to elongation.44
Both GTF2E2 homozygous mutations present in the children with TTD reported in this paper alter the WH2 domain (residues 142–207) of the TFIIE beta subunit (TFIIEβ) (Figure 4D). p.Ala150Pro lies within a leucine repeat motif and p.Asp187Tyr within a σ3 region that is similar to the bacterial σ factor subdomain 3.45 TFIIEβ binds to the WH domain of TFIIEα via its bHLH domain (basic region-helix-loop-helix motif, residues 193–240). The bHLH domain of TFIIEβ also binds to RNA pol II as well as to ssDNA, TFIIF, and TFIIB.46 TFIIEβ also binds XPB.47 The p.Ala150Pro change will destabilize the long alpha helix and cause it to bend or locally unfold (W. Yang, personal communication).
In confluent fibroblast cultures from all individuals with TTD studied here (but not from the individuals with XP with mutations in ERCC2), we observed reduced amount of the upper form of TFIIEα (Table 2 and Figures 6A, 7, and S1). This is in line with the observation that, being compatible with life, the transcriptional failure in TTD occurs only under certain circumstances and/or in specific cellular compartments.36, 48, 49, 50, 51, 52 In particular, high cell density is known to regulate the expression of specific target genes by triggering a cascade of phosphorylation-mediated pathways that ultimately result in more efficient transcription of specific genes (Orioli et al.9 and references therein).
Intriguingly, the reduced level of serine-phosphorylated TFIIEα is seen with GTF2E2 mutations leading to reduced TFIIE protein levels, as well as with ERCC2 mutations with normal TFIIE protein levels (Figures 6 and 7). In vitro studies have shown that the serine/threonine kinase CDK7, which is part of the CAK complex of TFIIH, can phosphorylate TFIIEα20 as well as TFIIEβ.53 In addition, TFIIE itself influences TFIIH activity by positively regulating CDK7 and XPD subunits.54 Overall, our findings support the causal link between CDK7 and TFIIEα phosphorylation (Figure 6G) and, therefore, the in vivo relevance of the crosstalk between TFIIE and TFIIH in transcription. In the two TTD-affected children with mutations in GTF2E2, several different scenarios could explain the reduced TFIIEα phosphorylation. The GTF2E2 mutations could affect the stability of the TFIIE complex, leading to degradation of both TFIIEα and TFIIEβ, which results in less TFIIEα substrate available for CDK7 phosphorylation and in turn to much less phosphorylated TFIIEα compared to normal cells. Additionally, reduced levels of total TFIIE could affect CDK7 function resulting in reduced TFIIEα phosphorylation. In the TTD-affected individuals with mutations in ERCC2, decreased TFIIEα phosphorylation could be a consequence of altered TFIIH integrity. As revealed by solution of the crystal structure of archaeal XPD,55, 56 the mutations found in TTD are predicted to decrease the stability of the XPD protein framework, according with the reduced level of the TFIIH content typically present in TTD cells.57 Furthermore, all the ERCC2 mutations found in individuals with TTD diminish the basal transcription activity of TFIIH.8
Our results suggest that TFIIEβ pathological changes could lead to altered RNA pol II-driven transcription via deregulated phosphorylation events, which might include TFIIEα phosphorylation seen in TTD-affected individuals with ERCC2 (XPD) mutations (Tables 2 and S1 and Figure S1). Reduced TFIIEα phosphorylation (Figures 6A and 7) could impair TFIIE function, which would affect the correct positioning of TFIIF, TFIIH, and RNA pol II at the PIC. Moreover, CDK7 controls the disengagement of TFIIE and recruits DRB sensitivity inducing factor (DSIF), thus allowing the pausing of RNA pol II to ensure gene-specific regulation and the recruitment of RNA-processing enzymes before elongation.53 Our data suggest that reduced phosphorylation of TFIIEα by CDK7 might affect TFIIE function on transcription initiation and elongation.
Individuals with (different) alterations in the XPD subunit of TFIIH might have the clinical phenotype of XP with markedly increased cancer risk on sunlight-exposed tissues or TTD with normal cancer risk and developmental abnormalities (Table 1 and Figure S1). Cultured cells from these individuals with XP or with TTD have impaired post-UV DNA repair and additionally have defective transcription.7, 8, 58 One explanation for the difference in phenotype is that the XP phenotype results from a predominance of the DNA repair defect whereas TTD is a consequence of a primary transcription abnormality. Studies in cells from TTD-affected individuals and mouse models support the relevance of transcriptional alterations to the TTD clinical outcome (reviewed in Stefanini et al.6). Besides defects in the basal transcription activity of TFIIH,8 several lines of evidence have highlighted gene expression deregulations in TTD3, 38, 49, 52, 59, 60, 61, 62 that result from altered signaling events, including the TFIIH-dependent activation of nuclear receptors as well as the TFIIH-mediated activation or displacement from the chromatin of specific transcription regulators.8, 9, 10, 11, 37, 51 The mutations leading to the TTD phenotype appear to alter the overall TFIIH structure, thus impairing TFIIH-dependent transcription.
The individuals with TTD we report have mutations in the basal transcription factor GTF2E2 with normal DNA repair (Table 2 and Figure S1). We also found decreased phosphorylation of TFIIEα in TTD cells with mutations in GTF2E2 or ERCC2 (which encodes the TFIIH subunit XPD) but not in XP cells with ERCC2 mutations. Collectively, our study underlines the role of TFIIE in transcription and its distinction from NER. The individuals with TTD with mutations in GTF2E2 highlight the importance of the direct interaction between TFIIH and TFIIE in the transcription process leading to a TTD clinical phenotype. Our results thus provide a link between the photosensitive and non-photosensitive forms of TTD and support the notion that the TTD clinical outcome is due to transcriptional defects, which in turn represent the major determinant distinguishing TTD from XP.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (C.K., K.H.K., J.J.D., S.G.K., D.T., D.L., L.B., R.S.), the Associazione Italiana per la Ricerca sul Cancro Grant IG 13537 (M.S.), and IG 17710 (D.O.) and Telethon Grant GEP13022 (E.B.). We thank Dr. Wei Yang (NIDDK, NIH) for the schematic image of TFIIE mutations, Russell Bandle (NCI) for helpful suggestions in the procedure of nuclear protein isolation, and Heather Fawcett (Sussex University) for technical assistance. We would also like to thank the Genomics Laboratory and CCR Sequencing Facility at Frederick National Laboratory for Cancer Research including Dan Soppett and Kristen Pike for DNA target preparation, Bao Tran for performing the sequencing, Ming Yi for help perfecting the GATK workflow, and Shashi Ratanayke and Keyur Talsania for assistance running the analysis.
Published: March 17, 2016
Footnotes
Supplemental Data include Supplemental Note of two case reports, six figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.008.
Contributor Information
Kenneth H. Kraemer, Email: kraemerk@nih.gov.
Miria Stefanini, Email: stefanini@igm.cnr.it.
Web Resources
The URLs for data presented herein are as follows:
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Human Genome Variation Society, http://www.hgvs.org/mutnomen/
Illumina, http://www.illumina.com/
Ingenuity Variant Analysis, http://www.ingenuity.com/products/variant-analysis
MutationTaster, http://www.mutationtaster.org/
NCBI HomoloGene, http://www.ncbi.nlm.nih.gov/homologene
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Variant Effect Predictor, http://useast.ensembl.org/Homo_sapiens/Tools/VEP
Supplemental Data
References
- 1.Giglia-Mari G., Coin F., Ranish J.A., Hoogstraten D., Theil A., Wijgers N., Jaspers N.G., Raams A., Argentini M., van der Spek P.J. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 2.Assfalg R., Lebedev A., Gonzalez O.G., Schelling A., Koch S., Iben S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012;40:650–659. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nonnekens J., Perez-Fernandez J., Theil A.F., Gadal O., Bonnart C., Giglia-Mari G. Mutations in TFIIH causing trichothiodystrophy are responsible for defects in ribosomal RNA production and processing. Hum. Mol. Genet. 2013;22:2881–2893. doi: 10.1093/hmg/ddt143. [DOI] [PubMed] [Google Scholar]
- 4.Compe E., Egly J.M. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 5.DiGiovanna J.J., Kraemer K.H. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefanini M., Botta E., Lanzafame M., Orioli D. Trichothiodystrophy: from basic mechanisms to clinical implications. DNA Repair (Amst.) 2010;9:2–10. doi: 10.1016/j.dnarep.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Ueda T., Compe E., Catez P., Kraemer K.H., Egly J.M. Both XPD alleles contribute to the phenotype of compound heterozygote xeroderma pigmentosum patients. J. Exp. Med. 2009;206:3031–3046. doi: 10.1084/jem.20091892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubaele S., Proietti De Santis L., Bienstock R.J., Keriel A., Stefanini M., Van Houten B., Egly J.M. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 9.Orioli D., Compe E., Nardo T., Mura M., Giraudon C., Botta E., Arrigoni L., Peverali F.A., Egly J.M., Stefanini M. XPD mutations in trichothiodystrophy hamper collagen VI expression and reveal a role of TFIIH in transcription derepression. Hum. Mol. Genet. 2013;22:1061–1073. doi: 10.1093/hmg/dds508. [DOI] [PubMed] [Google Scholar]
- 10.Arseni L., Lanzafame M., Compe E., Fortugno P., Afonso-Barroso A., Peverali F.A., Lehmann A.R., Zambruno G., Egly J.M., Stefanini M., Orioli D. TFIIH-dependent MMP-1 overexpression in trichothiodystrophy leads to extracellular matrix alterations in patient skin. Proc. Natl. Acad. Sci. USA. 2015;112:1499–1504. doi: 10.1073/pnas.1416181112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traboulsi H., Davoli S., Catez P., Egly J.M., Compe E. Dynamic partnership between TFIIH, PGC-1α and SIRT1 is impaired in trichothiodystrophy. PLoS Genet. 2014;10:e1004732. doi: 10.1371/journal.pgen.1004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botta E., Offman J., Nardo T., Ricotti R., Zambruno G., Sansone D., Balestri P., Raams A., Kleijer W.J., Jaspers N.G. Mutations in the C7orf11 (TTDN1) gene in six nonphotosensitive trichothiodystrophy patients: no obvious genotype-phenotype relationships. Hum. Mutat. 2007;28:92–96. doi: 10.1002/humu.20419. [DOI] [PubMed] [Google Scholar]
- 13.Heller E.R., Khan S.G., Kuschal C., Tamura D., DiGiovanna J.J., Kraemer K.H. Mutations in the TTDN1 gene are associated with a distinct trichothiodystrophy phenotype. J. Invest. Dermatol. 2015;135:734–741. doi: 10.1038/jid.2014.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakabayashi K., Amann D., Ren Y., Saarialho-Kere U., Avidan N., Gentles S., MacDonald J.R., Puffenberger E.G., Christiano A.M., Martinez-Mir A. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am. J. Hum. Genet. 2005;76:510–516. doi: 10.1086/428141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Tian Y., Chen Q., Chen D., Zhai Z., Shu H.B. TTDN1 is a Plk1-interacting protein involved in maintenance of cell cycle integrity. Cell. Mol. Life Sci. 2007;64:632–640. doi: 10.1007/s00018-007-6501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbett M.A., Dudding-Byth T., Crock P.A., Botta E., Christie L.M., Nardo T., Caligiuri G., Hobson L., Boyle J., Mansour A. A novel X-linked trichothiodystrophy associated with a nonsense mutation in RNF113A. J. Med. Genet. 2015;52:269–274. doi: 10.1136/jmedgenet-2014-102418. [DOI] [PubMed] [Google Scholar]
- 17.He Y., Fang J., Taatjes D.J., Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drapkin R., Reardon J.T., Ansari A., Huang J.C., Zawel L., Ahn K., Sancar A., Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y.C., Gralla J.D. Stimulation of the XPB ATP-dependent helicase by the beta subunit of TFIIE. Nucleic Acids Res. 2005;33:3072–3081. doi: 10.1093/nar/gki623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkuma Y., Roeder R.G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 21.Khan S.G., Muniz-Medina V., Shahlavi T., Baker C.C., Inui H., Ueda T., Emmert S., Schneider T.D., Kraemer K.H. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuschal C., DiGiovanna J.J., Khan S.G., Gatti R.A., Kraemer K.H. Repair of UV photolesions in xeroderma pigmentosum group C cells induced by translational readthrough of premature termination codons. Proc. Natl. Acad. Sci. USA. 2013;110:19483–19488. doi: 10.1073/pnas.1312088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanini M., Orecchia G., Rabbiosi G., Nuzzo F. Altered cellular response to UV irradiation in a patient affected by premature ageing. Hum. Genet. 1986;73:189–192. doi: 10.1007/BF00401225. [DOI] [PubMed] [Google Scholar]
- 24.Stefanini M., Giliani S., Nardo T., Marinoni S., Nazzaro V., Rizzo R., Trevisan G. DNA repair investigations in nine Italian patients affected by trichothiodystrophy. Mutat. Res. 1992;273:119–125. doi: 10.1016/0921-8777(92)90073-c. [DOI] [PubMed] [Google Scholar]
- 25.Stefanini M., Vermeulen W., Weeda G., Giliani S., Nardo T., Mezzina M., Sarasin A., Harper J.I., Arlett C.F., Hoeijmakers J.H. A new nucleotide-excision-repair gene associated with the disorder trichothiodystrophy. Am. J. Hum. Genet. 1993;53:817–821. [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botta E., Nardo T., Lehmann A.R., Egly J.M., Pedrini A.M., Stefanini M. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 30.Botta E., Nardo T., Orioli D., Guglielmino R., Ricotti R., Bondanza S., Benedicenti F., Zambruno G., Stefanini M. Genotype-phenotype relationships in trichothiodystrophy patients with novel splicing mutations in the XPD gene. Hum. Mutat. 2009;30:438–445. doi: 10.1002/humu.20912. [DOI] [PubMed] [Google Scholar]
- 31.Orioli D., Colaluca I.N., Stefanini M., Riva S., Dotti C.G., Peverali F.A. Rac3-induced neuritogenesis requires binding to Neurabin I. Mol. Biol. Cell. 2006;17:2391–2400. doi: 10.1091/mbc.E05-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh K.S., Khan S.G., Jaspers N.G., Raams A., Ueda T., Lehmann A., Friedmann P.S., Emmert S., Gratchev A., Lachlan K. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 33.Imoto K., Kobayashi N., Katsumi S., Nishiwaki Y., Iwamoto T.A., Yamamoto A., Yamashina Y., Shirai T., Miyagawa S., Dohi Y. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J. Invest. Dermatol. 2002;119:1177–1182. doi: 10.1046/j.1523-1747.2002.19514.x. [DOI] [PubMed] [Google Scholar]
- 34.Faghri S., Tamura D., Kraemer K.H., Digiovanna J.J. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J. Med. Genet. 2008;45:609–621. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura D., Merideth M., DiGiovanna J.J., Zhou X., Tucker M.A., Goldstein A.M., Brooks B.P., Khan S.G., Oh K.S., Ueda T. High-risk pregnancy and neonatal complications in the DNA repair and transcription disorder trichothiodystrophy: report of 27 affected pregnancies. Prenat. Diagn. 2011;31:1046–1053. doi: 10.1002/pd.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viprakasit V., Gibbons R.J., Broughton B.C., Tolmie J.L., Brown D., Lunt P., Winter R.M., Marinoni S., Stefanini M., Brueton L. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum. Mol. Genet. 2001;10:2797–2802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- 37.Boyle J., Ueda T., Oh K.S., Imoto K., Tamura D., Jagdeo J., Khan S.G., Nadem C., Digiovanna J.J., Kraemer K.H. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum. Mutat. 2008;29:1194–1208. doi: 10.1002/humu.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X., Khan S.G., Tamura D., Ueda T., Boyle J., Compe E., Egly J.M., DiGiovanna J.J., Kraemer K.H. Abnormal XPD-induced nuclear receptor transactivation in DNA repair disorders: trichothiodystrophy and xeroderma pigmentosum. Eur. J. Hum. Genet. 2013;21:831–837. doi: 10.1038/ejhg.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 40.Vermeulen W., Rademakers S., Jaspers N.G., Appeldoorn E., Raams A., Klein B., Kleijer W.J., Hansen L.K., Hoeijmakers J.H. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat. Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto T., Yamamoto S., Watanabe Y., Ohta T., Hanaoka F., Roeder R.G., Ohkuma Y. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEbeta subunit. J. Biol. Chem. 1998;273:19866–19876. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 42.Ohkuma Y., Hashimoto S., Wang C.K., Horikoshi M., Roeder R.G. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol. Cell. Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka A., Akimoto Y., Kobayashi S., Hisatake K., Hanaoka F., Ohkuma Y. Association of the winged helix motif of the TFIIEα subunit of TFIIE with either the TFIIEβ subunit or TFIIB distinguishes its functions in transcription. Genes Cells. 2015;20:203–216. doi: 10.1111/gtc.12212. [DOI] [PubMed] [Google Scholar]
- 44.Okuda M., Tanaka A., Arai Y., Satoh M., Okamura H., Nagadoi A., Hanaoka F., Ohkuma Y., Nishimura Y. A novel zinc finger structure in the large subunit of human general transcription factor TFIIE. J. Biol. Chem. 2004;279:51395–51403. doi: 10.1074/jbc.M404722200. [DOI] [PubMed] [Google Scholar]
- 45.Okuda M., Watanabe Y., Okamura H., Hanaoka F., Ohkuma Y., Nishimura Y. Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface. EMBO J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe T., Hayashi K., Tanaka A., Furumoto T., Hanaoka F., Ohkuma Y. The carboxy terminus of the small subunit of TFIIE regulates the transition from transcription initiation to elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:2914–2926. doi: 10.1128/MCB.23.8.2914-2926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grünberg S., Warfield L., Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Racioppi L., Cancrini C., Romiti M.L., Angelini F., Di Cesare S., Bertini E., Livadiotti S., Gambarara M.G., Matarese G., Lago Paz F. Defective dendritic cell maturation in a child with nucleotide excision repair deficiency and CD4 lymphopenia. Clin. Exp. Immunol. 2001;126:511–518. doi: 10.1046/j.1365-2249.2001.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanini M., Lagomarsini P., Arlett C.F., Marinoni S., Borrone C., Crovato F., Trevisan G., Cordone G., Nuzzo F. Xeroderma pigmentosum (complementation group D) mutation is present in patients affected by trichothiodystrophy with photosensitivity. Hum. Genet. 1986;74:107–112. doi: 10.1007/BF00282072. [DOI] [PubMed] [Google Scholar]
- 50.Stefanini M., Fawcett H., Botta E., Nardo T., Lehmann A.R. Genetic analysis of twenty-two patients with Cockayne syndrome. Hum. Genet. 1996;97:418–423. doi: 10.1007/BF02267059. [DOI] [PubMed] [Google Scholar]
- 51.Compe E., Malerba M., Soler L., Marescaux J., Borrelli E., Egly J.M. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 2007;10:1414–1422. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 52.Backendorf C., de Wit J., van Oosten M., Stout G.J., Mitchell J.R., Borgstein A.M., van der Horst G.T., de Gruijl F.R., Brouwer J., Mullenders L.H., Hoeijmakers J.H. Repair characteristics and differentiation propensity of long-term cultures of epidermal keratinocytes derived from normal and NER-deficient mice. DNA Repair (Amst.) 2005;4:1325–1336. doi: 10.1016/j.dnarep.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Larochelle S., Amat R., Glover-Cutter K., Sansó M., Zhang C., Allen J.J., Shokat K.M., Bentley D.L., Fisher R.P. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu H., Zawel L., Fisher L., Egly J.M., Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 55.Fan L., Fuss J.O., Cheng Q.J., Arvai A.S., Hammel M., Roberts V.A., Cooper P.K., Tainer J.A. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H., Rudolf J., Johnson K.A., McMahon S.A., Oke M., Carter L., McRobbie A.M., Brown S.E., Naismith J.H., White M.F. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehmann A.R. XPD structure reveals its secrets. DNA Repair (Amst.) 2008;7:1912–1915. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Egly J.M., Coin F. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst.) 2011;10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 59.de Boer J., de Wit J., van Steeg H., Berg R.J., Morreau H., Visser P., Lehmann A.R., Duran M., Hoeijmakers J.H., Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 60.Compe E., Drané P., Laurent C., Diderich K., Braun C., Hoeijmakers J.H., Egly J.M. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol. Cell. Biol. 2005;25:6065–6076. doi: 10.1128/MCB.25.14.6065-6076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang C., Kraemer K.H., Morris A., Schiffmann R., Price V.H., Menefee E., DiGiovanna J.J. Characterization of tiger-tail banding and hair shaft abnormalities in trichothiodystrophy. J. Am. Acad. Dermatol. 2005;52:224–232. doi: 10.1016/j.jaad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 62.Park J.Y., Cho M.O., Leonard S., Calder B., Mian I.S., Kim W.H., Wijnhoven S., van Steeg H., Mitchell J., van der Horst G.T. Homeostatic imbalance between apoptosis and cell renewal in the liver of premature aging Xpd mice. PLoS ONE. 2008;3:e2346. doi: 10.1371/journal.pone.0002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.